Abstract
Chlamydophila pneumoniae is a gram-negative obligate intracellular bacterial pathogen that causes pneumonia and bronchitis and may contribute to atherosclerosis. The developmental cycle of C. pneumoniae includes a morphological transition from an infectious extracellular elementary body (EB) to a noninfectious intracellular reticulate body (RB) that divides by binary fission. The C. pneumoniae genome encodes a type III secretion (T3S) apparatus that may be used to infect eukaryotic cells and to evade the host immune response. In the present study, Cpn0712 (CdsD), Cpn0704 (CdsQ), and Cpn0826 (CdsL), three C. pneumoniae genes encoding yersiniae T3S YscD, YscQ, and YscL homologs, respectively, were cloned and expressed as histidine- and glutathione S-transferase (GST)-tagged proteins in Escherichia coli. Purified recombinant proteins were used to raise hyper-immune polyclonal antiserum and were used in GST pull-down and copurification assays to identify protein-protein interactions. CdsD was detected in both EB and RB lysates by Western blot analyses, and immunofluorescent staining demonstrated the presence of CdsD within inclusions. Triton X-114 solubilization and phase separation of chlamydial EB proteins indicated that CdsD partitions with cytoplasmic proteins, suggesting it is not an integral membrane protein. GST pull-down assays indicated that recombinant CdsD interacts with CdsQ and CdsL, and copurification assays with chlamydial lysates confirmed that native CdsD interacts with CdsQ and CdsL. To the best of our knowledge, this is the first report demonstrating interactions between YscD, YscQ, and YscL homologs of bacterial T3S systems. These novel protein interactions may play important roles in the assembly or function of the chlamydial T3S apparatus.
Gram-negative bacterial pathogens such as Salmonella, Shigella, Yersinia, and Escherichia coli use type III secretion (T3S) to deliver toxins and effector proteins directly into the cytoplasm of host cells, resulting in bacterial uptake, survival, and virulence. The T3S apparatus is composed of approximately 20 to 25 functionally conserved proteins that assemble to form a unified structure containing oligomeric protein complexes in the bacterial cytoplasm (C ring), inner membrane (inner ring), periplasm (inner rod), outer membrane (outer ring; secretin), extracellular space (needle, needle extension, and ruler), and host cell membrane (translocons). This elongated needle complex is also known as the injectisome, and upon cellular contact, the translocons insert into the host cell plasma membrane, forming a conduit for the delivery of effector proteins into the host cytoplasm (10).
Chlamydophila pneumoniae is an obligate intracellular gram-negative pathogen with a unique biphasic developmental cycle. C. pneumoniae elementary bodies (EB) attach to host cells and are internalized via an unknown mechanism involving the activation of Rho family GTPases, actin polymerization, and the formation of microvilli (7, 9, 43). Once within cells, metabolically inert EB transform into reticulate bodies (RB) and divide by binary fission within membrane-bound intracellular inclusions. The developmental cycle culminates with the transformation of RB back into EB and the release of infectious EB from the host cell (22). T3S likely plays a key role in this infection process (16), and it has recently been suggested that chlamydial T3S may be involved in several virulence mechanisms, including the progression of the developmental cycle (45), the inhibition of host-cell apoptosis (37), and the prevention of immune system recognition of Chlamydia-infected cells (12, 25, 48).
Interestingly, phylogenetic analyses of four conserved proteins of the injectisome indicate that the Chlamydiae encode a distinct family of the T3S apparatus (18), and many T3S proteins of Chlamydia are homologous with the Yersinia secretion complex and flagellar T3S system proteins (10). Conclusive evidence of the use and presence of T3S structures on the surface of Chlamydia has yet to be reported; however, recent biochemical and genetic evidence suggests a functional T3S system in Chlamydia (16, 19). For instance, heterologous bacterial T3S systems are able to translocate chlamydial effectors, including the inclusion membrane proteins (14, 21, 41). Additionally, TARP (translocated actin recruiting phosphoprotein), CopN (the putative T3S plug protein), CopB/CopB2 (putative translocons), and CPAF (Chlamydia protease-like activity factor) are thought to be secreted into the host cytoplasm by Chlamydia (8, 14, 15, 28, 42, 48). In the absence of a genetic transformation system, biochemical characterization of the putative chlamydial T3S proteins provides important information about the roles of chlamydial T3S proteins.
The inner membrane ring of the T3S system is composed of approximately six proteins, including YscJ, YscR, YscS, YscT, YscU, and YscV (17, 46). Cytoplasmic and peripheral membrane proteins include YscQ, YscL, YscN, LcrE, YscK, and chaperones (10). YscD, a protein shown to be essential in T3S (30, 38), localizes to the inner membrane of Yersinia (38), although Pas, the YscD homolog of enterohemorrhagic E. coli, was located in both the cytoplasm and inner membrane (27). YscQ and YscL form cytoplasmic complexes in Yersinia (23), and crystallization of the YscQ homologs HrcQB-C and FliN (5, 13) revealed that YscQ tetramerizes in order to form the building blocks of the C ring of the T3S apparatus. YscL inhibits and tethers YscN, an ATPase, to the inner membrane ring of the Yersinia T3S apparatus (3) as well as interacts with YscQ, YscK, and YscN in cytoplasmic multiprotein T3S complexes (23). The precise role of these molecular interactions is not known. Three genes of C. pneumoniae, Cpn0704 (CdsQ), Cpn0826 (CdsL), and Cpn0712 (CdsD), encode Yersinia homologs YscQ, YscL, and YscD, respectively. Transcripts for CdsQ, CdsD, and CdsL were not detected in EB (29), although regulated transcription of these genes throughout the chlamydial developmental cycle has been shown (1, 2, 34, 40). CdsD has also been detected by mass spectrometry and immunoblot analysis, indicating that the protein is made in Chlamydia (20, 44). CdsD, CdsQ, and CdsL may therefore play important roles in the assembly and functioning of the putative chlamydial T3S apparatus.
Given the abundance of data supporting a functional T3S system in Chlamydia and the importance of interactions between protein subunits in the assembly and function of the injectisome in other bacteria, surprisingly little is known regarding the associations between proteins of the putative chlamydial T3S apparatus. To date, only a few studies have elucidated interactions between proteins predicted to be part of the chlamydial T3S system, and these have focused on identifying interactions between chaperones and effectors (14, 39). We report for the first time interactions between CdsD, CdsQ, and CdsL, proteins predicted to be related to the structure and function of the chlamydial T3S apparatus. Importantly, we identify CdsQ and CdsL as interacting partners of CdsD, two interactions never before demonstrated in bacterial T3S systems that may be important in the function of this apparatus in Chlamydia.
