Abstract
Wall teichoic acids are cell wall polymers that maintain the integrity of the cellular envelope and contribute to the virulence of Staphylococcus aureus. Despite the central role of wall teichoic acid in S. aureus virulence, details concerning the biosynthetic pathway of the predominant wall teichoic acid polymer are lacking, and workers have relied on a presumed similarity to the putative polyribitol phosphate wall teichoic acid pathway in Bacillus subtilis. Using high-resolution polyacrylamide gel electrophoresis for analysis of wall teichoic acid extracted from gene deletion mutants, a revised assembly pathway for the late-stage ribitol phosphate-utilizing enzymes is proposed. Complementation studies show that a putative ribitol phosphate polymerase, TarL, catalyzes both the addition of the priming ribitol phosphate onto the linkage unit and the subsequent polymerization of the polyribitol chain. It is known that the putative ribitol primase, TarK, is also a bifunctional enzyme that catalyzes both ribitol phosphate priming and polymerization. TarK directs the synthesis of a second, electrophoretically distinct polyribitol-containing teichoic acid that we designate K-WTA. The biosynthesis of K-WTA in S. aureus strain NCTC8325 is repressed by the accessory gene regulator (agr) system. The demonstration of regulated wall teichoic acid biosynthesis has implications for cell envelope remodeling in relation to S. aureus adhesion and pathogenesis.
Wall teichoic acids (WTA) are anionic, carbohydrate-based polymers that are covalently attached to the peptidoglycan matrix of many gram-positive bacteria (32, 41, 44). In Bacillus subtilis, WTA accounts for 30 to 60% of the total cell wall mass and has been implicated in a number of roles critical to maintaining the overall integrity of the cell envelope (21). The loss of cell surface charge balance, tensile strength, rigidity, porosity, and proper cell morphology along with misregulation of autolysins are all associated with mutations in WTA-related genes (32). In addition to a structural role, the WTA polymer itself may also serve as a phosphate reservoir for transition to growth in phosphate-depleted medium and in cation (Mg2+/Ca2+) assimilation/homeostasis (12).
While much is known about the structure and function of WTA in rod-shaped bacteria, comparatively little is known about the role of WTA in coccoid bacteria. In contrast to findings for B. subtilis (10), the growth rate and fitness of Staphylococcus aureus lacking WTA is not significantly impaired (11, 25, 45). S. aureus WTA has been shown, however, to play an essential role in adhesion to endothelial and epithelial tissues and to be critical for colonization in multiple infection models (2, 45, 46). Since adhesion is a key step in infection, WTA can be considered a quintessential virulence factor, making it a potential target for antimicrobial intervention. To exploit the WTA pathway as a novel drug target in S. aureus, it is necessary to define the activity of the enzymes in the pathway and to elucidate the mechanism of assembly.
The majority of S. aureus strains contain polyribitol phosphate (poly-RboP) WTA, whose biosynthetic pathway has been modeled using the prototype poly-RboP WTA-producing strain B. subtilis W23 (28). The proposed pathway has been accepted for S. aureus despite major differences in WTA gene organization between S. aureus and B. subtilis strain W23 (38). There are at least two separate polycistronic WTA gene clusters (in addition to the monocistronic tarO gene) in the S. aureus genome, whereas in B. subtilis W23 the WTA genes are in a single, divergently transcribed locus. In particular, the proposed poly-RboP WTA assembly model invokes two sequentially acting enzymes, an RboP primase (TarK) and an RboP polymerase (TarL). Qian et al. (38) noted that for this model to be correct for S. aureus, two enzymes (TarK and TarL) with more than 80% sequence identity would have to catalyze distinct reactions, namely, RboP priming and RboP polymerization. Since high sequence similarity is not uncommon among WTA polyol biosynthetic enzymes and a common donor (CDP-Rbo) is used by both enzymes, we sought to establish the pathway by experimentally addressing whether S. aureus has a unique three-enzyme pathway in which TarK and TarL are functionally redundant or whether indeed the enzymes have unique priming/polymerizing activities despite limited primary sequence divergence.
MATERIALS AND METHODS
Strains and growth conditions.
All of the S. aureus strains used are derivatives of the sequenced NCTC8325 reference strain (18). Plasmids were constructed in Escherichia coli Novablue (Novagen) cells and introduced into restriction-negative S. aureus strain RN4220 by electroporation (40). Plasmids were purified by pretreating cells with lysostaphin (50 μg/ml, 10 min, 37°C) prior to isolation using a standard plasmid miniprep protocol (Qiagen). Modified plasmids were then introduced into restriction-positive wild-type strains by electroporation. S. aureus was grown in either tryptic soy broth (TSB) or Luria-Bertani medium at 37°C unless otherwise noted. Antibiotic markers were selected with erythromycin (Em) (10 μg/ml), tetracycline (Tc) (2.5 μg/ml), and chloramphenicol (Cm) (for a single copy integrated into the genome, 5 μg/ml; for multicopy plasmid, 10 μg/ml). The bacterial strains used are listed in Table 1.
TABLE 1.
Bacterial strains
| Strain | Relevant genotype | Reference or source |
|---|---|---|
| S. aureus strains | ||
| RN450 | NCTC8325-4, prophage cured, rsbU agr+ | 33 |
| RN4220 | RN450 r− m+, partial agr defect | 27 |
| RN4220ΔtarO | RN4220 ΔtarO | 19 |
| RN6911 | RN450 agr::tetM null Tcr | 35 |
| SA113 | NCTC8325(φ11, φ12, φ13) r− m+agr | ATCC 35556 |
| SH1000 | RN450 rsbU+agr+ | 22 |
| JT200 | RN450(φ11) | This study |
| JT15 | RN4220 [geh::(pCL25int-PspanktarL) Emr] (SAOUHSC_00227) | This study |
| JT16 | JT15 tarL::tetL Tcr | This study |
| JT17 | RN4220 ΔtarI′J′ (SAOUHSC_00220-SAOUHSC_00221) | This study |
| JT18 | RN4220 ΔtarK (SAOUHSC_00222) | This study |
| JT19 | JT18(pMS74-PcadtarK) Emr | This study |
| JT20 | JT18 [geh::(pCL25int-PPentarK) Emr] | This study |
| JT21 | JT20 tarL::tetL Tcr | This study |
| JT22 | RN4220ΔtarO [geh::(pCL25int-PspanktarL) Emr] tarL::tetL Tcr by transduction (JT15 and then JT16 as donors) | This study |
| JT38 | RN4220(pCN33-agrA) Emr | This study |
| JT203 | RN450(pXen-1) Cmr | This study |
| JT204 | RN450(pXen-PtarI′J′K) Cmr | This study |
| JT205 | RN450(pXen-PtarIJL) Cmr | This study |
| JT207 | JT200 ΔtarK (SAOUHSC_00222) | This study |
| JT208 | JT207 [geh::(pCL25int-PPentarL) Emr] | This study |
| JT209 | JT207 [geh::(pCL25int-PPentarK) Emr] | This study |
| JT210 | JT207 [geh::(pCL25int-PPentarKBs) Emr] | This study |
| JT211 | JT207 [geh::(pCL25int-PPentarLBs) Emr] | This study |
| JT212 | JT207 [geh::(pCL25int-PPentarKLBs) Emr] | This study |
| JT213 | JT208 tarL::tetL Tcr by transduction (JT16 donor) | This study |
| JT214 | JT209 tarL::tetL Tcr by transduction (JT16 donor) | This study |
| JT215 | JT212 tarL::tetL Tcr by transduction (JT16 donor) | This study |
| JT220 | JT200 agr::tetM null Tcr by transduction (RN6911 donor) | This study |
| JT221 | JT207 agr::tetM null Tcr by transduction (RN6911 donor) | This study |
| JT302 | SH1000 agr::tetM null Tcr by transduction (RN6911 donor) | This study |
| JT409 | SA113 agr::tetM null Tcr by transduction (RN6911 donor) | This study |
| B. subtilis W23 | Prototype poly-RboP WTA strain | ATCC 23059 |
Construction of S. aureus WTA gene deletion strains.
