Abstract
Hjm and Hel308a are novel, RecQ-like DNA helicases recently identified in the euryarchaeotes Pyrococcus furiosus and Methanothermobacter thermautotrophicus, respectively. In this study, an Hjm/Hel308 homologue (designated StoHjm) from Sulfolobus tokodaii, a hyperthermophilic archaeon belonging to the Crenarchaeota subdomain of archaea, was cloned, purified, and characterized. Unlike Hjm and Hel308a, which unwind DNA in a 3′-to-5′ direction, StoHjm unwound DNA in both 3′-to-5′ and 5′-to-3′ directions. Remarkably, StoHjm exhibited structure-specific single-stranded-DNA-annealing and fork regression activities in vitro. In addition, gel filtration, affinity pulldown, and yeast two-hybrid analyses revealed that StoHjm physically interacted with StoHjc, the Holliday junction-specific endonuclease from S. tokodaii. This interaction may have functional significance, because the unwinding activity of StoHjm was inhibited by StoHjc in vitro. These results may suggest that the Hjm/Hel308 family helicases, in association with Hjc endonucleases, are involved in processing of stalled replication forks.
DNA is susceptible to numerous damaging agents of both endogenous and environmental origins (18). As a result, the replication progress is unavoidably disrupted and stopped by DNA lesions (27). To ensure accurate replication of the genetic materials and to maintain genome stability, the damaged DNA and stalled replication forks must be repaired and replication must be restarted (21, 23). In living cells, the nucleotide excision repair systems allow for removal of many types of DNA damage, reducing the chances of replication fork stalling (7). Stalled replication induced by lesions may be reinitiated through one of three proposed pathways: the noncleavage (template switching) pathway, the cleavage pathway, and the translesion synthesis pathway (23-25). In the noncleavage pathway, repair and restart of stalled replication may be error free. The cleavage (classical recombination repair) pathway may result in too much recombination, undermining genome stability. The translesion synthesis pathway is error prone due to the participation of low-fidelity DNA polymerases (21). The cleavage pathway is thought to be the dominant mode in bacteria, while replication repair (through template switching and translesion synthesis) is the dominant mode in T4 and eukaryotes (14).
In the cleavage and noncleavage pathways, two processes are considered to be critical, the formation of the Holliday junction, or the “chicken foot” structure, and the migration of the Holliday junction. During Holliday junction formation, the fork is regressed through annealing of nascent strands catalyzed by helicases. Holliday junctions have been observed by electron microscopy in the cells treated with DNA-damaging agents (33, 34), although it is unclear whether the fork regression process occurs in normal cells.
In Escherichia coli, the regression process is assumed to be initiated by the RecG helicase, which possesses both 3′-to-5′ and 5′-to-3′ helicase activities (22, 24, 25, 32). The homotetrameric RuvA and homohexameric RuvB proteins are believed to be involved in the migration of Holliday junctions (28). RuvC, the structure-specific endonuclease, interacts with the RuvAB complex and resolves Holliday junctions (6, 36). The 3′-to-5′ helicase RecQ is thought to act in conjunction with RecJ to target and process gapped and UV light-damaged DNA structures and to initiate the RecFOR recombination pathway (20). In eukaryotes, the enzymes involved in Holliday junction formation and migration are largely unknown. Several RecQ family DNA helicases have been identified. Some of these helicases have been linked to human diseases (4, 35), resulting in considerable interest in the RecQ family helicases in recent years. It has also been reported that, of the RecQ family helicases, only BLM, WRN, and RecQβ have fork regression activities toward model fork structures (15, 19, 26).
It is well established that archaea possess a simplified version of the eukaryotic protein apparatus for genetic information processing pathways (13). These include eukarya-like DNA recombination proteins, such as Mre11, Rad50, RadA, RPA, and Hef; however, many proteins in the recombination repair pathway have not yet been identified and characterized (13). There have been a few reports about DNA helicases that are involved in recombination repair in archaea (8, 9, 11, 13) but none regarding helicases that promote fork regression. Two helicases, Holliday junction migration DNA helicase (Hjm) from Pyrococcus furiosus and Hel308a from Methanothermobacter thermautotrophicus, have been identified recently (8, 9, 11). However, the real functions of Hjm/Hel308a helicases are unknown. In addition, an Hjm/Hel308a homologue from the Crenarchaeota subdomain of archaea has yet to be characterized.
Here, we cloned, purified, and characterized a homologue (designated StoHjm) of Hjm/Hel308a from Sulfolobus tokodaii, an archaeon belonging to Crenarchaeota. Unlike Hjm and Hel308a, which unwind DNA only in the 3′-to-5′ direction, StoHjm was able to unwind both 3′ overhang and 5′ overhang structures efficiently. StoHjm promoted structure-specific annealing of single-stranded DNA (ssDNA) and fork regression. Moreover, StoHrm was found to interact with StoHjc (the Holliday junction cleavage endonuclease from S. tokodaii). These findings suggest that StoHjm may have functional similarity to bacterial RecG in targeting replication forks and forming Holliday junctions.
MATERIALS AND METHODS
Chemicals, enzymes, bacterial strains, and vectors.
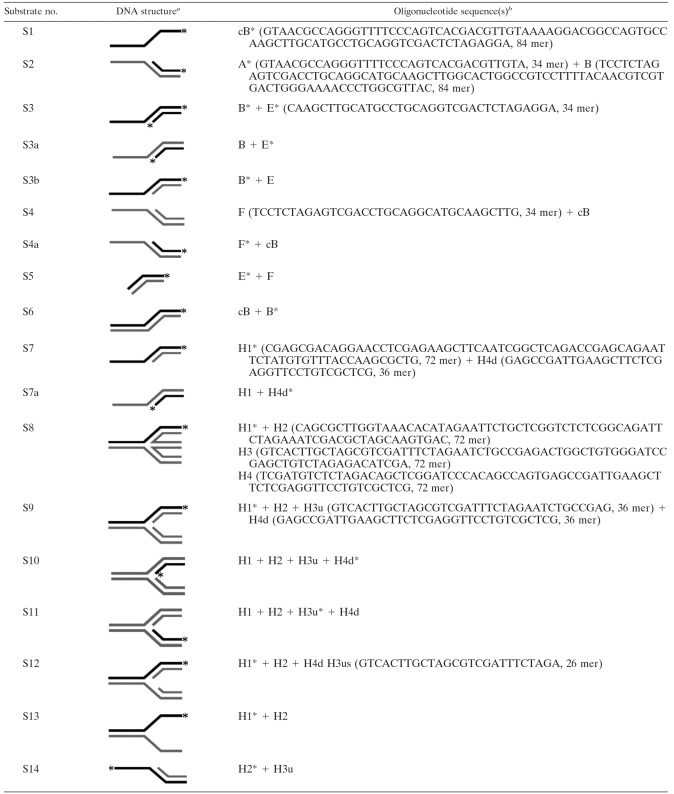
The oligonucleotides (Table 1) were synthesized by Invitrogen (Shanghai, China). Nickel nitrilotriacetic acid-agarose and [γ-32P]ATP were purchased from GE (Buckingham, United Kingdom). The pET15b vector was obtained from Merck (Shanghai, China). S. tokodaii strain 7 was purchased from Japan Collections of Microbes (catalog no. 10545). Restriction enzymes, the DNA ligase kit (version 2), T4 DNA kinase, and DNase I were purchased from Takara (Dalian, China).