MATERIALS AND METHODS
Construction of expression plasmids.
Genomic DNA was isolated from C. pneumoniae CWL029 (ATCC VR1310) (GenBank accession number AE001363) using the Sigma GenElute kit. attB-containing primers (Gateway; Invitrogen) specific to the full-length Cpn0826 open reading frame (ORF) (CdsL; amino acids 1 to 233), full-length Cpn0704 ORF (CdsQ; amino acids 1 to 371), full-length Cpn0324 ORF (CopN; amino acids 1 to 399), full-length Cpn0707 ORF (CdsN; amino acids 1 to 442), and full-length Cpn0478 ORF (HflX; amino acids 1 to 472) were used to amplify the genes with flanking attB sites. The full-length Cpn0712 ORF (CdsD; amino acids 1 to 845), the forkhead-associated 2 (FHA-2) domain of CdsD (FHA-2; amino acids 398 to 547), full-length Cpn0095 (PknD; amino acids 1 to 932), and the PknD kinase domain (KD) (amino acids 1 to 293) were cloned previously (24). The amplified products were cloned into pDONR201 (Gateway; Invitrogen) to generate pENT vectors. The pENT vectors were subcloned into pDEST15 or pDEST17 to generate the expression vectors pEX15CdsQ, pEX15CdsL, pEX17CdsQ, pEX17CdsL, pEX15CopN, pEX15CdsN, and pEX17HflX (pEX17CdsD, pEX17FHA-2, pEX17PknD, and pEX15KD were from the previous study). All constructs were verified by sequencing.
Separation of EB into soluble and integral membrane protein fractions.
Phase separation of EB proteins was carried out essentially as described previously (4, 16). Briefly, chlamydial EB were treated with 5 mM dithiothreitol and pelleted for 20 min at 4°C. EB were resuspended in 50 mM Tris-HCl, pH 7.4, 100 mM KCl, and 1% Triton X-114, incubated on ice for 10 min, and then incubated at 37°C for 10 min. The sample was pelleted at 16,000 × g in a microcentrifuge, and the top phase (soluble proteins) was removed to a separate tube and treated with Triton X-114, while the bottom phase (containing integral membrane proteins) was treated with buffer without Triton X-114. This cycle was repeated four times, and proteins from each phase were precipitated overnight with 10% trichloroacetic acid.
Production of recombinant protein.
E. coli Rosetta(pLysS) or BL21(DE3) was transformed with protein expression vectors (see above) and plated on Luria-Bertani (LB) plates containing 100 μg/ml ampicillin with or without 34 μg/ml chloramphenicol. Three medium-sized colonies from each plate were pooled into 5 ml LB broth containing the appropriate antibiotics and grown overnight at 37°C. Overnight cultures were inoculated into 750 ml LB broth containing antibiotics, incubated at 37°C at 270 rpm until the optical density at 600 nm was 0.4 to 0.6, and then cooled on ice to 20°C. The production of recombinant protein was initiated with the addition of 0.2 mM isopropyl-β-d-thiogalactopyranoside, and cultures were incubated at room temperature (23°C) at 270 rpm for 2 h. Cultures were then centrifuged at 6,000 × g for 20 min, and pellets were washed with 200 ml ice-cold mtPBS buffer consisting of 140 mM NaCl, 16 mM Na2HPO4, and 4 mM NaH2PO4. Pellets were resuspended in 10 ml ice-cold mtPBS (containing 1× complete EDTA-free protease inhibitors) for purification on glutathione agarose and frozen overnight at −20°C.
Purification of GST proteins.
Culture suspensions were thawed on ice and sonicated (as described above), and insoluble material was removed by centrifugation at 20,000 × g for 20 min at 4°C. Supernatants were filtered through 0.2-μm acrodisc filters (Pall Corporation) onto 250 μl glutathione agarose (Sigma) and rotated on a rocking platform for 16 h at 4°C. Beads were collected by centrifugation at 500 × g for 10 min at 4°C and washed four times with mtPBS. Glutathione S-transferase (GST) fusion proteins on glutathione agarose beads were stored at 4°C for use in the pull-down assays.
Production and affinity purification of rabbit polyclonal antibody to CdsD.
Hyper-immune guinea pig and rabbit antisera were raised against CdsD and the FHA-2 domain of CdsD, respectively. Briefly, His-CdsD and His-FHA-2 were expressed in Rosetta(pLysS) and purified on Ni nitrilotriacetic acid (NTA) agarose (Qiagen) according to established procedures (24), and 100 μg or 500 μg was delivered to Cocalico Biologicals, Inc. (Reamstown, PA), as a Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel slice. Three rounds of immunizations with the adjuvant Titermax were employed. Immunoglobulin G (IgG) molecules were precipitated from the hyper-immune antisera with 50% saturated ammonium sulfate solution, resuspended in cold phosphate-buffered saline (PBS), and dialyzed into PBS. Activated CH-Sepharose beads (Sigma) were coupled with 10 mg His-FHA-2 and used to affinity purify the rabbit anti-FHA-2 IgG. Briefly, antibody was incubated with the His-FHA-2-conjugated beads overnight at 4°C with rocking and washed with ice-cold PBS until the A280 was less than 0.02. Glycine (100 mM, pH 3.0) was used to elute the rabbit anti-FHA-2 IgG molecules in 1-ml fractions into Eppendorf tubes containing 10 μl of 1.5 M Tris, pH 8.8. Fractions were analyzed by probing Western blots containing chlamydial EB lysate, E. coli lysate containing overexpressed His-CdsD, and E. coli lysate containing His-PknD.
Indirect immunofluorescence.