Genes were deleted using a modified protocol for the pKOR1 E. coli-S. aureus shuttle vector (4). The pKOR1 plasmid has a temperature-sensitive S. aureus replicon and a Tc-inducible antisense secY cassette for counterselection. In-frame gene deletion cassettes with flanking homology arms were constructed by overlap assembly PCR, using primer pairs P1/P2 and P3/P4 to amplify ∼1 kb upstream and downstream of the targeted gene. Fragments were assembled by a second round of PCR to obtain a deletion cassette containing the first and last 90 bp of the targeted gene. The cassette was digested (ApaI) and ligated into a restricted vector (ApaI/EcoRV) to obtain integration plasmids pMS17 and pMS18 (Table 2). A marked Tcr deletion cassette was constructed by ligation of tetL (amplified from pCL25 with primers tetL-for and tetL-rev) into the P1/P4 tarL PCR insert (BstBI/MluI) to obtain pMS21. Primers and gene deletion vectors are listed in Table 2.
TABLE 2.
Plasmids and primers
| Plasmid or primer | Description or sequencea | Reference or source |
|---|---|---|
| Plasmids | ||
| pKOR1 | E. coli-S. aureus shuttle vector, ori(Ts) inducible, secY antisense counterselection, Apr Cmr | 4 |
| pCN33 | E. coli-S. aureus shuttle vector, Apr Emr | 7 |
| pCN59 | E. coli-S. aureus shuttle vector, PcadcadC blaZ transcriptional terminator, Apr Emr | 7 |
| pCN68 | E. coli-S. aureus shuttle vector, PblaZ GFP blaZ transcriptional terminator, Apr Emr | 7 |
| pXen-1 | E. coli-S. aureus shuttle vector, luxABCDE Cmr | Xenogen |
| pCL25 | Phage L54a attP-attB (geh) integration vector, Specr Tcr | 30 |
| pMS13 | pKOR1 tarO::tetL allelic replacement vector | This study |
| pMS17 | pKOR1 ΔtarI′J′ allelic replacement vector | This study |
| pMS18 | pKOR1 ΔtarK allelic replacement vector | This study |
| pMS21 | pKOR1 tarL::tetL allelic replacement vector | This study |
| pCL25int | pCL25 with phage 54a integrase and ermC cassette replacing tetL, Specr Emr | This study |
| pCL25intPspank | pCL25int with Pspank IPTG-inducible promoter PPen, lacI | This study |
| pCL25intPPen | pCL25int with PPen constitutive promoter, blaZ transcriptional terminator | This study |
| pMS74 | pCN59 with low-copy-number pI258 replicon, Apr Emr | This study |
| pMS80 | E. coli-S. aureus shuttle vector pLI50 with tarO, Apr Cmr | This study |
| Primers | ||
| P1tarK | GATTACATGGGCCCGATCTAGTGGTTATG | |
| P2tarK | CTGGTCATGATAAACAAATAAATATTTATC | |
| P3tarK | GTTTATCATGACCAGGAAAAAGTTGTGCCA | |
| P4tarK | GATTACATCCCGGGCGATGTCGAATATGG | |
| P1tarI′J′ | GATTACATGGGCCCGCAACATCATTGGGCG | |
| P2tarI′J′ | AATCTCATTAAATTCACTCACTAAAATGAAC | |
| P3tarI′J′ | GAATTTAATGAGATTAATGTATGTACGATG | |
| P4tarI′J′ | TGGTGCCTTTAAAACAGTTGG | |
| P1tarL | GATTACATGGGCCCCCTAGACAATTTGAAG | |
| P4tarL | CATCTACAATATCAAAACGGTAGTATTGACC | |
| tetL-for | GCTTAGTTCGAATGGGGAAAGCTTCACAGA | |
| tetL-rev | GCTTGAACGCGTTTAAGTCTAACACACTAGAC | |
| tarLspank-for | GGCTATGTCGACCAGTATTAAAATGGATTATG | |
| tarLspank-rev | GCTACAGCTAGCACTTTGACTACTATATAAAC | |
| agrA-F | TGTTATGCATGCCTGGCCTACGTGATTATTTC | |
| agrA-R | TGTTCGAATTCACGCGTCATATTTAATTTTG | |
| TarO-F | GCTATCTGGGCCCGATTAATAATAATGC | |
| TarO-R | GCTATCTCGTACGCCTAAAATATACTCATAGC | |
| ProtarI′J′K-for | GCTTATGAATTCGTATGCACCTTTGAGGACG | |
| ProtarI′J′K-rev | GCTTATGGATCCGTTGTCCTCCATTCTGTC | |
| ProtarIJL-for | GCTTATGAATTCGTGATAACTTAAAAAC | |
| ProtarIJL-rev | GCTTATGGATCCAAAATACTTCCTCCATTC | |
| Ppenfor | GACCTGCAGGCATGCAAGCTAATTCC | |
| Ppenrev | GACGGTCGTCGACAATATTTGATTGATCGTAACCAG | |
| tarLpen-for | CGTAGAATTCGAGGAGGAGTAAAAGTATGGTTAAAAGTAAGAT | |
| tarLpen-rev | GGTCCGCGGCGCGCCTTAGCTACCAAATAAATTTC | |
| tarKpen-for | CGTAGAATTCGAGGAGGAGTAAAAGTATGACAAAAACGAAACAAGC | |
| tarKpen-rev | GACCGTCGAGCTCCCAACATTAGCGTCTAAACAAATC | |
| tarLBs pen-for | CCTATGGTCGACGAGGAGGAGTAAAAGTATGAAGCTGGCCAG | |
| tarLBs pen-rev | GGTCCGCGGCGCGCCTTATCTTTTAAGGACTTTATC | |
| tarKBs pen-for | CCTATGGTCGACGAGGAGGAGTAAAAGTATGAAAACATTCCTTAC | |
| tarKBs pen-rev | GGTCCGCGGCGCGCCCTAATCGGCACCAGATG |
Restriction sites are italicized, synthetic ribosome binding sites are underlined, and gene coding sequences are indicated by bold type.
Plasmids were electroporated into S. aureus RN4220 and plated on TSB agar containing 10 μg/ml Cm at 30°C. Plasmid-containing clones were then inoculated into 50 ml of prewarmed TSB and grown at 42°C for 12 to 15 generations (∼4 h) without selection to reduce plasmid copy number and favor integration. Cm (5 μg/ml) was then added, and growth continued overnight. Cultures were streaked to obtain single colonies (42°C, 5 μg/ml Cm), and clones were screened for plasmid integration by PCR using primers P1/P4 to verify cointegrates. Clones from which wild-type and mutant alleles were amplified at approximately equal intensities were resolved by overnight growth in TSB at 30°C, followed by pKOR1 vector counterselection on tryptic soy agar with anhydrotetracycline (1 μg/ml). Gene deletion in Cms clones was confirmed by PCR analysis using primers that anneal outside the P1-P4 deletion cassette.
Plasmid construction.
The low-copy-number (four or five copies per cell), cadmium-inducible complementation vector pMS74 was constructed by ligating the pI258 replicon from pRN8298 into the parent pCN59 vector (7) using the NarI/ApaI restriction sites. The tarK gene was then amplified (with primers tarKpen-for and tarKpen-rev) to incorporate flanking restriction sites and ligated into pMS74 (SalI/AscI). The accessory gene regulator (agr) complementation vector was made by PCR amplification of the wild-type agrA allele along with the P1 promoter and terminator (with primers agrA-F and agrA-R [42]) and subsequently ligated into pCN33 using EcoRI/SphI. The promoter probe plasmids pXen-PtarI′J′K and pXen-PtarIJL luxABCDE were made by ligating the intergenic DNA sequences directly upstream of the respective start codons for tarI′ (528 bp; primers ProtarI′J′K-for and ProtarI′J′K-rev) and tarI (279 bp; primers ProtarIJL-for and ProtarIJL-rev) into pXen-1 (EcoRI/BamHI). Plasmid pMS80 was assembled by cloning the tarO gene (with primers TarO-F and TarO-R) into pLI50 (29).