TABLE 1.
Substrates used in this study
Substrates labeled at their 5′ ends are denoted with an asterisk. Bold lines indicate labeled strands, and shaded lines indicate unlabeled strands.
Oligonucleotide sequences (A, B, cB [strand complementary to B], E, F, H1, H2, H3, H3u, H3us, H4, and H4d) are shown in a 5′-to-3′ direction. Various combinations of oligonucleotides were used to generate substrates, as indicated.
Gene cloning and plasmid construction.
The gene encoding StoHjm (ST0590, NP376477) was amplified by PCR using S. tokodaii strain 7 genomic DNA as a template, the upstream primer 5′-TCCAGTTTCCATATGGAGACCATTTCTATTGACGATTTGCCG-3′, and the downstream primer 5′-GGATGATGTCGACTCAAGCAATAGTTCTTGCAGCTTCTCTG-3′ (underlined sequences indicate the NdeI and SalI sites in the upstream and downstream primers, respectively). Amplified fragments of the StoHjm gene were digested with NdeI and SalI and inserted into a modified pET15b vector (31) to create pET15b/His-StoHjm. The gene was also cloned into a modified pET15b vector (lacking the His tag) to express native StoHjm. The gene encoding StoHjc (ST1444, NP377404) was amplified using the upstream primer 5′-GCCGCGCATATGTATATTGTGAATTCCA-3′ and the downstream primer 5′-GCGGCGGTCGACTCATAAGAAAGAATCTAAG-3′. The amplified fragments were digested and inserted into the modified pET15b vector to create pET15b/His-StoHjc. The nucleotide sequences of the inserted StoHjm and StoHjc genes were confirmed by sequencing (Invitrogen, Shanghai, China).
Expression and purification of recombinant proteins.
Recombinant His-tagged StoHjm proteins were produced in E. coli strain BL21 (DE3)-CodonPlus-RIL grown in 1,000 ml Luria-Bertani (LB) medium containing ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml). The cells were grown until they reached an optical density at 600 nm of 0.4 at 37°C, and then expression was induced with IPTG (isopropyl-β-d-thiogalactopyranoside) (1 mM) for 16 h at 16°C. The cells were harvested and disrupted by sonication in buffer A (50 mM Tris-HCl, pH 8.0, and 200 mM NaCl). The sample was incubated at 80°C for 30 min and centrifuged at 10,000 × g for 10 min. The soluble heat-resistant fraction was precipitated with 80% saturated ammonium sulfate. The precipitated protein was resuspended in buffer A and dialyzed against buffer B (50 mM Tris-HCl, pH 8.0, and 100 mM NaCl) to remove the ammonium sulfate. After dialysis, the sample was loaded onto a 5-ml HiTrap Q column, which was preequilibrated with buffer B. The fractions eluted at 350 to 450 mM NaCl were pooled and loaded onto a 1-ml nickel nitrilotriacetic acid-agarose column. The column was washed with 10 column volumes of buffer A containing 40 mM imidazole and eluted with 3 column volumes of elution buffer containing 500 mM imidazole. The eluted fractions were pooled, concentrated, and separated on a gel filtration column (Sephacryl S-200). The expression and purification of native StoHjm were the same as for His-tagged StoHjm, except that the nickel affinity chromatography step was omitted.
Recombinant His-tagged StoHjc protein was expressed and purified under the same conditions as for His-tagged StoHjm, except that StoHjc was eluted with 380 to 470 mM NaCl during anion exchange chromatography, and these fractions were pooled for subsequent purification. Protein concentrations were measured by the Bradford method.
Preparation of DNA substrates.
Oligonucleotides (Table 1) were labeled at the 5′ ends by using T4 polynucleotide kinase according to the manufacturer's instructions. The final concentration of the labeled oligonucleotide in the labeling mixture was 1,000 nM. The annealing experiment was conducted with a 50-μl mixture containing 40 mM Tris-acetate (pH 7.8), 0.5 mM magnesium acetate, 200 nM unlabeled oligonucleotides, and 100 nM labeled oligonucleotides. The mixtures were heated at 95°C for 5 min, gradually cooled to room temperature, and stored at 4°C for further use. Various DNA substrates were constructed using various combinations of oligonucleotides (Table 1).
ATPase assay.
For the ATPase assay, purified StoHjm was treated with DNase I to remove a hint amount of DNA in the enzyme sample. ATPase activity was assayed at 50°C for 30 min with a 20-μl mixture containing 20 mM Tris-HCl (pH 8.0), 5 mM magnesium chloride, 1 mM dithiothreitol (DTT), 1 nM ssDNA (34 mer), 500 μM ATP, 0.2 μCi labeled [γ-32P]ATP, and the indicated concentration of StoHjm. The reactions were terminated by the addition of EDTA to give a final concentration of 50 mM. Two-microliter aliquots were spotted onto polyethylenimine-cellulose plates (Merck, Germany). ATP and released Pi were then separated chromatographically in 1 M formic acid and 0.5 M LiCl.
DNA binding assay.
Standard DNA binding assays were performed with a 20-μl mixture containing 20 mM Tris-HCl (pH 8.0), 5 mM magnesium chloride, 1 mM DTT, 5 mM ATP, 1 nM DNA substrate, 12% glycerol, and the indicated enzyme. The mixtures were incubated at 25°C for 30 min and loaded on a 6% polyacrylamide gel in TBE buffer (45 mM Tris-borate, 1 mM EDTA). Samples were electrophoresed at 120 V for 60 min.
Helicase assay.