HeLa cells at 60 to 70% confluence on 1.1 cm2 coverslips were infected with C. pneumoniae CWL029 at a multiplicity of infection of 1 and grown for 48 h at 37°C in the presence of 1 μg/ml cycloheximide in minimal essential medium (with l-glutamine and Earle's salts) and a 5% CO2 atmosphere. Infected HeLa cells were washed three times with cold PBS and fixed for 10 min in 3.7% formaldehyde in PBS. Coverslips were then washed three times with cold PBS and stored at 4°C for up to 2 weeks. Cells were permeabilized with 0.05% Tween-20 in PBS for 5 min, washed three times with PBS, and blocked with 1% bovine serum albumin (BSA) in PBS for 1 h at room temperature. Primary antibody solutions were prepared by diluting either rabbit preimmune serum to a dilution of 1:1,250 (32 μg/ml total serum protein) or affinity-purified rabbit anti-FHA-2 IgG to a dilution of 1:25 (16 μg/ml final concentration) in normal mouse gamma globulin direct fluorescent-antibody assay reagent (Diagnostics Hybrids) containing Evans blue counterstain. Antibody was incubated on the coverslips overnight at 4°C with shaking. Coverslips were then washed three times with PBS, and fluorescein isothiocyanate-conjugated sheep anti-rabbit IgG (Sigma) was added at a dilution of 1:200 in 1% BSA in PBS for 1 h at room temperature. Coverslips were washed three times in PBS and mounted on a 1:1 PBS-glycerol solution and visualized using an Olympus BX51 fluorescent microscope. Images were captured at ×400 magnification with a mounted camera (Q-Color5; Olympus) using the software QCapture.
GST pull-down assays.
In order to investigate protein-protein interactions between predicted components of the chlamydial T3S system, pull-down assays were carried out essentially as described previously (35). Briefly, 10 μl glutathione agarose beads bound to 20 micrograms of prey protein (GST or CdsQ, CdsL, CopN, CdsN, or the PknD KD fused with GST) (see the description of the purification of GST proteins above) were blocked overnight in 4% BSA Tris-buffered saline with Tween 20. The agarose beads were then briefly centrifuged at 4°C, and the supernatant was discarded. The agarose beads were then resuspended and incubated with E. coli lysate containing bait protein (His-CdsD, His-FHA-2, His-CdsQ, or His-CdsL) in binding buffer (50 mM potassium phosphate, pH 7.2; 150 mM KCl; 1 mM MgCl2) containing 2% (vol/vol) glycerol and 0.2% (vol/vol) Triton X-100 for 3 h at 4°C. Beads were collected by brief centrifugation and washed four times with 50 mM Tris-HCl, pH 7.45, containing either 0, 200, or 500 mM NaCl. Each wash step was completed within 5 min; essentially the beads were resuspended in wash buffer, and the tubes were inverted three times and then allowed to settle on ice prior to a brief centrifugation and the removal of the supernatants. Washed beads were resuspended in 20 μl 2× SDS-PAGE loading buffer and boiled for 5 min prior to SDS-PAGE, Western blotting, and enhanced chemiluminescence (ECL).
Copurification assays.
Copurification assays were carried out essentially as described previously (23) in order to investigate the interaction of CdsD from RB with His-CdsQ and His-CdsL. Briefly, 42 h postinfection, RB were purified on a discontinuous gradient (6) and lysed with a freeze-thaw cycle and incubated on ice for 30 min in PBS containing 1% Triton X-100. RB lysates were precleared by microcentrifugation at 16, 000 × g for 30 min and then incubated with E. coli lysate containing equal amounts of His-CdsQ, His-CdsL, or His-HflX GTPase (negative control) in binding buffer for 6 h. Twenty microliters of Ni NTA agarose beads were then added to each tube overnight. Beads were collected by brief centrifugation, washed four times with binding buffer containing 20 mM imidazole, and boiled in 2× SDS-PAGE loading buffer. Copurified CdsD was detected by Western blotting and ECL using hyper-immune guinea pig anti-CdsD antiserum.
RESULTS
Sequence analysis of CdsD, CdsQ, and CdsL.
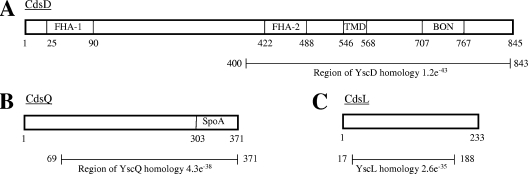
CdsD (gene identifier Cpn0712) consists of 845 amino acids with a predicted molecular mass of approximately 93.2 kDa. CdsD contains two FHA domains, one located near the N terminus of the molecule and the other within the 444 C-terminal amino acids that are homologous with YscD (expectation value [E value] of 1.2e−43; see www.tigr.org) (Fig. 1A). Putative transmembrane and phospholipid binding domains are also located within the region of YscD homology. CdsQ (gene identifier Cpn0704) consists of 371 amino acids with a predicted molecular mass of approximately 41.2 kDa. Amino acids 69 to 371 are homologous with YscQ (E value of 4.3e−38) (Fig. 1B), amino acids 303 to 371 are homologous with the “surface presentation of antigens” (SpoA) domain (e = 0.0019), and amino acids 309 to 371 are homologous to FliN (E value of 7.9e−06), a flagellar motor switch of the flagellar T3S apparatus. CdsL (gene identifier Cpn0826) consists of 233 amino acids with a predicted molecular mass of approximately 25.8 kDa. Amino acids 17 to 188 are homologous with YscL (E value of 2.6e−35) (Fig. 1C), and amino acids 13 to 201 are homologous with FliH (e = 0.00084), a flagellar assembly protein.
FIG. 1.
Domain organization of CdsD, CdsQ, and CdsL. (A) CdsD contains a predicted transmembrane domain (TMD), phospholipid binding domain (BON), and two forkhead-associated (FHA) domains. The region of YscD homology (E value of 1.2e−43) is shown. (B) CdsQ contains a “surface presentation of antigens” (SpoA) domain. The region of YscQ homology (E value of 4.3e−38) is shown. (C) The region of YscL homology (E value of 2.6e−35) is shown. Amino acid numbers are given.
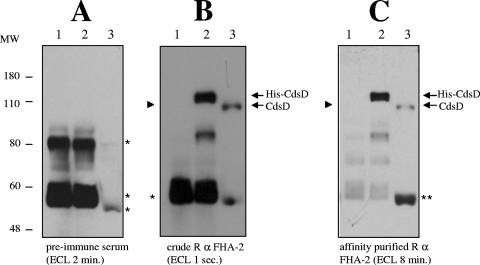
Detection of full-length CdsD in chlamydial EB.