Site-specific integration plasmids based on the bacteriophage L54a attP recombination system were constructed by ligation of the int-attP fragment (ClaI/BamHI) of pCL55 into pCL25 (ClaI/BglII) (29). The tetL marker (partial SpeI/ClaI digest) was then replaced with the ermC marker from pCN59 (AvrII/ClaI) by ligation to make pCL25int. An isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter (Pspank) was installed by ligation of the Pspank-MCS-PPenlacI cassette of pDR110 (D. Rudner, Harvard Medical School) into pCL25int with EcoRI/BamHI to obtain pCL25intPspank. A version containing the strong, constitutive PPen promoter was constructed by PCR amplification of this module from pDR110 (with primers Ppenfor and Ppenrev) and then swapping the PPen promoter for the constitutive PblaZ promoter of pCN68 (7) using SphI/SalI restriction sites. In our hands, particularly if used with a multicopy plasmid, the PPen promoter was considerably more stable and less prone to rearrangement than the PblaZ promoter of pCN68. Genes were cloned into the modified vector using amplified PCR products containing EcoRI/SalI and AscI restriction sites. The PPen-MCS-gene-blaZ transcriptional terminator cassettes were then excised from the pCN68 vector backbone (SphI/SfoI) and ligated into the multiple cloning site of pCL25int (partial SphI/SmaI digest) to obtain pCL25int-PPen. Integration plasmids were electroporated into RN4220 and then moved by transduction into RN450 (see below). Plasmids and primers are listed in Table 2.
WTA extraction.
WTA was isolated from a 20-ml culture of S. aureus stationary-phase cells grown in TSB overnight at 37°C. Cells were collected by centrifugation (2,000 × g, 10 min), washed once with 30 ml of buffer 1 [50 mM 2-(N-morpholino)ethanesulfonic acid (MES), pH 6.5], and resuspended in 30 ml of buffer 2 (4% [wt/vol] sodium dodecyl sulfate [SDS], 50 mM MES; pH 6.5). Samples were placed in a boiling water bath for 1 h, and then the cells were collected by centrifugation (10,000 × g, 10 min). The pellet was resuspended in buffer 2, transferred to a 2-ml microcentrifuge tube, and sedimented (14,000 × g, 10 min). Next, the pellet was washed once with buffer 2, once with buffer 3 (2% NaCl, 50 mM MES, pH 6.5), and finally with buffer 1. After the last wash, samples were treated with proteinase K (20 mM Tris-HCl [pH 8.0], 0.5% [wt/vol] SDS, and 20 μg of proteinase K in 1 ml) and incubated at 50°C for ∼4 h. Following digestion, samples were washed once with buffer 3 and then at least three times with distilled H2O to remove the SDS. Samples were thoroughly resuspended in 1 ml of 0.1 M NaOH and shaken at room temperature for 16 h to hydrolyze WTA (13). Insoluble cell wall debris was removed by centrifugation (14,000 × g, 10 min), and the supernatant containing the hydrolyzed WTA was either directly analyzed by polyacrylamide gel electrophoresis (PAGE) or processed further for long-term storage. Samples were neutralized with 0.1 M acetic acid, extensively dialyzed against distilled H2O (molecular mass cutoff, 1 kDa), and then lyophilized to obtain a white powder. Approximately 500 μg of crude WTA was typically isolated from a 20-ml culture.
WTA PAGE analysis.
The WTA PAGE protocol of Pollack and Neuhaus (37) used for B. subtilis poly-GroP WTA analysis was modified to improve S. aureus poly-RboP WTA resolution. A Bio-Rad Protean II xi electrophoresis cell (20 by 16 cm by 0.75 mm) was used for separation. The separating gel (20% total acrylamide [T]; percentage of T that was the cross-linker bisacrylamide [C], 6%) was cast by mixing 20 ml of Tris-HCl buffer (3 M Tris-HCl, pH 8.5) with 40 ml of an acrylamide stock solution (30% T, 6% C). The solution was polymerized using 600 μl of 10% ammonium persulfate along with 60 μl of tetramethylethylenediamine. The stacking gel (3% T, 0.26% C) was ∼1 cm long and was cast using a mixture of 1 ml acrylamide stock solution (30% T, 0.26% C), 3 ml of Tris-HCl buffer (3 M Tris-HCl, pH 8.5), and 6 ml of distilled H2O. The stacking gel was polymerized with 100 μl of 10% ammonium persulfate and 10 μl of tetramethylethylenediamine. WTA samples were diluted 1:3 in loading buffer (50% glycerol in running buffer with a trace of bromophenol blue) to obtain a final volume no greater than 10 μl. Typically, ∼200 ng of crude WTA sample was loaded for each sample. Gels were developed at 4°C for ∼18 h using constant current (80 mA with two gels) and a Tris-Tricine running buffer (0.1 M Tris base, 0.1 M Tricine, pH 8.2 [39]). WTA bands were visualized using the alcian blue-silver staining protocol (37).
Luciferase promoter reporter assay.
Overnight cultures of RN450 carrying the pXen plasmid with promoter fragments were diluted 1:200 into fresh TSB containing 10 μg/ml Cm and incubated at 37°C with shaking (200 rpm). Aliquots were withdrawn at 30-min intervals, and cells were pelleted by centrifugation (5,000 × g, 2 min). Cells were resuspended in phosphate-buffered saline and transferred to a 96-well microplate, and the optical density at 532 nm (OD532) (Perkin Elmer HTS 7000) and luminescence (Perkin Elmer MicroBeta Trilux) were determined to estimate promoter activity.
Phage infection and transduction.
The numbers of PFU for various host strains were determined by mixing phage (∼500 PFU) with exponentially dividing cells (OD600, 0.5) grown in TSB with 200 μg/ml CaCl2 (TSB-Ca). The mixtures were incubated at 30°C for 20 min, suspended in TSB-Ca top agar (0.5% agar), and then poured over TSB-Ca agar plates (1.5% agar). Plaques were counted after 16 h of growth at 30°C.
Donor lysates were obtained by plating as described above, except that the generalized transducing phage φ11 was used at a concentration that resulted in confluent top agar phage lawns. Phage were eluted by addition of TSB and gentle shaking overnight at room temperature. Lysates were filter sterilized (0.45 μm) and checked for a lack of viable donor bacteria by plating. Recipient bacteria were grown in TSB to mid-logarithmic phase (2-ml cultures), at which point CaCl2 was added to 0.5 mM. Donor lysate was added, and the tubes were incubated at 30°C for 15 min. For phage-sensitive recipient strains, mixtures were pelleted and washed three times with TSB containing 20 mM citrate before addition of top agar. Otherwise, 10 ml of TSB top agar (0.5% agar) was added directly, and the mixture was poured over TSB agar plates. Inducing amounts of antibiotics (0.05 μg/ml Em or Tc) were included in the top agar to facilitate phenotypic expression during incubation at 37°C for 1 h. The plates were then overlaid with 15 ml of TSB top agar containing an antibiotic at a selective concentration (12.5 μg/ml Tc or 10 μg/ml Em). Transductants were scored after 48 h of incubation at 37°C.
To transduce markers into the ΔtarO background (JT22), tarO was provided on a plasmid (pMS80) to temporarily restore the phage WTA receptor. After transduction, the plasmid was cured by protoplast regeneration to restore the ΔtarO genotype (20).