Standard helicase activity assays were carried out with a 20-μl mixture containing 20 mM Tris-HCl (pH 8.0), 5 mM magnesium chloride, 1 mM DTT, 5 mM ATP, 1 nM DNA substrate, and the indicated concentration of StoHjm. The mixtures were incubated at 50°C for 30 min, and the reactions were stopped by the addition of 10 μl of stop buffer containing 20 mM protease K, 50 mM EDTA, 0.5% sodium dodecyl sulfate (SDS), 25% glycerol, and 0.025% bromophenol blue. The mixtures were then incubated at room temperature for 20 min, loaded onto a 10% polyacrylamide gel in TBE buffer, and electrophoresed at 150 V for 90 min.
Standard annealing activity assay.
Standard strand-annealing activity was assayed with a mixture (20 μl) containing 20 mM Tris-HCl (pH 8.0), 5 mM magnesium chloride, 1 mM DTT, 1 nM of each substrate, and the indicated amount of StoHjm. The mixtures were incubated at 55°C, 40°C, or 25°C for 30 min, and the reactions were terminated by the addition of 10 μl of stop buffer containing 20 mM protease K, 50 mM EDTA, 0.5% SDS, 25% glycerol, and 0.025% bromophenol blue. The mixtures were then incubated at room temperature for 30 min, loaded on a 10% polyacrylamide gel in TBE buffer, and electrophoresed at 150 V for 90 min. Since spontaneous annealing was observed at 55°C and the activity of StoHjm was undetectable at 25°C (data not shown), 40°C was chosen as the standard reaction temperature.
Endonuclease assay.
Several DNA structures (S7a, S8, S9, and S13) (Table 1) were used to test structure-specific cleavage of StoHjc. The reaction was carried out with a mixture (20 μl) containing 20 mM Tris-HCl (pH 8.0), 1 mM DTT, 5 mM magnesium chloride, 1 nM labeled DNA substrate, and 50 nM StoHjc. The mixture was incubated at 50°C for 30 min. The reactions were stopped by the addition of an equal amount of a stop buffer (95% formamide, 10 mM EDTA, and 0.1 mg/ml bromophenol blue). The samples were boiled for 5 min, immediately placed on ice, and then electrophoresed on a 15% polyacrylamide gel containing urea.
Fork reversal activity assay in the absence of StoHjc.
A 3′ overhang (S3) (Table 1) that was labeled on both strands and a 5′ overhang (S4, unlabeled) were used to test the fork reversal activity of StoHjm. The reaction mixture (20 μl) contained 20 mM Tris-HCl (pH 8.0), 1 mM DTT, 5 mM magnesium chloride, and 1 nM of each DNA substrate. The mixtures were incubated at 40°C for 30 min. The reactions were stopped by the addition of 10 μl of stop buffer containing 20 mM protease K, 50 mM EDTA, 0.5% SDS, 25% glycerol, and 0.025% bromophenol blue. The samples were separated by polyacrylamide gel electrophoresis (PAGE).
Fork reversal activity assay in the presence of StoHjc.
A 3′ overhang and a 5′ overhang that were labeled on the recessed strands and had complementary single-strand regions (S3a and S4a) (Table 1) were first used as the substrates. The 3′ and 5′ overhangs with each of the four strands labeled were also used for analysis. The reaction mixture (20 μl) contained 20 mM Tris-HCl (pH 8.0), 1 mM DTT, 5 mM magnesium chloride, 1 nM of each DNA substrate, 64 nM StoHjm, and 50 nM StoHjc. The reaction was stopped by the addition of an equal volume of a stop buffer containing 95% formamide, 10 mM EDTA, and 0.025% bromophenol blue. The samples were boiled for 5 min, immediately placed on ice, and separated on a 15% polyacrylamide gel containing urea.
Gel filtration.
To analyze complex formation, 1 mg StoHjm and 1 mg StoHjc were combined to give a final volume of 2 ml. The tube was then incubated at 0°C for 30 min. The mixture (2 ml) was loaded onto a Sephacryl S-200 HR column (GE) preequilibrated with a buffer made up of 50 mM Tris-HCl (pH 8.0) and 100 mM NaCl. The column was eluted at a constant flow rate of 1 ml/min. StoHjm (1 mg in 2 ml) and StoHjc (1 mg in 2 ml) were also loaded onto the gel separately and used as controls.
Pulldown assay.
His-tagged StoHjc (50 μg) and an equal amount of native StoHjm were mixed in a buffer containing 50 mM Tris-HCl (pH 8.0) and 500 mM NaCl on ice for 30 min. The mixture was then combined with 20 μl of nickel nitrilotriacetic acid-agarose slurry preequilibrated in 50 mM Tris-HCl (pH 8.0) and 500 mM NaCl. The agarose beads were collected by centrifugation at 500 × g for 1 min and washed five times at room temperature in 500 μl wash buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 40 mM imidazole). Next, 50 μl of SDS-PAGE loading buffer (250 mM Tris-HCl, 0.5% SDS, 50% glycerol, 0.5% bromophenol blue, and 500 mM DTT) was added to the samples, which were then boiled at 100°C for 15 min and separated on a 15% SDS polyacrylamide gel. The gel was stained with Coomassie bright blue. His-tagged StoHjc and StoHjm were also loaded separately as the controls.
Yeast two-hybrid analysis.
Matchmaker GAL4 Two-Hybrid System 3 (Clontech) was used for yeast two-hybrid analysis according to the manufacturer's instructions. Saccharomyces cerevisiae AH109 was used as the reporter strain. Genes encoding StoHjm and StoHjc were fused to the GAL4 activation domain (AD) in pGADT7 and the GAL4 DNA binding domain (BD) in pGBKT7. The resultant plasmids were named AD-StoHjm, AD-StoHjc, BD-StoHjm, and BD-StoHjc. The plasmids were cotransformed into yeast AH109 by the polyethylene glycol/lithium acetate method. Interactions were assessed on two types of media. Synthetic dropout (SD) medium lacking Leu and Trp (SD/−Leu/−Trp) was used as a control to select transformants, and SD/−His/−Leu/−Trp (Clontech), along with 10 mM 3-amino-1,2,4-triazole (3-AT), was used for screening.
Data collection and quantification.
Gels and polyethylenimine-cellulose plates were exposed to a phosphor screen (GE) for 4 h. Phosphor screens were then imaged using a Typhoon 9410 scanner (GE). The results were analyzed using Image Quant 5.0 software (GE). All the results were based on at least three independent tests.
RESULTS
Cloning, expression, and purification of StoHjm and StoHjc.