The presence of proteins in metabolically inert but infectious EB may indicate a role for these proteins early in the chlamydial developmental cycle. In order to determine if C. pneumoniae EB contain CdsD, we separated EB and RB by differential centrifugation on a discontinuous gastrografin gradient (6), separated the EB proteins by SDS-PAGE, and carried out Western blotting and ECL detection with hyper-immune rabbit anti-CdsD antiserum. Preimmune rabbit serum did not detect CdsD in EB or His-tagged CdsD in E. coli (Fig. 2A, lanes 2 and 3), but rabbit antiserum raised against the FHA-2 domain of CdsD detected a protein of ∼100 kDa in an EB lysate (Fig. 2B, lane 3), which correlates well with the predicted size of CdsD (93 kDa). Antibody reactivity toward lower molecular mass proteins was seen in both the preimmune and immune sera (Fig. 2A and B), indicating the presence of nonspecific cross-reactive antibodies. In order to remove these antibodies, we affinity purified the rabbit anti-FHA-2 antiserum on CH-Sepharose beads cross-linked with His-FHA-2. The affinity-purified antibody retained the ability to detect CdsD in both chlamydial EB lysate and E. coli lysate containing His-CdsD and exhibited minimal cross-reactivity with lower molecular mass proteins (Fig. 2C). The affinity purified antibody did not cross-react with His-PknD and did not yield a signal in the molecular mass range of CdsD in an E. coli lysate containing His-PknD, demonstrating the specificity of the affinity-purified antiserum for CdsD (Fig. 2C, compare lanes 1 and 2). These results demonstrate that the affinity-purified antibody specifically reacts with CdsD in Chlamydia.
FIG. 2.
Detection of full-length CdsD in C. pneumoniae EB. (A) Preimmune rabbit serum does not detect CdsD in EB by Western blot analysis. (B) Hyper-immune rabbit anti-FHA-2 antiserum detects His-CdsD in E. coli lysate and CdsD in chlamydial EB lysate. (C) Affinity-purified rabbit anti-FHA-2 IgG detects His-CdsD in E. coli and CdsD in chlamydial EB and exhibits minimal cross-reactivity with lower-molecular-weight species. Lane 1, E. coli lysate containing His-PknD (migrates at approximately 108 kDa; location marked with an arrow head); lane 2, E. coli lysate containing His-CdsD; lane 3, chlamydial EB lysate. Asterisks in panels A and B indicate the locations of nonspecific antibody cross-reactivity. Double asterisks in panel C indicate a probable CdsD degradation product. MW, molecular weight (in thousands).
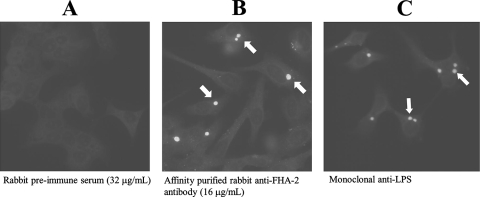
Detection of CdsD in chlamydial RB.
Indirect immunofluorescence of Chlamydia-infected HeLa cells at 48 h postinfection was used to visualize CdsD in RB in infected cells. Affinity-purified rabbit anti-FHA-2 IgG uniformly stained whole chlamydial inclusions 48 h postinfection (Fig. 3B), suggesting the presence of CdsD in chlamydial RB. Rabbit preimmune serum did not react with chlamydial inclusions (Fig. 3A). Monoclonal antibody to chlamydial lipopolysaccharide produced a similar staining pattern as the anti-CdsD antibody (Fig. 3C). These results suggest that CdsD is present in chlamydial RB, which predominate in inclusions 48 h postinfection.
FIG. 3.
Detection of CdsD within C. pneumoniae inclusions at 48 h postinfection using indirect immunofluorescence. (A) Unstained inclusions; rabbit preimmune serum used at 32 μg/ml as the primary antibody. (B) Stained inclusions; affinity-purified rabbit anti-FHA-2 antibody used at 16 μg/ml as the primary antibody. (C) Stained inclusions; fluorescein isothiocyanate-conjugated monoclonal anti-chlamydial lipopolysaccharide antibody was used to directly stain C. pneumoniae inside inclusions. Evans blue was used as a counterstain to visualize HeLa cells, and images were captured at ×400 magnification. Arrows in panels B and C indicate stained chlamydial inclusions.
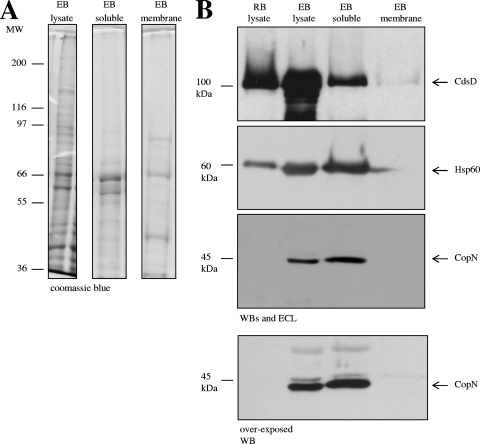
Localization of CdsD with cytoplasmic proteins in Chlamydia.
In order to determine the localization of CdsD in Chlamydia, soluble and integral membrane protein fractions of EB were prepared based on differential solubility in Triton X-114 (4, 16); the protein fractions were then separated by SDS-PAGE and stained with Coomassie blue (Fig. 4A). Western blotting followed by ECL detection of CdsD with anti-CdsD antibody demonstrated that CdsD is predominantly in the soluble fraction of Triton X-114-solubilized EB (Fig. 4B, top panel). A fraction of CdsD molecules was also detected in the integral membrane protein fraction (Fig. 4B, top panel), although this could be due to a minor contamination with soluble proteins. Two other chlamydial proteins, Hsp60 and CopN, not thought to be membrane proteins, were not detected in the integral membrane protein fraction even after extended exposure of the Western blot to film (Fig. 4B, bottom panels). Together the results suggest CdsD is predominantly soluble in EB but may localize to the inner membrane, potentially as a peripheral membrane protein. CdsD is also present in RB (Fig. 4B).
FIG. 4.
CdsD localizes with cytoplasmic EB proteins. (A) Chlamydial EB proteins were separated into cytoplasmic (soluble) and integral membrane (membrane) protein fractions using Triton X-114 and stained with Coomassie blue. MW, molecular weight (in thousands). (B) Fractions shown in panel A were tested for the presence of CdsD (top row) by Western blot analysis using guinea pig anti-CdsD antiserum. CdsD was found in both RB and EB lysates and predominantly localized with cytoplasmic EB proteins, although a small portion was found in the membrane fraction. The Western blot was reprobed for chlamydial Hsp60 (row 2), which was not present in the integral membrane fraction (note: the signal representing Hsp60 in the “EB soluble” lane is seen encroaching into the “EB membrane” lane). An identical Western blot was probed for CopN (a Chlamydia protein predicted to be cytoplasmic) using guinea pig anti-CopN antibody (shown in the row below the Hsp60 Western blot). CopN was detected in EB lysate and in the soluble fraction but was not detected in RB lysate or the EB integral membrane fraction, even after overexposing the membrane to the film (bottom row). The absence of CopN in RB is consistent with secretion of CopN by Chlamydia. The sizes of the molecular mass markers (in thousands) are indicated on the left.