Phenotypic analysis of agr expression.
Because of the inherent instability of the accessory gene regulator (agr) locus (42), the expression of agr was explicitly determined for each strain used in this study. The production of δ-hemolysin was scored on plates containing TSA with 5% sheep blood (TSAII; BD) as described previously (1). Extracellular protein fractions were obtained from stationary-phase cultures grown overnight in Luria-Bertani medium by centrifugation to remove bacteria and then filter sterilized (0.45 μm). Protein was concentrated by precipitation using a modified sodium deoxycholate-trichloroacetic acid protocol (5) and then separated by SDS-PAGE (12% acrylamide). Protein samples were either stained with Coomassie blue to determine the total protein content or blotted onto nitrocellulose membranes. An amount of extracellular protein corresponding to 1 ml of culture (OD600 adjusted to 1) was loaded for each sample for total protein staining, while ∼5 μg of extracellular protein per sample was loaded for gels that were to be probed by Western blotting. Membranes were developed by overnight blocking in Tris-buffered saline with Tween 20 containing bovine serum albumin (5%) and then probed with sheep polyclonal anti-α-hemolysin—horseradish peroxidase conjugate (Abcam ab15949) antibody using the manufacturer's standard protocol.
RESULTS
tarL gene is an essential WTA gene, while the tarI′J′K genes are dispensable.
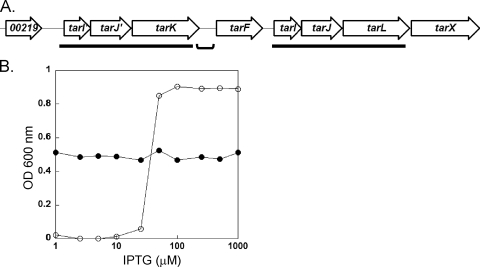
The WTA gene cluster of all S. aureus genomes sequenced to date contains two ∼3.4-kb gene clusters (tarI′J′K and tarIJL) with over 75% DNA sequence identity (Fig. 1A) (38). These clusters are putatively involved in the biosynthesis of the poly-RboP repeat, through the formation of CDP-Rbo (tarI′J′/tarIJ) and subsequent polymerization (RboP priming [tarK] and RboP polymerization [tarL]) onto the linkage unit acceptor. To examine whether these clusters have distinct activities or simply contain duplicated genes, in-frame gene deletion vectors were constructed using the temperature-sensitive replication vector pKOR1 (4). While numerous excisants derived from tarK and tarI′J′ cointegrates retained the ΔtarK and ΔtarI′J′ mutant alleles, respectively, tarL and tarIJ cointegrates that were resolved using the same procedure consistently failed to yield deletion mutants. This suggests that the tarI′J′K cluster is dispensable, while the tarIJL cluster is essential in S. aureus RN4220.
FIG. 1.
Genetic organization of WTA determinants in S. aureus. (A) The WTA locus of S. aureus is predicted to contain at least three transcripts [tarI′J′K, tarF, and tarIJL(X)] (43). The ∼3.4-kb duplication regions are underlined, while the bracket indicates the strain-dependent intergenic region that separates tarI′J′K from the rest of the locus. (B) Titration of TarL levels with the IPTG inducer confirms that tarL is an essential gene for growth of JT16 in vitro (○). A stationary-phase culture of JT16 grown in TSB supplemented with 1 mM IPTG was diluted (1:200) into fresh TSB containing various amounts of IPTG and incubated for 24 h at 30°C before culture turbidity was measured. Blocking the first step of WTA biosynthesis by deleting tarO relieved growth dependence on TarL in the otherwise isogenic strain JT22 (•).
Regulated ectopic expression coupled with deletion of the wild-type allele is an established method for testing gene essentiality in S. aureus (14). The Spank system was chosen since the basal level is low enough to generate conditionally lethal phenotypes in S. aureus (23, 48). To confirm that the tarL gene is indeed essential in wild-type cells and that the lack of viable deletion mutants was not attributable to polar effects, a tarL depletion strain (JT16) was constructed by placing a second chromosomal copy of tarL in the phage L54a integration site under the control of the IPTG-inducible Pspank promoter. The native tarL gene was then deleted using the standard pKOR1 procedure, except that the IPTG inducer was included in the medium during the cointegrate resolution step. Growth analysis showed that there was an absolute dependence on IPTG and hence tarL expression, indicating that TarL is an essential enzyme in the WTA pathway (Fig. 1B). When the first gene in the pathway (tarO) was also deleted in the same background (JT22), the tarL gene was no longer necessary for growth. This conditional dispensability is consistent with the results of a WTA gene deletion study performed by D'Elia et al., who observed a similar pattern with WTA linkage unit genes (11). However, the nonessential nature of tarK, which encodes an enzyme whose activity is predicted to occur prior to the activity encoded by tarL, contradicts the proposed RboP primase-RboP polymerase model (11).
S. aureus synthesizes two distinct poly-RboP WTA polymers.
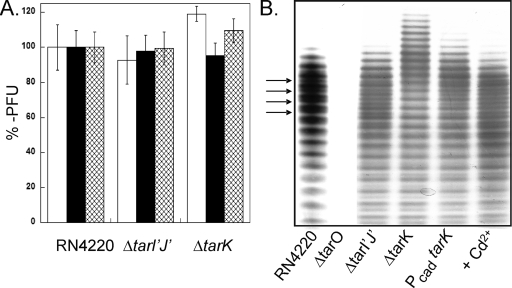
Since it was possible that the RboP primase-RboP polymerase model could still be correct if the putative truncated WTA of a ΔtarK deletion strain (and possibly a ΔtarI′J′ deletion strain) is somehow nontoxic compared to the predicted RboP-primed WTA precursor of the ΔtarL strain, the presence of mature poly-RboP WTA was examined using a panel of bacteriophages. WTA is the primary receptor for the majority of gram-positive bacteriophages, including the poly-RboP WTA-specific phages of S. aureus (3). Thus, the abilities of a panel of phages to infect the mutant strains were determined to probe for structural and/or compositional WTA alterations due to deletion of genes in the tarI′J′K cluster. However, all three phages tested formed plaques with efficiencies comparable to the wild type (Fig. 2A). Collectively, the data suggest that both TarK and TarL can catalyze poly-RboP TA chain polymerization using a common pool of CDP-ribitol donors (tarI′J′ and tarIJ).
FIG. 2.
Deletion of the tarI′J′K genes does not influence phage binding but does influence WTA composition. (A) Plaque-forming efficiency of φ11 (open bars), φ80α (filled bars), and φ85 (cross-hatched bars) measured using wild-type strain RN4220, ΔtarI′J′ strain JT17, and ΔtarK strain JT18 as the recipient strains. PFU values were normalized to the number of plaques observed using wild-type RN4220, and the bars indicate standard errors from three separate trials. (B) PAGE analysis of isolated WTA extracts visualized with alcian blue-silver staining. RN4220, wild-type strain RN4220; ΔtarO, strain RN4220ΔtarO; ΔtarI′J′, strain JT17; ΔtarK, strain JT18; PcadtarK, strain JT19 with no inducer; + Cd2+, strain JT19 with 1 μM Cd2+. The arrows indicate regularly spaced intervening K-WTA bands.