Hjm/Hel308a family DNA helicases have been characterized only in the Euryarchaeota subdomain of archaea, and there is accumulating evidence that the protein machineries for DNA replication and repair differ greatly in the Euryarchaeota and Crenarchaeota subdomains. To understand the properties of the Hjm/Hel308a family DNA helicases in Crenarchaeota, we searched for and identified an Hjm/Hel308a homologue in the genome of Sulfolobus tokodaii, a hyperthermophilic archaeon belonging to Crenarchaeota. A comparison of homologues from several archaeal species is shown in Fig. S1A in the supplemental material. The S. tokodaii homologue, designated StoHjm (ST0590, 81 kDa), had amino acid sequence identities of 35% with Hjm from Pyrococcus furiosus, 29% with Hel308a from M. thermautotrophicus, 38% with the Archaeoglobus fulgidus homologue, 65% with the Sulfolobus solfataricus homologue, and 31% with the Aeropyrum pernix homologue. For further functional analysis of StoHjm (see below), a homologue of the Holliday junction endonuclease from S. tokodaii (StoHjc, ST1444, 14 kDa) was also identified. Amino acid sequence alignment of Hjc homologues from several archaea is shown in Fig. S1B in the supplemental material. StoHjc had sequence identities of 21% with the P. furiosus homologue, 31% with the M. thermautotrophicus homologue, 27% with the A. fulgidus homologue, 70% with the S. solfataricus homologue, and 31% with the A. pernix homologue.
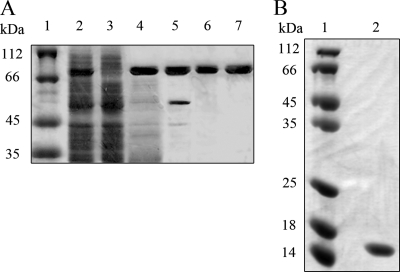
The gene encoding StoHjm was cloned, and two resultant plasmids, named pET15b/His-StoHjm and pET15b/StoHjm, were transformed into E. coli to express N-terminal His-tagged and native proteins, respectively. The proteins were strongly expressed and highly thermostable (Fig. 1A, lanes 1 to 4). Highly purified protein was obtained by anion exchange, nickel affinity, and gel filtration chromatography (Fig. 1A, lanes 5 to 7). The gene encoding StoHjc was also cloned, and the resultant plasmid, pET15b/His-StoHjc, was transformed into E. coli for expression. N-terminal His-tagged StoHjc was obtained by heat treatment, anion exchange, nickel affinity, and gel filtration chromatography (Fig. 1B). Bands with sizes expected for both StoHjm and StoHjc were present on the SDS-PAGE gels.
FIG. 1.
SDS-PAGE of StoHjm and StoHjc during purification. (A) Purification of StoHjm (81 kDa). Lanes: 1, molecular size markers; 2, total cell protein after sonication; 3, total cell protein of untransformed E. coli (as a control); 4, heat-treated supernatant (80°C, 30 min); 5, protein eluted from ion-exchange column; 6, protein after purification by nickel affinity chromatography; 7, protein purified by gel filtration. (B) Purification of StoHjc (14 kDa). StoHjc was purified by heat treatment, ammonium sulfate precipitation, anion exchange, Ni2+-nitrilotriacetic acid affinity chromatography, and gel filtration. Lanes: 1, molecular size markers; 2, purified StoHjc. All samples were separated on 15% denatured polyacrylamide gels. The gels were stained with Coomassie bright blue.
StoHjm is an ATP- and magnesium-dependent DNA helicase.
Next, we investigated the unwinding activity of StoHjm by using a double-stranded DNA (dsDNA) with a 3′ overhang as a substrate (S3a, 34/84-mer) (Table 1). The effects of increasing concentrations of magnesium and ATP on unwinding activity are shown in Fig. S2A and S2B in the supplemental material, and the effect of increasing amounts of StoHjm on ATPase activity are shown in Fig. S2C in the supplemental material. The results revealed that StoHjm is a magnesium-dependent ATPase and DNA helicase, with optimal magnesium and ATP concentrations for helicase activity being around 5 mM (see Fig. S2 in the supplemental material). Therefore, these concentrations were used in subsequent helicase assays. We found that the ATPase activity of StoHjm was stimulated by ssDNA (34-mer, as low as 1 nM) and dsDNA (34 bp, as low as 1 nM) (data not shown).
StoHjm DNA binding preferences.
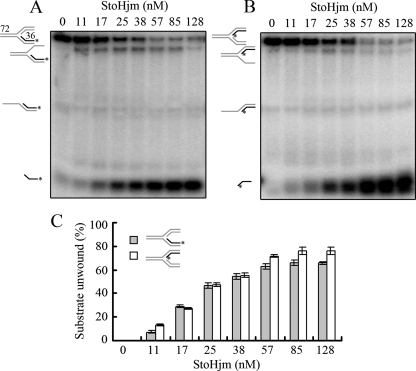
To investigate the DNA binding preferences of StoHjm, we conducted gel mobility shift assays using several DNA substrates (Table 1). Comparison of the affinity of StoHjm for each substrate is shown in Fig. 2. The affinity of StoHjm for ssDNA (S1, 84-mer) (Table 1 and Fig. 2A) was stronger than that for blunt-ended dsDNA (S6, 84 bp) (Fig. 2B). StoHjm binding to 3′ or 5′ overhangs (S3b and S4a, 34-mer/84-mer) was stronger than binding to ssDNA and blunt-ended dsDNA (Fig. 2C, D, and H), indicating that ssDNA/dsDNA junctions were preferentially bound by StoHjm. The affinity of StoHjm to replication forks with a single-stranded region (S12, 10 mer) (Table 1) on the leading parent strand was stronger than its affinity to forked DNA lacking this gap (S9) (Fig. 2E, F, and H). The binding of StoHjm to the Holliday junction was also strong (S8) (Table 1 and Fig. 2G). These results indicate that StoHjm preferentially binds to ssDNA, overhang structures, and branched DNA but not blunt-ended DNA. The DNA binding properties of StoHjm are similar to those of Hjm from P. furiosus and Hel308a from M. thermautotrophicus (9, 11).
FIG. 2.
StoHjm preferentially binds to ssDNA and branched DNA. The binding of StoHjm (0 to 128 nM) to ssDNA (S1, 84 mer) (A), dsDNA (S6) (B), a 3′ overhang (S3a) (C), a 5′ overhang (S2) (D), a replication fork (S9) (E), a replication fork with a ssDNA (10 mer) region on the leading strand (S12) (F), or a Holliday junction (S8) (G) was assayed. (H) Quantification of results in panels A to G. Binding is expressed as percent loss of free DNA (total DNA minus the remaining free DNA substrate) against the total DNA level in the control. The lengths of the DNA strands (in nucleotides) are indicated.