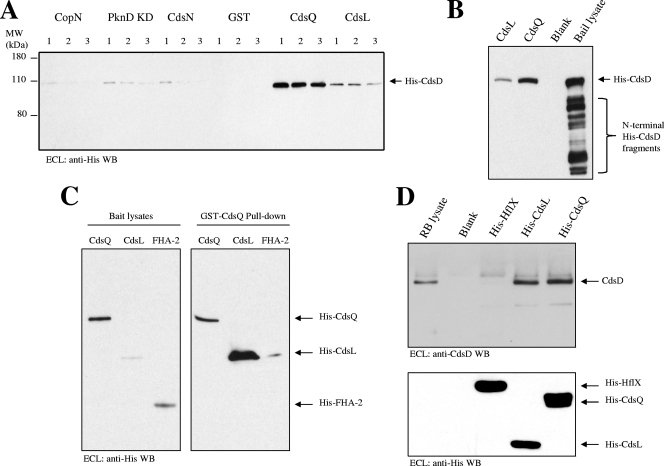
Interaction of CdsD, CdsQ, and CdsL in GST pull-down assays.
GST pull-down assays were used to determine if putative chlamydial T3S proteins interact. When incubated with an E. coli lysate containing His-CdsD as “bait,” GST fusion “prey” proteins CdsQ and CdsL, but not GST alone, bound His-CdsD (Fig. 5A). In order to test the avidity of the interactions, three tubes containing each bait-prey combination were set up and washed with increasing amounts of salt. Each tube was washed with 50 mM Tris buffer containing 0, 200, or 500 mM NaCl (Fig. 5A, lanes 1, 2 and 3, respectively). CopN, CdsN, and the PknD kinase domain exhibited weak and presumably nonspecific interactions with CdsD under low-salt concentrations, and these interactions were readily prevented with quick washes containing 500 mM salt. Both CdsQ and CdsL remained associated with CdsD after washing with 500 mM NaCl. C-terminally truncated His-CdsD present in the E. coli lysate and visible by ECL with anti-His antibody (the His-tag is located on the N terminus) did not bind to CdsQ or CdsL (Fig. 5B), suggesting that the C-terminal region of CdsD is essential for binding to CdsQ and CdsL. Pull-down assays with GST-CdsQ as prey and E. coli lysates containing the bait proteins His-CdsQ, His-CdsL, and His-FHA-2 (Fig. 5C, left panel) revealed CdsQ-CdsQ and CdsQ-CdsL interactions (Fig. 5C, right panel). Interactions between CdsQ and the FHA-2 domain of CdsD were not observed and served as a negative control. These results suggest that the FHA-2 region of CdsD alone does not interact with CdsQ, but that full-length CdsQ, CdsL, and CdsD form stable intermolecular interactions.
FIG. 5.
Interaction between CdsD, CdsQ, and CdsL in GST pull-down and copurification assays. (A) CdsD binds with CdsQ and CdsL but not CopN, PknD KD, CdsN, or GST in a GST pull-down assay. E. coli lysate containing His-CdsD as bait was incubated with 20 μg of GST or 20 μg of the indicated GST chlamydial fusion proteins (CopN, PknD KD, CdsN, CdsQ, or CdsL) on glutathione agarose beads. Beads were collected by centrifugation and washed three times with either 50 mM Tris-HCl, pH 7.45 (lane 1), 50 mM Tris-HCl, pH 7.45, containing 200 mM NaCl (lane 2), or 50 mM Tris-HCl, pH 7.45, containing 500 mM NaCl (lane 3), and proteins were eluted in 2× SDS loading buffer. Binding of CdsD with the prey proteins was revealed by anti-His Western blotting. (B) GST-CdsL and GST-CdsQ interact with full-length His-CdsD but not with C-terminally truncated His-CdsD in a GST pull-down assay. (C) CdsQ-CdsQ and CdsQ-CdsL interactions identified by GST pull-down assay. E. coli lysates containing the bait proteins His-tagged CdsQ, CdsL, and FHA-2 are shown in the left panel, and the results from the pull-down are shown in the right panel. CdsQ did not bind with the FHA-2 domain of CdsD. (D) CdsD from a chlamydial RB lysate copurifies with His-CdsQ and His-CdsL but not with His-HflX on Ni NTA agarose as revealed by Western blotting using anti-CdsD antibody (top). The nitrocellulose membrane was reprobed with anti-His antibody and demonstrates equivalent amounts of His-tagged proteins purified and loaded in each lane (bottom).
Copurification of CdsD with CdsQ and CdsL.
Copurification assays were used to determine if native CdsD binds with His-tagged CdsQ or CdsL. Briefly, E. coli lysates containing equivalent amounts of His-tagged CdsQ, CdsL, or HflX were incubated with a chlamydial RB lysate prepared from RB 42 h postinfection. Ni NTA agarose beads were added overnight, and the beads were collected and washed with binding buffer containing 20 mM imidazole. CdsD copurified with His-CdsQ and His-CdsL but not with His-HflX (a chlamydial GTPase presumably not related to T3S) as detected by Western blot analysis using anti-CdsD antibody (Fig. 5D, top panel). The blot was reprobed with anti-His antibody to visualize the purified recombinant proteins (Fig. 5D, bottom panel). The results corroborate the GST pull-down assays and demonstrate that CdsD interacts with CdsQ and CdsL.
DISCUSSION
In the present study, we investigated the expression and localization of CdsD in C. pneumoniae and identified novel interactions between components of the putative chlamydial T3S apparatus. CdsD was expressed in chlamydial EB and RB as an ∼105-kDa protein, localized with cytoplasmic proteins in EB after Triton X-114 phase partitioning, and was evenly distributed throughout the inclusion body at 48 h postinfection based on a consolidated staining pattern. We demonstrated interactions between CdsQ molecules and CdsQ and CdsL and for the first time identify CdsQ and CdsL as molecular binding partners of CdsD. These results suggest CdsD, CdsQ, and CdsL may interact in Chlamydia to form part of the chlamydial T3S apparatus.