To determine whether the clusters were simply duplicated or if there were more subtle differences between the strains that did not alter the susceptibility to phage infection, WTA was directly analyzed by extraction and separation by PAGE. Numerous standard extraction conditions were tested, including acid-catalyzed (5% trichloroacetic acid), base-catalyzed (0.1 N NaOH), and nucleophile-mediated WTA cleavage at neutral pH using an aqueous solution of saturated lithium thiocyanate. While all of the methods produced comparable WTA profiles (data not shown), base-mediated cleavage performed as described by Endl et al. consistently provided the best WTA yield (13). The PAGE protocol that produced well-spaced, single-band resolution for poly-GroP WTA of B. subtilis 168 (37) did not give satisfactory results for S. aureus poly-RboP WTA. Accordingly, the PAGE protocol was systematically adjusted (see Materials and Methods) until acceptable resolution was obtained (Fig. 2B). Unlike the WTA-depleted ΔtarO control sample, the ΔtarI′J′ strain yielded both a WTA quantity and a profile similar to those of the wild-type RN4220 strain. The ΔtarK WTA chains actually lengthened by 7 to 10 units (Fig. 2A, lane 4), in contrast to the truncated WTA chains that would be expected with the TarK RboP primase model. The shift to a longer average repeat was accompanied by a decrease in WTA heterogeneity, with a series of single repeat-spaced bands seen in the wild-type sample disappearing (Fig. 2B). The conversion of heterogeneous, short WTA (wild type) to long, homogeneous WTA (ΔtarK) could be controlled stepwise by placing TarK under the control of a Cd2+ promoter (Fig. 2B, lanes 5 and 6). The banding pattern indicates that S. aureus makes two distinct WTA polymers, a primary TarL-directed polymer (designated L-WTA) and a secondary TarK-directed polymer (designated K-WTA). The retention of K-WTA in the ΔtarI′J′ strain suggests that tarIJ can provide the CDP-ribitol donor needed for biosynthesis of both teichoic acid polymers.
tarI′J′K promoter is significantly weaker than the tarIJL promoter.
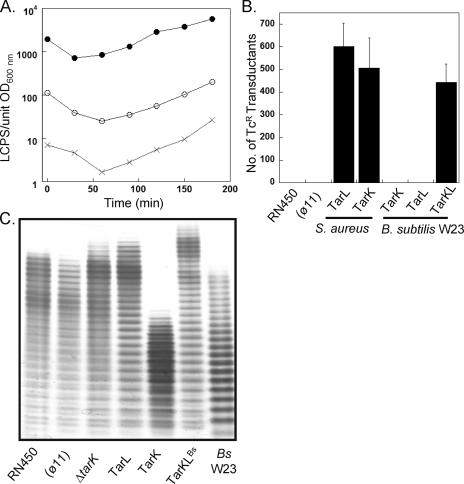
The apparent inability to obtain cross-complementation with TarK and TarL despite the finding that both enzymes apparently make a poly-Rbo WTA raised the possibility that there is differential expression from the respective native promoters. The putative upstream promoter regions from the tarI′J′K (528 bp) and tarIJL (279 bp) clusters were cloned into the luciferase-based promoter probe plasmid pXen1 (17). Both promoters drove expression of the luxABCDE cassette compared to the empty vector background control (Fig. 3A). The tarIJL promoter, however, produced ∼30-fold more luminescence than the tarI′J′K promoter, which suggests that L-WTA is the predominant form of WTA in S. aureus.
FIG. 3.
TarK and TarL promoter analysis and WTA gene complementation. (A) Expression of luxABCDE from pXen-1 using the DNA promoter region fragments (JT204 tarI′JK [○], JT205 tarIJL [•], and JT203 control [×]) measured during early log growth. LCPS, relative luminescence counts per second. (B) Efficiency of φ11-mediated transduction of the tarL::tetL allele into recipient strains overexpressing WTA genes determined by counting viable Tcr colonies after 48 h of incubation. (φ11), strain JT200; S. aureus TarL, strain JT208; S. aureus TarK, strain JT209; B. subtilis W23 TarK, strain JT210; B. subtilis W23 TarL, strain JT211; B. subtilis W23 TarKL, strain JT212. (C) WTA was extracted from the complemented tarL::tetL transductants and analyzed by PAGE. (φ11), strain JT200; ΔtarK, strain JT207; TarL, strain JT213; TarK, strain JT214; TarKLBs, strain JT215; Bs W23, B. subtilis W23 WTA.
TarL and TarK of S. aureus are bifunctional enzymes.
The substantial difference in promoter strength between the tarI′J′K and tarIJL clusters suggested that TarK may be able to complement TarL if it is overexpressed from a strong constitutive promoter. To explore this possibility, various WTA genes were placed under control of the gram-positive PPen promoter and stably integrated into the chromosome of recipient strains. A tarL::tetL allele phage donor strain (JT16) was constructed to test complementation by scoring the efficiency of transduction into recipient strains. Wild-type strain RN450 was used instead of RN4220 since the latter strain has been mutagenized to accept foreign DNA and may harbor unmapped mutations (27). In addition to overexpression of a given gene, the chromosomal tarK gene of each recipient strain was deleted so that the presence of Tcr transductants signaled that the given gene alone could complement TarL in trans. Otherwise, the tarL::tetL allele would be lethal, as it is in the wild-type recipient (Fig. 3B). Indeed, TarK from S. aureus could replace TarL provided that it was sufficiently expressed. A transduction-mediated complementation experiment was also performed with recipients harboring integrated copies of TarK and/or TarL from B. subtilis W23 (TarKBs, TarLBs, and TarKLBs). While TarKBs and TarLBs alone could not complement, transductants were obtained at efficiencies comparable to those of the S. aureus TarL positive control recipient when the two B. subtilis genes (TarKLBs) were supplied simultaneously. Thus, both TarL and TarK of S. aureus are bifunctional enzymes with activities (i.e., RboP priming and polymerization) equivalent to those of the two monofunctional B. subtilis W23 RboP enzymes, TarKBs and TarLBs.
To confirm restoration of WTA biosynthesis in the complemented strains, WTA was extracted and analyzed by PAGE (Fig. 3C). All strains synthesized high-molecular-weight WTA polymer, including the S. aureus-B. subtilis W23 hybrid pathway. The WTA profile of the φ11-lysogenized strain (JT200) was identical to that of wild-type strain RN450, indicating that phage integration does not influence WTA composition. The WTA extracted from the strain overexpressing TarL (JT213) was nearly identical to the WTA of both the lysogenized wild-type control strain (JT200) and the ΔtarK strain (JT207) (see below), consisting of long, homogeneous polymers of L-WTA with little or no K-WTA. In comparison, overexpression of TarK (JT214) without any TarL present resulted in a average polymer length that was ∼10 repeats shorter. While K-WTA was the predominant species, a series of intervening bands with diminished intensity were clearly discernible. The banding pattern of the hybrid S. aureus-B. subtilis W23 WTA pathway strain (JT215) was less complex, consisting of a homogeneous ladder comprised of polymers whose migration distance was identical to that of S. aureus L-WTA.
agr influences WTA length through TarK.
The WTA profile of the RN450 strains (Fig. 3C, lanes 1 and 2) is similar to that of RN4220 (ΔtarK) (Fig. 2B, lane 4) in terms of both length and lack of K-WTA heterogeneity. Further, deletion of tarK from RN450 did not lead to a longer WTA (Fig. 3C, lanes 2 and 3) as observed in RN4220, hinting that tarK regulation is different in the two strains. This is surprising considering that RN4220 was directly derived from the parent RN450 strain (27). While no known RN4220-specific mutations map to WTA-related genes, RN4220 is a partial agr mutant due to a frameshift mutation in agrA (42). The agr system is a two-component quorum-sensing network that attenuates expression of numerous virulence-related cell surface features (8, 34) and was thus a potential cause of the observed strain-dependent expression of K-WTA.