StoHjm preferentially unwinds nascent strands of the replication fork.
We next tested the unwinding activities of StoHjm toward several DNA structures, specifically S5, S3a, S4a, S9, and S13 (Fig. 3A to E and Table 1). StoHjm unwound blunt-ended dsDNA with a very low efficiency (Fig. 3A, F, and G). Interestingly, the unwinding activities of StoHjm toward dsDNA substrates with 3′ and 5′ extensions (overhangs) increased considerably (Fig. 3B, C, F, and G). This result suggests that the overhang structure facilitates unwinding by StoHjm. The unwinding substrate preference of StoHjm was in good agreement with its DNA binding preference (Fig. 2B to D).
FIG. 3.
Structure-specific unwinding by StoHjm. Unwinding assays were conducted with blunt-ended dsDNA (S5) (A), dsDNA with a 3′ overhang (S3a) (B), dsDNA with a 5′ overhang (S2) (C), a replication fork containing nascent strands (S9) (D), or Y-structured DNA (S13) (E). (F) Quantification of the results in panels A to E. (G) Time courses of the helicase activities of StoHjm assayed using 3′ overhang (open circles), 5′ overhang (open squares), and blunt-ended (open triangles) substrates. Percent unwinding was calculated as the ratio of unwound DNA (total DNA substrate minus the remaining substrate) to total labeled DNA. The lengths of the DNA strands (in nucleotides) are labeled. P1, P2, P3, and P4 in panel D indicate the unwinding products observed when fork DNA was used.
The unwinding activity of StoHjm toward replication forks was also assayed using substrate S9, which had 36-bp parental dsDNA and equal lengths of dsDNA on the nascent strands (Fig. 3D and Table 1). Products in which the nascent strand was absent (products P1 and P2) (Fig. 3D) were generated more rapidly than those composed of one parent strand and its annealed nascent strand (products P3 and P4) (Fig. 3D and F), indicating that StoHjm unwinds the nascent strands of the replication fork more efficiently than the parent strands. To confirm this, we assayed the unwinding of Y-structured DNA (S13, a replication fork lacking nascent strands) (Table 1 and Fig. 3E). Substrate reduction was greater with replication forks than with Y-structured DNA (Fig. 3D, E, and F), showing that unwinding is more rapid if the substrate contains nascent strands. These results demonstrate that StoHjm preferentially unwinds the nascent strands of the replication fork.
To investigate which nascent strand (i.e., the leading or lagging strand) is more efficiently unwound by StoHjm, we prepared substrates in which the nascent leading strand or lagging strand was labeled (S10 and S11) (Table 1). As shown in Fig. 4A to C, StoHjm unwound the nascent leading and lagging strands with almost the same efficiency. Unwinding efficiency was only slightly higher with the nascent lagging strand at StoHjm concentrations of 57 nM or higher (Fig. 4C). These results demonstrate that StoHjm is able to unwind both nascent strands on the replication fork efficiently. The unwinding efficiencies with nascent leading and lagging strands were similar to those with overhang structures (Fig. 3B and C), perhaps due to the fact that the leading nascent strand of the replication fork resembles a 5′ overhang structure, while the lagging nascent strand of the replication fork resembles a 3′ overhang structure. This unwinding activity of StoHjm differs from those reported for Hjm from P. furiosus and for Hel308a from M. thermautotrophicus (9, 11), as these enzymes unwind only the lagging strand of the replication fork and the 3′ overhang structure. The difference in unwinding activity between StoHjm and Hjm/Hel308a may be due to evolutional divergence between Euryarchaeota and Crenarchaeota and may indicate divergent cellular functions of Hjm/Hel308a helicases in these two major archaeal subdomains.
FIG. 4.
Unwinding of the nascent leading and lagging strands of the replication fork by StoHjm. A replication fork containing a labeled leading strand (S11) (A) or a labeled lagging strand (S10) (B) was used as a substrate. (C) Quantification of the results in panels A and B. Percent unwinding was calculated as the ratio of unwound DNA (total DNA substrate minus the remaining substrate) to total labeled DNA. The lengths of the DNA strands (in nucleotides) are labeled.
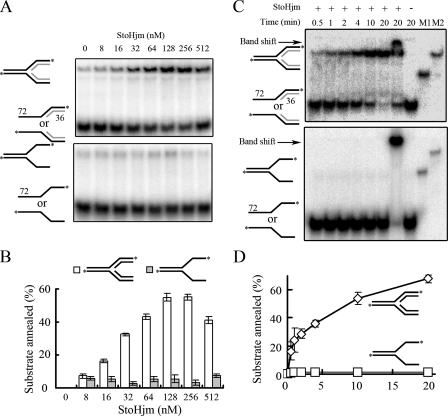
Structure-specific annealing activity of StoHjm.
During the unwinding assays, we noticed that the substrates were not completely unwound, even though the reaction mixtures were incubated for over 1 hour and the enzyme concentration was 1,000 nM (data not shown). This suggested that StoHjm may have a strand-annealing activity. A similar phenomenon has also been reported for human BLM (2). To test if StoHjm has annealing activity, we used two synthesized DNA strands (Table 1) (H1 and H2, 72-mer) with partial complementary regions (36-mer) that, upon annealing, could form a Y-structured DNA (S13) (Table 1). A very weak annealing product was observed in the presence of StoHjm (Fig. 5A and C, lower panels, and B and D). However, when we used DNA substrates that could form a pseudo-replication fork (S7 and S14) (Table 1), remarkably increased amounts of products (replication fork structure) were generated with increasing amounts of StoHjm (8 to 512 nM) (Fig. 5A, upper panel, and B) and with increasing reaction times (Fig. 5C, upper panel, and D). Only a very small amount of fork DNA structure could be spontaneously produced in the absence of StoHjm (Fig. 5A, lane 1). We also used substrates that could form 3′ overhang or 5′ overhang dsDNA to test the annealing ability of StoHjm; however, no product could be detected under these conditions (data not shown). These results demonstrate that StoHjm promotes fork-specific strand annealing.
FIG. 5.
Structure-specific annealing activity of StoHjm. (A) Activity as a function of enzyme concentrations. Upper panel, single strands with partial complementary sequences that have the potential to form a Y-structure DNA (H1 and H2). Lower panel, 3′ and 5′ overhangs that have the potential to form a pseudo-replication fork containing nascent strands (S7 and S14). The reaction time was 30 min. (B) Quantification of the results in panel A. The percentage of annealed substrate was calculated as the ratio of the annealed product to the total labeled DNA. (C) Annealing activity as a function of reaction time. The concentration of StoHjm used was 100 nM. “M1” and “M2” indicate markers representing Y-shaped and fork DNAs, respectively. The arrows indicate gel shift DNAs. (D) Quantification of the results in panel B.