Virulence and flagella-associated T3S systems have a base located in the bacterial cytoplasm that contains over 100 molecules of FliN (26, 47), a YscQ homolog that forms part of a concentric structure known as the C ring (5). Structural analysis of HrcQB-C, the conserved C-terminal region of a YscQ homolog in Pseudomonas syringae, revealed a tetrameric “dimer of dimers” complex and two clusters of conserved residues that may mediate interactions with other proteins (13). Spa33, a Shigella YscQ homolog, interacts with multiple T3S proteins, including structural components of the basal body, MxiN (a YscL homolog), Spa47 (a YscN homolog), Spa32 (a molecular ruler determining the needle length), and several effectors (32). Spa33 has been localized to the C ring of the T3S apparatus using electron microscopy, was shown to be required for injectisome formation and protein secretion, and may act as a recruiting platform or scaffold for the concentration of effector molecules prior to secretion (32). Yeast three-hybrid assays demonstrated that YscQ simultaneously interacts with YscK and YscL, suggesting that separate regions on YscQ may mediate binding to different proteins in Yersinia (23). Similarly, YscL, a negative regulator of the ATPase YscN (3), has been shown to bring together YscN and YscQ (23). Based on the data, it was suggested that interaction between YscQ, YscL, YscN, and YscK may be important in the assembly and/or function of the Y. pestis T3S apparatus (23). We have shown, using a GST pull-down assay, interactions between CdsQ molecules, consistent with the requirement for YscQ dimerization in creating building blocks of the T3S C ring (5, 13). Furthermore, intermolecular associations were identified between CdsQ and CdsL, mirroring the interactions between their counterpart proteins in Yersinia and Shigella and suggesting that these two proteins may be important in chlamydial T3S. Using both pull-down and copurification assays with chlamydial lysates, we identified interactions between CdsQ and CdsD and between CdsL and CdsD, indicating that CdsD interacts with both CdsQ and CdsL and is likely a component of the T3S system. Given that recombinant CdsQ and CdsL were used in the pull-down and copurification assays, the interaction of CdsD with CdsQ and CdsL remains to be demonstrated in Chlamydia. Weaker interactions were observed between CdsD and CopN and the PknD KD and CdsN, but these interactions were eliminated in the presence of 500 mM NaCl, and therefore, further study is required to determine their specificity. Interestingly, CdsQ did not pull down a 150-amino-acid fragment encompassing the FHA-2 domain of CdsD, and both CdsQ and CdsL were unable to pull down C-terminal truncations of CdsD, collectively suggesting that the C terminus of CdsD plays a critical role in mediating stable protein-protein interactions with CdsQ and CdsL. Studies are currently under way to assess these interactions in C. pneumoniae and to determine the domains responsible for mediating interactions between CdsD, CdsQ, and CdsL.
Determining the localization of T3S proteins is important in order to elucidate and understand their roles in secretion. To date, strong evidence for the localization of various YscD homologs has not been forthcoming. EscD, the enteropathogenic E. coli YscD homolog, was shown to interact with EscC in a yeast two-hybrid assay (11); mass spectrometry was used to identify EscC as a major component of the T3S outer membrane ring, and the interaction between EscD and EscC was corroborated using a GST pull-down assay (35). Together with the presence of a putative transmembrane domain in EscD, it was suggested that EscD localizes as an integral membrane protein in order to interact with EscC in the outer membrane. Biochemical evidence supporting membrane localization of YscD comes from a study in Yersinia where it was shown that YscD localizes to the inner membrane in cell fractionation experiments (38). Conversely, CdsD has been detected in the Sarkosyl-insoluble outer-membrane fraction by mass spectrometry (44) and shown to transiently exist in the inclusion membrane by immunofluorescence (20) and to be in the soluble protein fraction of Chlamydia (this report). Tanzer and Hatch recognized that the localization of CdsD to the outer membrane of C. trachomatis is likely an artifact of the Sarkosyl extraction method (44). CdsD was not labeled when intact bacteria were treated with the lipophilic, photoactivable chemical [125I]TID, in contrast with the labeling of canonical outer membrane proteins, such as the major outer membrane protein (44). Additionally, treatment of Chlamydia with trypsin prior to cell fractionation with Sarkosyl did not result in a reduction in the size of the band representing CdsD in the Sarkosyl-insoluble outer-membrane fraction. Together the data indicate CdsD is not surface exposed. Recently a single immunofluorescent image was provided as evidence that CdsD is secreted into the inclusion membrane at 20 h postinfection (20). Closer inspection of the image, however, reveals that one may interpret the image as CdsD-laden chlamydial RB directly associated with the inclusion membrane. The authors also present Western blot data contradicting the secretion of CdsD into the inclusion membrane by showing the accumulation of CdsD in Chlamydia throughout the developmental cycle and report that immunofluorescent images from other time points (0.5, 6, 48, and 72 h postinfection) do not reveal inclusion membrane staining for CdsD but do reveal intraluminal staining (suggesting CdsD is found within the bacteria). In addition, the 84-kDa CdsD protein detected by Western blot analysis brings into question antibody specificity, as Tanzer and Hatch detected CdsD as a 120-kDa protein using mass spectrometry (44) and we detected CdsD migrating slower than a 100-kDa reference protein. Given the discrepancy of the localization of YscD homologs in the literature, we prepared an affinity-purified antibody to the FHA-2 domain of CdsD and detected full-length CdsD in both EB and RB lysates by Western blot analysis. We detected CopN, a putative T3S plug protein, in EB but not in RB lysates, consistent with CopN secretion by Chlamydia early in the replication cycle (15). By analogy, the presence of CdsD in both RB and EB suggests that it is not secreted, consistent with a structural T3S system component. Phase separation of chlamydial EB proteins into cytoplasmic and integral membrane protein fractions based on differential solubility in Triton X-114 revealed that CdsD is predominantly soluble in EB. It could be that chlamydial CdsD is a peripheral membrane protein with a high hydrophilic character. Cytoplasmic localization of CdsD would be consistent with the interactions of CdsD with the putative chlamydial C-ring protein CdsQ and with the tethering protein CdsL, both presumably cytoplasmic proteins.