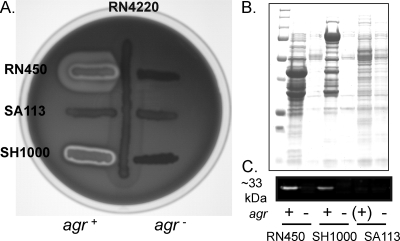
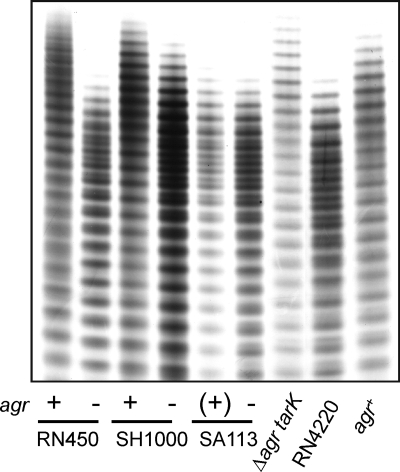
The agr locus is genetically unstable, particularly during cultivation outside the host (42), and it was therefore necessary to explicitly link the agr phenotype to the WTA profile using paired isogenic strains. The agr::tetM null allele was transduced into three commonly used NCTC8325 strains, JT200 [RN450(φ11)], SA113 [NCTC8325(φ11, φ12, φ13])], and SH1000 (RN450 rsbU +). The agr+ phenotype was evaluated by comparison to the corresponding agr::tetM transductant using the following criteria: (i) positive δ-hemolysin (RNAIII) typing on TSA blood agar, (ii) a high level of extracellular protein production, and (iii) secretion of α-hemolysin. The RN450 (JT200) and SH1000 strains used in this study were clearly agr+, producing RNAIII-encoded δ-hemolysin (the enhanced zone of clearing in the inner halo at the cross streak intersection due to synergistic hemolysis with RN4220 secreted β-hemolysin [1] [Fig. 4A]) and a comparatively large quantity of extracellular proteins (Fig. 4B), a high percentage of which was α-hemolysin (upregulated by agr) (Fig. 4C). In comparison, SA113 was nonhemolytic and presumably an agr. The WTA profiles of the strains corresponded without exception to the agr phenotype (Fig. 5) (i.e., long, homogeneous L-WTA polymers in agr+ strains and short, heterogeneous polymers in agr strains). The relationship is independent of the expression level of the alternative sigma factor (σB) as SH1000 behaved like rsbU mutant strains. The effect of deleting agr on WTA composition was not seen in a ΔtarK background (JT221), indicating that agr specifically modulated WTA through tarK and that the chain shortening was not due to a general pleiotropic mechanism, such as alteration of the growth rate (Fig. 5). Further, complementation of the agr defect in RN4220 (JT38) repressed K-WTA synthesis and lengthened WTA, matching the profile of the parent RN450 strain. The WTA expression profile of RN4220 is therefore the result of a single lesion in the agr locus and indicates that the agr global regulatory system represses synthesis of K-WTA.
FIG. 4.
Analysis of the agr phenotype. (A) Secretion of hemolysins as qualitatively assessed on TSA blood agar plates. The inner halo of clearing is due to the agr RNAIII transcript which encodes δ-hemolysin (1). (B) Extracellular protein fraction obtained from an equal number of cells separated by SDS-PAGE and stained with Coomassie blue. (C) Western blot of extracellular protein fractions probed with anti-α-hemolysin antibody. Approximately 5 μg of protein was loaded for each sample.
FIG. 5.
agr system affects WTA length and heterogeneity: PAGE profile of WTA extracts from wild-type strains and the corresponding agr::tetM null deletion strains. Repair of the agr defect in RN4220 repressed K-WTA and increased the average chain length of L-WTA. Δagr tarK, strain JT221; agr+, strain JT38.
DISCUSSION
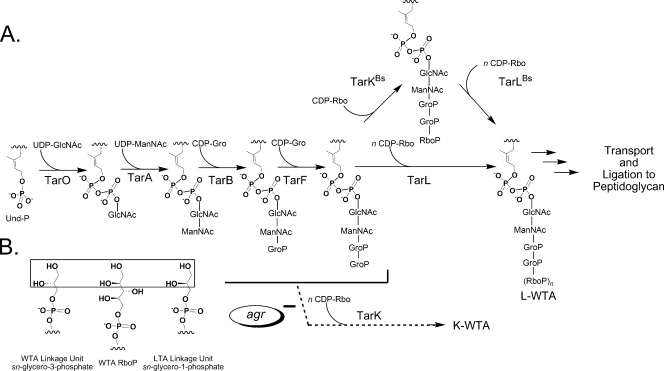
It has been suggested that the tarK and tarL genes of S. aureus each have a unique enzymatic activity, namely, RboP priming (TarK) of the linkage unit and the subsequent RboP polymerization (TarL) to complete the teichoic acid chain (11). This model was based on the proposed B. subtilis W23 poly-RboP pathway (28), although there is limited experimental evidence concerning the enzymes involved in RboP addition for either bacterial strain. The validity of extending the B. subtilis W23 two-enzyme RboP primase-polymerase model to S. aureus has been questioned based on a computational analysis of the gene clusters, which revealed not only different genetic organizations but also an apparent three-gene duplication specific to S. aureus (tarI′J′K and tarIJL) (Fig. 1A) (38). For the two-enzyme RboP primase-polymerase model to hold true in S. aureus, two proteins, TarK and TarL (80% identity and 90% similarity), would have to catalyze distinct reactions. Qian et al. suggested that different activities were possible given the sequence divergence between the N termini of TarK and TarL (38). They also suggested a second possibility, that the two clusters may actually be functionally redundant, which would mean that there is no RboP priming enzyme in the pathway. This study suggests a third scenario, in which both TarL and TarK are indeed bifunctional RboP priming and RboP polymerizing WTA enzymes yet remain functionally nonredundant. We propose that TarL and TarK direct the synthesis of two distinct cell wall WTA polymers (a primary L-WTA and a secondary K-WTA, respectively) in a revised poly-RboP WTA pathway (Fig. 6). We further suggest that the sequence divergence in the N termini of TarK and TarL actually reflects different linkage unit substrate specificity and not a shift between RboP priming and polymerization.
FIG. 6.
(A) Revised model of poly-RboP WTA biosynthesis in S. aureus incorporating a bifunctional TarL RboP priming and polymerization enzyme. The validated two-enzyme model for B. subtilis W23 is shown for comparison. The branch point for the agr repressed K-WTA pathway is arbitrary as the structure is unknown (see text). The addition of a single GroP unit is shown for TarF to correlate with the observed activity of recombinant enzyme (6). Und-P, undecaprenyl phosphate. (B) Chemical structure of the acceptor substrate during addition of CDP-Rbo to GroP in the WTA linkage unit, to RboP in the WTA chain, and to GroP in lipoteichoic acid (LTA). Note the different stereochemistry of the secondary alcohol at the α-carbon in the in vivo (GroP WTA and RboP WTA) and in vitro acceptors (enclosed in a box).
A regulated ectopic expression system was used to show that TarL is an essential gene (Fig. 1B) but that a ΔtarK mutant is viable. The ΔtarK strain makes full-length WTA RboP polymer, thus ruling out the possibility that TarK of S. aureus has a dedicated RboP primase activity in vivo. Second, the WTA heterogeneity determined by high-resolution PAGE directly corresponds to expression of TarK. Strains expressing only TarL are homogeneous with respect to extractable WTA, whereas strains expressing both enzymes have a second set of intervening bands that is dependent upon TarK. The unique electrophoretic mobility suggests that K-WTA has a different structure than L-WTA and that S. aureus NCTC8325 has at least two WTA polymer types. While the structure of the newly identified K-WTA is not known, all phages tested infected equally well whether K-WTA was produced at basal levels (RN4220), not present (JT18), or overexpressed in the presence (JT209) or in the absence (JT214) of TarL (Fig. 2A and data not shown). Since poly-RboP WTA is the receptor for phage attachment (3) and strains expressing only tarK are susceptible, K-WTA is equivalent to L-WTA with regard to phage binding and is therefore likely composed of RboP repeats. This is consistent with the retention of K-WTA when the tarI′J′ genes are deleted, as the tarIJ products (characterized CDP-Rbo-synthesizing enzymes [36]) can supply the common donor substrate pool for both L-WTA and K-WTA. The difference between the two structures may stem from a poly-RboP chain modification or from differences within the linkage unit. The two known WTA modifications in S. aureus are glycosylation with d-GlcNAc and esterification with d-alanine residues (32). However, deletion of the genes encoding two putative glycosyl transferases located within the WTA clusters (SAOUHSC_00644 and SAOUHSC_00228) does not affect the amount of K-WTA in RN4220 (T. C Meredith, J. G. Swoboda, and S. Walker, unpublished), and d-alanine teichoic acid esters are highly labile under the basic conditions used for extraction (31). Alkaline hydrolysis of WTA occurs between C-4 of ManNAc and the proximal GroP within the linkage unit (26). Barring the existence of unknown WTA modifications and/or an alternative connection to the linkage unit disaccharide core, this localizes the structural difference between K-WTA and L-WTA to the linkage unit region distal to ManNAc but prior to the poly-RboP repeat. A potential candidate is the number of GroP units, as both (GroP)2- and (GroP)3-ManNAc-GlcNAc-PP-undecaprenyl have been shown to serve equally well as acceptors for RboP units in S. aureus H membrane preparations (47).