In vitro regression of a synthetic replication fork by StoHjm.
Since StoHjm catalyzed unwinding of nascent leading and lagging strands and promoted annealing of parent strands (Fig. 4 and 5), we wondered if unwinding of both nascent strands occurs simultaneously and if this unwinding event occurs simultaneously with annealing of the unwound parent strands. If concurrent, these activities would impart replication fork regression activity, as has been reported for the RecG helicase in E. coli and the BLM helicase in humans (19, 23, 25, 26).
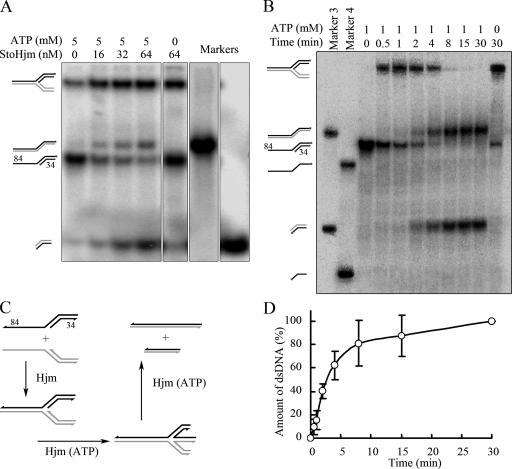
To examine if StoHjm has fork regression activity, we used S3 and S4 as substrates which could form an authentic replication fork. S3 was labeled on both nascent and parental lagging strands, while S4 was not labeled (Fig. 6C and Table 1). Because unwinding requires ATP hydrolysis, we tested if StoHjm has regression activity in the presence of 5 mM ATP (Fig. 6A and C). In the absence of StoHjm, only a small proportion of substrates spontaneously annealed to form a fork, even though the reaction mixture contained 5 mM ATP (Fig. 6A). Spontaneous migration may have also occurred, since faint bands corresponding to long and short dsDNA were detected; however, the amount of spontaneous migration product was small (Fig. 6A). By contrast, when StoHjm was added without ATP, much more annealed fork product was present, but neither long nor short dsDNA could be detected (Fig. 6A and B). This result indicates that in the absence of ATP, StoHjm was able to promote annealing and prevent spontaneous migration. In the presence of ATP and increased amounts of StoHjm, increasing amounts of products corresponding to annealed parent strands (84/84-mer) and annealed nascent strands were generated (Fig. 6A). Increasing amounts of products were also generated with increasing reaction times (Fig. 6B and D).
FIG. 6.
Replication fork regression activity of StoHjm in the absence of StoHjc. The annealing and unwinding activities of StoHjm were assayed with a 3′ overhang (S3) (Table 1), which was labeled on both strands, and an unlabeled 5′ overhang (S4) with a complementary single-strand region. All the samples and the markers were separated on a single gel. (A) Activity as a function of enzyme concentration. The reaction time was 30 min. (B) Schematic illustration of reactions shown in panel A. (C) Activity as a function of reaction time. The concentration of StoHjm was 100 nM. (D) Quantification of the results in panel B. The percentage of products was the ratio of 84/84- and 34/34-bp dsDNA in combination to the total DNA substrates.
The annealed parent strands (84/84-mer) and annealed nascent strands were possibly generated by fork regression. In this process, two nascent strands were unwound from template strands at the same time and annealed to form a short double strand. The parent strands also annealed to form a long double strand simultaneously (Fig. 6C). Another possibility is that the nascent strands were generated from the annealed fork separately, and the long duplex DNA was formed by strand displacement. Still another is that the generation of duplex DNAs was due to unwinding of the unannealed overhang substrates and subsequent spontaneous annealing. However, because we did not see bands of the ssDNAs in the gel profiles (Fig. 6A and B), we assume that in the presence of StoHjm, annealing of the two complementary ssDNA regions of the substrates might be much stronger than unwinding of 3′ or 5′ overhangs, and the substrates were preferentially annealed to form a replication fork. The unwinding of nascent strands from the parent strands and their subsequent annealing to form complementary duplex DNAs might occur simultaneously. Accordingly, neither the 84-mer nor the 34-mer ssDNA would be produced. The results from the experiment may suggest a replication fork regression activity in vitro; however, more evidence is needed to verify the existence of the regression activity (see below).
Fork regression activity generates a Holliday junction, or “chicken foot” structure, during fork reversal. Since Hjc is the Holliday junction-specific endonuclease in archaea, we tested whether the addition of StoHjc to the reaction mixture might lead to cleavage of “chicken foot” structures (17, 30). To investigate if Holliday junctions were generated, we first tested the cleavage activity of StoHjc. As shown in Fig. S3 in the supplemental material, StoHjc specifically cleaved the Holliday junction structure. Next, StoHjc was included in the putative fork regression reaction mixture along with StoHjm (Fig. 7A). In this experiment, S3a (with the lagging nascent strand labeled) and S4a (with the leading nascent strand labeled) (Table 1) were used. Electrophoresis of the denatured reaction products revealed that neither StoHjm nor StoHjc alone could generate cleavage products, even if ATP was present (Fig. 7A). A similar result was obtained when StoHjm and StoHjc were incubated together in the absence of ATP (Fig. 7A). However, in the presence of StoHjm, StoHjc, and ATP, bands of various discrete lengths (<36-mer) appeared (Fig. 7A and B, lanes 9 and 10). The shorter DNA fragments are assumed to be the StoHjc-cleaved products on both nascent strands of “chicken foot” structures formed by StoHjm.
FIG. 7.
Fork reversal activity of StoHjm in the presence of StoHjc. (A) Cleavage by StoHjc (50 nM) in association with StoHjm (64 nM) and in the presence of ATP (5 mM). Both nascent strands were labeled, while the parent strands were not labeled. Labeled 48-, 36-, 26-, and 20-mer strands were used as markers. (B) Cleavages observed when substrates with each of the four strands labeled were used. The substrates used are shown above lanes 1 to 8. The substrates with either nascent strand labeled were used as controls (lanes 9 and 10). StoHjm (100 nM) was used in the reactions. (C) Schematic illustration of reactions shown in panels A and B.