A consistent and essential role for YscD homologs in T3S and pathogenicity has been well documented. E. coli ΔescD mutants were unable to produce the T3S apparatus (35), Yersina yscD was shown to be necessary for the secretion of the effector Yop proteins (30, 38), and Pseudomonas aeruginosa ΔpscD mutants were attenuated in a caterpillar model of infection (31). CdsD, however, has an additional 400 amino acids relative to YscD homologs and contains two FHA domains that may mediate binding to phosphorylated proteins, suggesting CdsD may have an additional role in T3S. The presence of FHA domains on CdsD has led to the recent suggestion that it may bind phosphorylated chaperone-substrate complexes of the T3S system (36). A report on Pseudomonas aeruginosa, however, identified a T6S apparatus protein with an FHA domain that was phosphorylated by the protein kinase PpkA, triggering protein secretion; it was proposed that after kinase autophosphorylation, the FHA domain of this scaffolding protein is recruited to and binds the kinase, resulting in FHA domain phosphorylation and the induction of protein secretion (33). We have recently shown that the membrane-localized C. pneumoniae protein kinase PknD autophosphorylates and phosphorylates CdsD on both its FHA-1 and FHA-2 domains (24). In addition to potentially recognizing phosphorylated chaperone-substrate complexes of the T3S system as proposed previously (36), the FHA domains of CdsD may mediate transient interactions with PknD as an important step in chlamydial T3S. It is tempting to speculate that the phosphorylation of CdsD FHA domains by PknD may be an activating or triggering event in chlamydial T3S.
Acknowledgments
We thank Ken Fields for providing the phase separation protocol and for helpful technical advice and discussions. We thank Brian Coombes for editing the manuscript. We wish to acknowledge Rami El-Sebai for his contribution in constructing pENT-CdsQ and pENT-CdsL and the members of the Mahony lab for helpful discussions on this project.
D.L.J. and C.B.S. are supported by a grant to J.B.M. from the Canadian Institutes of Health Research. This work was funded in part by a grant from the Canadian Institutes of Health Research.
Footnotes
Published ahead of print on 15 February 2008.
REFERENCES
- 1.Belland, R. J., D. E. Nelson, D. Virok, D. D. Crane, D. Hogan, D. Sturdevant, W. L. Beatty, and H. D. Caldwell. 2003. Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc. Natl. Acad. Sci. USA 10015971-15976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belland, R. J., G. Zhong, D. D. Crane, D. Hogan, D. Sturdevant, J. Sharma, W. L. Beatty, and H. D. Caldwell. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. USA 1008478-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaylock, B., K. E. Riordan, D. M. Missiakas, and O. Schneewind. 2006. Characterization of the Yersinia enterocolitica type III secretion ATPase YscN and its regulator, YscL. J. Bacteriol. 1883525-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 2561604-1607. [PubMed] [Google Scholar]
- 5.Brown, P. N., M. A. Mathews, L. A. Joss, C. P. Hill, and D. F. Blair. 2005. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J. Bacteriol. 1872890-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldwell, H. D., J. Kromhout, and J. Schachter. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 311161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carabeo, R. A., S. S. Grieshaber, A. Hasenkrug, C. Dooley, and T. Hackstadt. 2004. Requirement for the Rac GTPase in Chlamydia trachomatis invasion of non-phagocytic cells. Traffic 5418-425. [DOI] [PubMed] [Google Scholar]
- 8.Clifton, D. R., K. A. Fields, S. S. Grieshaber, C. A. Dooley, E. R. Fischer, D. J. Mead, R. A. Carabeo, and T. Hackstadt. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. USA 10110166-10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coombes, B. K., and J. B. Mahony. 2002. Identification of MEK- and phosphoinositide 3-kinase-dependent signalling as essential events during Chlamydia pneumoniae invasion of HEp2 cells. Cell. Microbiol. 4447-460. [DOI] [PubMed] [Google Scholar]
- 10.Cornelis, G. R. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4811-825. [DOI] [PubMed] [Google Scholar]
- 11.Creasey, E. A., R. M. Delahay, S. J. Daniell, and G. Frankel. 2003. Yeast two-hybrid system survey of interactions between LEE-encoded proteins of enteropathogenic Escherichia coli. Microbiology 1492093-2106. [DOI] [PubMed] [Google Scholar]
- 12.Dong, F., J. Sharma, Y. Xiao, Y. Zhong, and G. Zhong. 2004. Intramolecular dimerization is required for the chlamydia-secreted protease CPAF to degrade host transcriptional factors. Infect. Immun. 723869-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fadouloglou, V. E., A. P. Tampakaki, N. M. Glykos, M. N. Bastaki, J. M. Hadden, S. E. Phillips, N. J. Panopoulos, and M. Kokkinidis. 2004. Structure of HrcQB-C, a conserved component of the bacterial type III secretion systems. Proc. Natl. Acad. Sci. USA 10170-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fields, K. A., E. R. Fischer, D. J. Mead, and T. Hackstadt. 2005. Analysis of putative Chlamydia trachomatis chaperones Scc2 and Scc3 and their use in the identification of type III secretion substrates. J. Bacteriol. 1876466-6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fields, K. A., and T. Hackstadt. 2000. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol. Microbiol. 381048-1060. [DOI] [PubMed] [Google Scholar]
- 16.Fields, K. A., D. J. Mead, C. A. Dooley, and T. Hackstadt. 2003. Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol. Microbiol. 48671-683. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh, P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68771-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gophna, U., E. Z. Ron, and D. Graur. 2003. Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events. Gene 312151-163. [DOI] [PubMed] [Google Scholar]
- 19.Hefty, P. S., and R. S. Stephens. 2007. Chlamydial type III secretion system is encoded on ten operons preceded by sigma 70-like promoter elements. J. Bacteriol. 189198-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrmann, M., A. Schuhmacher, I. Muhldorfer, K. Melchers, C. Prothmann, and S. Dammeier. 2006. Identification and characterization of secreted effector proteins of Chlamydophila pneumoniae TW183. Res. Microbiol. 157513-524. [DOI] [PubMed] [Google Scholar]