Transduction efficiency analysis yielded clear results, showing that TarK could complement TarL if it was provided at levels much higher than those produced from its relatively weak endogenous promoter (Fig. 3A and 3B). The WTA profile when only TarK was expressed from an unregulated strong promoter (JT214) not only increased the amount of K-WTA, as anticipated (shorter chain), but also produced heterogeneity despite the fact that TarL was absent (Fig. 3C). This suggests that TarK can synthesize L-WTA when it is overexpressed from a strong promoter, albeit much less efficiently than K-WTA. The accumulation of WTA intermediates is lethal in S. aureus (11). The need for high-level TarK expression to support viability in a ΔtarL background may reflect the need to utilize an alternative linkage unit acceptor substrate to alleviate the buildup of WTA intermediates.
The S. aureus TarK and TarL results raised the possibility that the B. subtilis W23 model may also be incorrect. However, the heterologous complementation results support the two-enzyme RboP priming-RboP polymerization pathway in B. subtilis W23 as proposed (28) since both the TarKBs and TarLBs enzymes are needed to replace the bifunctional S. aureus TarL enzyme (Fig. 5 and 6). The hybrid WTA pathway strain makes a single WTA species, suggesting that the putative K-WTA linkage unit acceptor is not recognized by TarKLBs.
Both TarK and TarL of S. aureus are bifunctional RboP priming and polymerization enzymes, implying that there is active site plasticity. While the CDP-Rbo donor is the same, each activity requires the utilization of a primary hydroxyl acceptor with an α-carbon secondary alcohol of differing stereochemistry (Fig. 6B). Active site plasticity may explain why lipoteichoic acid, a GroP polyol with different stereochemistry than GroP of the WTA linkage unit (32), can serve as an acceptor in vitro but does not occur in vivo due to lipoteichoic acid terminus accessibility (15, 16).
The biological significance of having K-WTA in addition to the primary L-WTA is unknown at present. However, the observation that in all S. aureus strains the K-WTA cluster is separated from the main WTA cluster by an intergenic gap of variable length suggests a conserved role (38). PAGE analysis confirmed that the average poly-RboP chain length increased by approximately 7 to 10 repeat units either when tarK was deleted or when the agr system was functional (Fig. 2B and 5). The average chain length of poly-RboP WTA in RN450 as determined by compositional analysis has previously been estimated to be ∼22 RboP repeats (24), indicating that the agr system can change the average length of WTA by up to 50%. This alone would be expected to significantly influence both the bacterial surface and the degree of surface accessibility for intracellular contacts. While details concerning the mechanism of agr regulation of K-WTA are not known, a clear agr- and tarK-dependent effect on WTA length and composition in S. aureus NCTC8325 has been demonstrated (Fig. 5). The quorum-sensing agr system is a global regulator of numerous virulence determinants that is most active during the transition from exponential to postexponential growth (8, 34). The agr locus generally upregulates expression of exoproteins (including α-hemolysin), while it represses the synthesis of cell surface adhesins. The net effect is to change the bacterial surface from a “proadhesion” state to a “low-adhesion” state in order to facilitate dissemination once a local cell density has been reached (8, 34). Given the role of WTA in adhesion to mammalian cells (45, 46), it is possible to speculate that agr either directly or indirectly represses K-WTA in order to reduce the total WTA surface density. Further, the shorter WTA chain length that results when agr is not active (i.e., early exponential growth) may facilitate surface exposure of other bona fide bacterial surface protein-based adhesins collectively known as MSCRAMMs (microbial surface components recognizing adhesive matrix molecules) (9). This raises the possibility that not only WTA length but also WTA density may be part of the agr regulon and that S. aureus has the ability to dynamically modulate its WTA.
Acknowledgments
This research was supported by the NIH (grant GM078477 to S.W. and grant F3178727 to J.G.S.) and by training grant T32-AI07061-30 to T.C.M.
We thank the following individuals for generously providing materials: Olaf Schneewind (University of Chicago), who provided pKOR1, RN4220, and RN4220ΔtarO; Jean C. Lee (Harvard Medical School), who provided phages φ80α, φ85, and pLI50; Richard P. Novick (New York University), who provided pCN33, pCN59, pCN68, and pRN8298; David Rudner (Harvard Medical School), who provided pDR110; Chia Lee (University of Arkansas), who provided pCL25 and pCL55; Mark S. Smeltzer (University of Arkansas), who provided RN6911; Ambrose L. Cheung (Dartmouth College), who provided SH1000; and Francis Arhin (Targanta Therapeutics, Quebec, Canada), who provided RN450 and φ11.
Footnotes
Published ahead of print on 15 February 2008.