To reveal the cleavage of StoHjm/SotHjc in more detail, we prepared overhang substrates in which only one of the four strands was labeled (Fig. 7B, lanes 1 to 8). The cleavage activities toward these substrates were examined. When the nascent leading strand was labeled (lanes 1 and 2), a product of about 20 nucleotides (nt) was generated. Similarly, when the parent lagging strand was labeled, a product with the same size was generated (lanes 7 and 8). It is hypothesized that one cleavage event occurred across the nascent leading strand and the parent lagging strand (Fig. 7C). By contrast, when the lagging nascent was labeled, two products, with sizes of around 25 and 15 nt, were observed (lanes 3 and 4). This could indicate that two cleavage sites existed across the labeled nascent lagging strand in the generated Holliday junction (Fig. 7C). If this were the case when the leading parent strand was labeled, it would be expected that two products, one with a size of about 75 nt and another with a size of about 65 nt, should be generated. Indeed, a band around the 72-nt marker was visible (Fig. 7B, lanes 5 and 6), although under our electrophoresis conditions, the products might not have properly separated. Our results not only support the assumption that StoHjm generates “chicken foot” structures from the replication fork but also suggest that there exist three cleavage sites in the Holliday junctions generated by StoHjm/StoHjc.
StoHjm physically interacts with StoHjc.
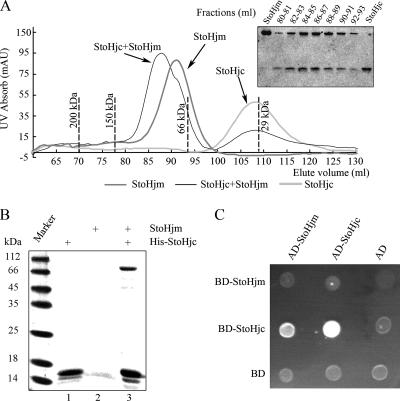
To determine if StoHjm and StoHjc form a complex, we conducted gel filtration analysis (Fig. 8A). When the two proteins were combined and loaded onto a Sephacryl S-200 HR column, the elution peak was shifted to a position with a molecular mass larger than that of either StoHjm (69 kDa) or StoHjc (29 kDa). SDS-PAGE analysis showed that the peak contained both StoHjm and StoHjc, and the amounts of StoHjc in the peak fractions differed according to the levels of StoHjm (Fig. 8A). This result demonstrates that StoHjm and StoHjc form a complex in solution. Based on the elution volumes of either StoHjm or StoHjc alone or StoHjm and StoHjc in combination, we assumed that StoHjm and StoHjc formed monomers and dimers, respectively, and that the StoHjm-StoHjc complex was composed of one StoHjm molecule and two StoHjc molecules.
FIG. 8.
Physical interaction between StoHjm and StoHjc. (A) Gel filtration analysis of StoHjm and StoHjc, either alone or combined. The peaks corresponding to StoHjm, StoHjc, and the StoHjm/StoHjc mixtures are indicated by arrows. The elution volumes of protein markers are indicated by dashed lines. SDS-PAGE analysis of the peak fractions is shown in the inset of the figure. (B) Pulldown assay of StoHjc and StoHjm. Lane 1, His-tagged StoHjc (16 kDa); lane 2, non-His-tagged StoHjm (79 kDa); lane 3, His-tagged StoHjc combined with StoHjm. (C) Yeast two-hybrid analysis. The transformants were cultured on SD/−His/−Leu/−Trp plus 10 mM 3-AT. AU, atomic units.
Next, we carried out a pulldown assay. As shown in Fig. 8B, His-Tagged StoHjc was able to pull down StoHjm, which has no His tag. Thus, StoHjm and StoHjc strongly interact, leading to the formation of a stable StoHjm-StoHjc complex in vitro.
Yeast two-hybrid analysis was also conducted to confirm the interaction between StoHjm and StoHjc. As shown in Fig. 8C, cotransformation of AD-StoHjm and BD-StoHjc allowed cells to grow well on SD/−His/−Leu/−Trp, indicating an in vivo interaction between both proteins. Interestingly, AD-StoHjc and BD-StoHjc cotransformants grew well on SD/−His/−Leu/−Trp, indicating that StoHjc forms an oligomer in vivo.
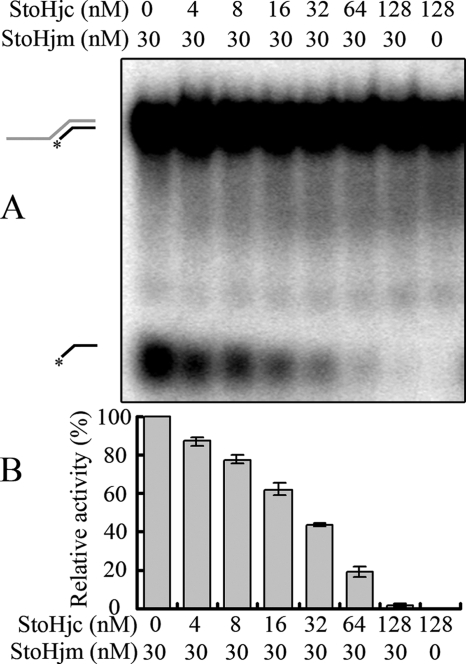
The unwinding activity of StoHjm is inhibited by StoHjc.
To understand the function of StoHjm-StoHjc interactions, we performed unwinding assays in the presence of increasing amounts of StoHjc, using a dsDNA substrate with a 3′ overhang (S3a, 84/34-mer) (Table 1). We found that StoHjc inhibited the unwinding activity of StoHjm (Fig. 9). Interestingly, when the molar ratio of StoHjc to StoHjm was increased to about two and above, unwinding was completely inhibited (Fig. 9). This may indicate that, in the putative StoHjc-StoHjm complex, StoHjc exists as a dimer, while StoHjm is monomeric.
FIG. 9.
The unwinding activity of StoHjm is inhibited by StoHjc. (A) A 3′ overhang DNA (S3a) (Table 1) was used to test the effect of increasing concentrations of StoHjc on the unwinding activity of StoHjm (30 nM). The assay conditions were the same as those for Fig. 3. (B) Quantification of the results in panel A. Activity in the presence of StoHjc was divided by activity in the absence of StoHjc to determine the relative unwinding activity. The activity was expressed as the ratio of unwound substrate (labeled 34-mer ssDNA) to total substrate.