- 21.Ho, T. D., and M. N. Starnbach. 2005. The Salmonella enterica serovar Typhimurium-encoded type III secretion systems can translocate Chlamydia trachomatis proteins into the cytosol of host cells. Infect. Immun. 73905-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hybiske, K., and R. S. Stephens. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. USA 10411430-11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson, M. W., and G. V. Plano. 2000. Interactions between type III secretion apparatus components from Yersinia pestis detected using the yeast two-hybrid system. FEMS Microbiol. Lett. 18685-90. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, D. L., and J. B. Mahony. 2007. Chlamydophila pneumoniae PknD exhibits dual amino acid specificity and phosphorylates Cpn0712, a putative type III secretion YscD homolog. J. Bacteriol. 1897549-7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawana, K., A. J. Quayle, M. Ficarra, J. A. Ibana, L. Shen, Y. Kawana, H. Yang, L. Marrero, S. Yavagal, S. J. Greene, Y. X. Zhang, R. B. Pyles, R. S. Blumberg, and D. J. Schust. 2007. CD1d degradation in Chlamydia trachomatis-infected epithelial cells is the result of both cellular and chlamydial proteasomal activity. J. Biol. Chem. 2827368-7375. [DOI] [PubMed] [Google Scholar]
- 26.Khan, I. H., T. S. Reese, and S. Khan. 1992. The cytoplasmic component of the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 895956-5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kresse, A. U., K. Schulze, C. Deibel, F. Ebel, M. Rohde, T. Chakraborty, and C. A. Guzman. 1998. Pas, a novel protein required for protein secretion and attaching and effacing activities of enterohemorrhagic Escherichia coli. J. Bacteriol. 1804370-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lugert, R., M. Kuhns, T. Polch, and U. Gross. 2004. Expression and localization of type III secretion-related proteins of Chlamydia pneumoniae. Med. Microbiol. Immunol. 193163-171. [DOI] [PubMed] [Google Scholar]
- 29.Maurer, A. P., A. Mehlitz, H. J. Mollenkopf, and T. F. Meyer. 2007. Gene expression profiles of Chlamydophila pneumoniae during the developmental cycle and iron depletion-mediated persistence. PLoS Pathog. 3e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michiels, T., J. C. Vanooteghem, C. Lambert de Rouvroit, B. China, A. Gustin, P. Boudry, and G. R. Cornelis. 1991. Analysis of virC, an operon involved in the secretion of Yop proteins by Yersinia enterocolitica. J. Bacteriol. 1734994-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyata, S., M. Casey, D. W. Frank, F. M. Ausubel, and E. Drenkard. 2003. Use of the Galleria mellonella caterpillar as a model host to study the role of the type III secretion system in Pseudomonas aeruginosa pathogenesis. Infect. Immun. 712404-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morita-Ishihara, T., M. Ogawa, H. Sagara, M. Yoshida, E. Katayama, and C. Sasakawa. 2006. Shigella Spa33 is an essential C-ring component of type III secretion machinery. J. Biol. Chem. 281599-607. [DOI] [PubMed] [Google Scholar]
- 33.Mougous, J. D., C. A. Gifford, T. L. Ramsdell, and J. J. Mekalanos. 2007. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat. Cell Biol. 9797-803. [DOI] [PubMed] [Google Scholar]
- 34.Nicholson, T. L., L. Olinger, K. Chong, G. Schoolnik, and R. S. Stephens. 2003. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J. Bacteriol. 1853179-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogino, T., R. Ohno, K. Sekiya, A. Kuwae, T. Matsuzawa, T. Nonaka, H. Fukuda, S. Imajoh-Ohmi, and A. Abe. 2006. Assembly of the type III secretion apparatus of enteropathogenic Escherichia coli. J. Bacteriol. 1882801-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peters, J., D. P. Wilson, G. Myers, P. Timms, and P. M. Bavoil. 2007. Type III secretion a la Chlamydia. Trends Microbiol. 15241-251. [DOI] [PubMed] [Google Scholar]
- 37.Pirbhai, M., F. Dong, Y. Zhong, K. Z. Pan, and G. Zhong. 2006. The secreted protease factor CPAF is responsible for degrading pro-apoptotic BH3-only proteins in Chlamydia trachomatis-infected cells. J. Biol. Chem. 28131495-31501. [DOI] [PubMed] [Google Scholar]
- 38.Plano, G. V., and S. C. Straley. 1995. Mutations in yscC, yscD, and yscG prevent high-level expression and secretion of V antigen and Yops in Yersinia pestis. J. Bacteriol. 1773843-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slepenkin, A., L. M. de la Maza, and E. M. Peterson. 2005. Interaction between components of the type III secretion system of Chlamydiaceae. J. Bacteriol. 187473-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slepenkin, A., V. Motin, L. M. de la Maza, and E. M. Peterson. 2003. Temporal expression of type III secretion genes of Chlamydia pneumoniae. Infect. Immun. 712555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subtil, A., C. Delevoye, M. E. Balana, L. Tastevin, S. Perrinet, and A. Dautry-Varsat. 2005. A directed screen for chlamydial proteins secreted by a type III mechanism identifies a translocated protein and numerous other new candidates. Mol. Microbiol. 561636-1647. [DOI] [PubMed] [Google Scholar]
- 42.Subtil, A., C. Parsot, and A. Dautry-Varsat. 2001. Secretion of predicted Inc proteins of Chlamydia pneumoniae by a heterologous type III machinery. Mol. Microbiol. 39792-800. [DOI] [PubMed] [Google Scholar]
- 43.Subtil, A., B. Wyplosz, M. E. Balana, and A. Dautry-Varsat. 2004. Analysis of Chlamydia caviae entry sites and involvement of Cdc42 and Rac activity. J. Cell Sci. 1173923-3933. [DOI] [PubMed] [Google Scholar]
- 44.Tanzer, R. J., and T. P. Hatch. 2001. Characterization of outer membrane proteins in Chlamydia trachomatis LGV serovar L2. J. Bacteriol. 1832686-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson, D. P., P. Timms, D. L. McElwain, and P. M. Bavoil. 2006. Type III secretion, contact-dependent model for the intracellular development of chlamydia. Bull. Math. Biol. 68161-178. [DOI] [PubMed] [Google Scholar]
- 46.Yip, C. K., and N. C. Strynadka. 2006. New structural insights into the bacterial type III secretion system. Trends Biochem. Sci. 31223-230. [DOI] [PubMed] [Google Scholar]
- 47.Zhao, R., N. Pathak, H. Jaffe, T. S. Reese, and S. Khan. 1996. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J. Mol. Biol. 261195-208. [DOI] [PubMed] [Google Scholar]
- 48.Zhong, G., P. Fan, H. Ji, F. Dong, and Y. Huang. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 193935-942. [DOI] [PMC free article] [PubMed] [Google Scholar]