REFERENCES
- 1.Adhikari, R. P., S. Arvidson, and R. P. Novick. 2007. A nonsense mutation in agrA accounts for the defect in agr expression and the avirulence of Staphylococcus aureus 8325-4 traP::kan. Infect. Immun. 754534-4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aly, R., H. R. Shinefield, C. Litz, and H. I. Maibach. 1980. Role of teichoic acid in the binding of Staphylococcus aureus to nasal epithelial cells. J. Infect. Dis. 141463-465. [DOI] [PubMed] [Google Scholar]
- 3.Archibald, A. R. 1980. Phage receptors in Gram positive bacteria, p. 5-26. In L. Randall, L. Philipson, and K. Lonberg-Holm (ed.), Virus receptors (receptors and recognition), series B, vol. 7. Chapman and Hall, London, United Kingdom. [Google Scholar]
- 4.Bae, T., and O. Schneewind. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 5558-63. [DOI] [PubMed] [Google Scholar]
- 5.Bensadoun, A., and D. Weinstein. 1976. Assay of proteins in the presence of interfering materials. Anal. Biochem. 70241-250. [DOI] [PubMed] [Google Scholar]
- 6.Brown, S., Y. H. Zhang, and S. Walker. 2008. A revised pathway proposed for Staphylococcus aureus wall teichoic acid biosynthesis based on in vitro reconstitution of the intracellular steps. Chem. Biol. 1512-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charpentier, E., A. I. Anton, P. Barry, B. Alfonso, Y. Fang, and R. P. Novick. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 706076-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y. Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 401-9. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, S. R., and S. J. Foster. 2006. Surface adhesins of Staphylococcus aureus. Adv. Microb. Physiol. 51187-224. [DOI] [PubMed] [Google Scholar]
- 10.D'Elia, M. A., K. E. Millar, T. J. Beveridge, and E. D. Brown. 2006. Wall teichoic acid polymers are dispensable for cell viability in Bacillus subtilis. J. Bacteriol. 1888313-8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Elia, M. A., M. P. Pereira, Y. S. Chung, W. Zhao, A. Chau, T. J. Kenney, M. C. Sulavik, T. A. Black, and E. D. Brown. 2006. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J. Bacteriol. 1884183-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellwood, D. C., and D. W. Tempest. 1972. Effects of environment on bacterial wall content and composition. Adv. Microb. Physiol. 783-116. [Google Scholar]
- 13.Endl, J., H. P. Seidl, F. Fiedler, and K. H. Schleifer. 1983. Chemical composition and structure of cell wall teichoic acids of staphylococci. Arch. Microbiol. 135215-223. [DOI] [PubMed] [Google Scholar]
- 14.Fan, F., R. D. Lunsford, D. Sylvester, J. Fan, H. Celesnik, S. Iordanescu, M. Rosenberg, and D. McDevitt. 2001. Regulated ectopic expression and allelic-replacement mutagenesis as a method for gene essentiality testing in Staphylococcus aureus. Plasmid 4671-75. [DOI] [PubMed] [Google Scholar]
- 15.Fischer, W., H. U. Koch, P. Rosel, and F. Fiedler. 1980. Alanine ester-containing native lipoteichoic acids do not act as lipoteichoic acid carrier. Isolation, structural and functional characterization. J. Biol. Chem. 2554557-4562. [PubMed] [Google Scholar]
- 16.Fischer, W., H. U. Koch, P. Rosel, F. Fiedler, and L. Schmuck. 1980. Structural requirements of lipoteichoic acid carrier for recognition by the poly-(ribitol phosphate) polymerase from Staphylococcus aureus H. A study of various lipoteichoic acids, derivatives, and related compounds. J. Biol. Chem. 2554550-4556. [PubMed] [Google Scholar]
- 17.Francis, K. P., D. Joh, C. Bellinger-Kawahara, M. J. Hawkinson, T. F. Purchio, and P. R. Contag. 2000. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect. Immun. 683594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillaspy, A. F., V. Worrell, J. Orvis, B. A. Roe, D. W. Dyer, and J. J. Iandolo. 2006. The Staphylococcus aureus NCTC8325 genome, p. 381-412. In V. Fischetti, R. Novick, J. Ferretti, D. Portnoy, and J. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, DC.
- 19.Grundling, A., and O. Schneewind. 2006. Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. J. Bacteriol. 1882463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruss, A., and R. Novick. 1986. Plasmid instability in regenerating protoplasts of Staphylococcus aureus is caused by aberrant cell division. J. Bacteriol. 165878-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hancock, I. C., and J. Baddiley. 1985. Biosynthesis of the bacterial envelope polymers teichoic acid and teichuronic acid, p. 279-307. In A. N. Martonosi (ed.), The enzymes of biological membranes, 2nd ed., vol. 2. Plenum Press, New York, NY. [Google Scholar]
- 22.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 1845457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jana, M., T. T. Luong, H. Komatsuzawa, M. Shigeta, and C. Y. Lee. 2000. A method for demonstrating gene essentiality in Staphylococcus aureus. Plasmid 44100-104. [DOI] [PubMed] [Google Scholar]
- 24.Jenni, R., and B. Berger-Bachi. 1998. Teichoic acid content in different lineages of Staphylococcus aureus NCTC8325. Arch. Microbiol. 170171-178. [DOI] [PubMed] [Google Scholar]
- 25.Kaito, C., and K. Sekimizu. 2007. Colony spreading in Staphylococcus aureus. J. Bacteriol. 1892553-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kojima, N., Y. Araki, and E. Ito. 1985. Structure of the linkage units between ribitol teichoic acids and peptidoglycan. J. Bacteriol. 161299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305709-712. [DOI] [PubMed] [Google Scholar]
- 28.Lazarevic, V., F. X. Abellan, S. B. Moller, D. Karamata, and C. Mauel. 2002. Comparison of ribitol and glycerol teichoic acid genes in Bacillus subtilis W23 and 168: identical function, similar divergent organization, but different regulation. Microbiology 148815-824. [DOI] [PubMed] [Google Scholar]
- 29.Lee, C. Y., S. L. Buranen, and Z. H. Ye. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103101-105. [DOI] [PubMed] [Google Scholar]
- 30.Luong, T. T., and C. Y. Lee. 2007. Improved single-copy integration vectors for Staphylococcus aureus. J. Microbiol. Methods 70186-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirelman, D., B. D. Beck, and D. R. Shaw. 1970. The location of the d-alanyl ester in the ribitol teichoic acid of Staphylococcus aureus. Biochem. Biophys. Res. Commun. 39712-717. [DOI] [PubMed] [Google Scholar]
- 32.Neuhaus, F. C., and J. Baddiley. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67686-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novick, R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33155-166. [DOI] [PubMed] [Google Scholar]
- 34.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 481429-1449. [DOI] [PubMed] [Google Scholar]
- 35.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 123967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pereira, M. P., and E. D. Brown. 2004. Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase activities in Haemophilus influenzae and Staphylococcus aureus. Biochemistry 4311802-11812. [DOI] [PubMed] [Google Scholar]
- 37.Pollack, J. H., and F. C. Neuhaus. 1994. Changes in wall teichoic acid during the rod-sphere transition of Bacillus subtilis 168. J. Bacteriol. 1767252-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian, Z., Y. Yin, Y. Zhang, L. Lu, Y. Li, and Y. Jiang. 2006. Genomic characterization of ribitol teichoic acid synthesis in Staphylococcus aureus: genes, genomic organization and gene duplication. BMC Genomics 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166368-379. [DOI] [PubMed] [Google Scholar]
- 40.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73133-138. [DOI] [PubMed] [Google Scholar]
- 41.Shockman, G. D., and J. F. Barrett. 1983. Structure, function, and assembly of cell walls of gram-positive bacteria. Annu. Rev. Microbiol. 37501-527. [DOI] [PubMed] [Google Scholar]
- 42.Traber, K., and R. Novick. 2006. A slipped-mispairing mutation in AgrA of laboratory strains and clinical isolates results in delayed activation of agr and failure to translate delta- and alpha-haemolysins. Mol. Microbiol. 591519-1530. [DOI] [PubMed] [Google Scholar]
- 43.Wang, L., J. D. Trawick, R. Yamamoto, and C. Zamudio. 2004. Genome-wide operon prediction in Staphylococcus aureus. Nucleic Acids Res. 323689-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward, J. B. 1981. Teichoic and teichuronic acids: biosynthesis, assembly, and location. Microbiol. Rev. 45211-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weidenmaier, C., J. F. Kokai-Kun, S. A. Kristian, T. Chanturiya, H. Kalbacher, M. Gross, G. Nicholson, B. Neumeister, J. J. Mond, and A. Peschel. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10243-245. [DOI] [PubMed] [Google Scholar]
- 46.Weidenmaier, C., A. Peschel, Y. Q. Xiong, S. A. Kristian, K. Dietz, M. R. Yeaman, and A. S. Bayer. 2005. Lack of wall teichoic acids in Staphylococcus aureus leads to reduced interactions with endothelial cells and to attenuated virulence in a rabbit model of endocarditis. J. Infect. Dis. 1911771-1777. [DOI] [PubMed] [Google Scholar]
- 47.Yokoyama, K., T. Miyashita, Y. Araki, and E. Ito. 1986. Structure and functions of linkage unit intermediates in the biosynthesis of ribitol teichoic acids in Staphylococcus aureus H and Bacillus subtilis W23. Eur. J. Biochem. 161479-489. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, L., F. Fan, L. M. Palmer, M. A. Lonetto, C. Petit, L. L. Voelker, A. St John, B. Bankosky, M. Rosenberg, and D. McDevitt. 2000. Regulated gene expression in Staphylococcus aureus for identifying conditional lethal phenotypes and antibiotic mode of action. Gene 255297-305. [DOI] [PubMed] [Google Scholar]