DISCUSSION
Hjm was originally identified by screening cell extracts and the genomic library of the euryarchaeon P. furiosus (8). It has been shown to dissolve four-way junctions in an ATP-dependent manner (8). In a subsequent study, the Hjm gene was transformed into E. coli strain DnaE486, which harbors a mutation of the DNA polymerase α subunit and accumulates stalled replication forks. These transformants had a phenotype similar to that of RecQ transformants. Also, Hjm was shown to have 3′-to-5′ helicase activity (9). Based on these findings, Hjm was assumed to be a RecQ-like helicase (9). Hel308a, an Hjm homologue in another euryarchaeon, M. thermautotrophicus, was identified through genetic screening for helicase genes by using the same E. coli system (11). Hel38a also brought about the same phenotype in E. coli strain DnaE486 cells and RecQ. Like Hjm, Hel308a has a 3′-to-5′ helicase activity, and these homologues share 31% amino acid identity (9, 11). Despite these biochemical and genetic investigations, the real functions of the two DNA helicases are still unclear.
In this study, we cloned and characterized a homologue of Hjm and Hel308a from the crenarchaeon S. tokodaii. Although the replication fork binding and unwinding properties of StoHjm are quite similar to those of Hjm from P. furious and Hel308a from M. thermautotrophicus (Fig. 2 to 4) (9, 11), the unwinding polarity of StoHjm differs from that of Hjm and Hel308a. This difference may have arisen as a result of divergent evolution and may indicate that the homologues in Euryarchaeota and Crenarchaeota play different roles, as seen in differences in their DNA transactions.
In an attempt to define the functions of StoHjm, we conducted in vitro assays using substrates that can form pseudo- or authentic replication forks through annealing. Our results revealed that StoHjm promotes structure-specific annealing. In the presence of ATP, the enzyme probably further catalyzes annealing of the authentic replication fork, resulting in four-way junctions and migration to produce long parent strands and, eventually, short nascent strands. Based on these findings, we conclude that StoHjm has in vitro fork reversal activity similar to that of the reported bacterial RecG protein. So far, several substrates have been used to examine fork reversal activities of helicases in other organisms (14, 15, 26). Although the substrate that we have used is authentic and simple, spontaneous migration may occur. For this reason, a more complicated substrate, such as a plasmid-sized replication fork, might be needed for further analysis (3, 17).
We have also found that StoHjm interacts with StoHjc. Hjc, the Holliday junction-specific endonuclease found in archaea (3, 17), reportedly interacts with proliferating cell nuclear antigen (PCNA) and RadB (5, 12, 16) in P. furiosus. Hjm from P. furiosus has also been reported to interact with PCNA, and this interaction stimulates Hjm helicase activity (9). Therefore, Hjc, PCNA, and Hjm in P. furiosus may form a complex. In this study, we found that StoHjm physically interacts with StoHjc, as shown by gel filtration, pulldown, and yeast two-hybrid analyses. This interaction is likely to be functional, since possible four-way junctions were generated and cleaved efficiently in the presence of both enzymes. Moreover, the addition of StoHjc resulted in inhibition of StoHjm unwinding activity when a 3′ overhang was used as a substrate. Although this inhibition may be due to competitive DNA binding between StoHjc and StoHjm, it is possible that StoHjc inhibits the enzymatic activity of StoHjm in the presence of unfavorable substrates, such as a dsDNA substrate with a 3′ overhang, through protein-protein interactions. Alternatively, a negative regulation mechanism involving StoHjc may account for this inhibition of StoHjm. This mechanism may act to reduce cleavage and recombination events in the cells. Further experiments are needed to determine if fork regression reactions mediated by StoHjc are inhibited. In addition, Hjc from S. solfataricus has been reported to physically interact with PCNA (5). It will be interesting to investigate whether StoHjm, StoPCNA, and StoHjc form a protein complex. If so, the interactions among Hjm, PCNA, and Hjc might be conserved in archaea.
Based on our results, we propose that StoHjm, the Hjm/Hel308 homologue, functions like RecG in E. coli, generating four-way junctions, or “chicken foot” structures, to repair stalled replication forks. We also propose that the structure-specific endonuclease Hjc acts coordinately with the Hjm/Hel308a homologue to process the DNA structures and that this coordination of functions occurs through direct interaction between these enzymes. It will be interesting to examine whether Hjm/Hel308a, and even mammalian Hel308 proteins, have similar fork-specific annealing and fork reversal activities. Indeed, strand-annealing activities have also been observed in some of the RecQ family helicases that have 3′-to-5′ helicase activities, such as BLM, RecQ1, and RecQ5β (2, 10, 29). On the other hand, since cleavage tends to generate gene recombination, it will be interesting to investigate if StoHjm, in coordination with other proteins, catalyzes template switching, which does not cause recombination to occur. Further experiments will be necessary to unravel the mechanism by which Hjm mediates recombinational repair in conjunction with replication and recombination proteins, including Hjc, PCNA, RadA, and RadB.
During the preparation of this report, the structure of Hel308 from A. fulgidus was published (1). A mechanism accounting for the preference of Hel308 for branched DNA was proposed based on this structure analysis. This structure will provide a framework for understanding of the functions of the newly identified Hjm/Hel308a DNA helicase family.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (2004CB719604 to Y.S.) and the National Natural Science Foundation of China (30470386 to Y.S. and 30700011 to D.S.).
Footnotes
Published ahead of print on 22 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Buttner, K., S. Nehring, and K. P. Hopfner. 2007. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat. Struct. Mol. Biol. 14647-652. [DOI] [PubMed] [Google Scholar]
- 2.Cheok, C. F., L. Wu, P. L. Garcia, P. Janscak, and I. D. Hickson. 2005. The Bloom's syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 333932-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daiyasu, H., K. Komori, S. Sakae, Y. Ishino, and H. Toh. 2000. Hjc resolvase is a distantly related member of the type II restriction endonuclease family. Nucleic Acids Res. 284540-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhillon, K. K., J. Sidorova, Y. Saintigny, M. Poot, K. Gollahon, P. S. Rabinovitch, and R. J. Monnat. 2007. Functional role of the Werner syndrome RecQ helicase in human fibroblasts. Aging Cell 653-61. [DOI] [PubMed] [Google Scholar]
- 5.Dorazi, R., J. L. Parker, and M. F. White. 2006. PCNA activates the Holliday junction endonuclease Hjc. J. Mol. Biol. 364243-247. [DOI] [PubMed] [Google Scholar]
- 6.Dunderdale, H. J., G. J. Sharples, R. G. Lloyd, and S. C. West. 1994. Cloning, overexpression, purification, and characterization of the Escherichia coli RuvC Holliday junction resolvase. J. Biol. Chem. 2695187-5194. [PubMed] [Google Scholar]
- 7.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, DC.
- 8.Fujikane, R., K. Komori, H. Shinagawa, and Y. Ishino. 2005. Identification of a novel helicase activity unwinding branched DNAs from the hyperthermophilic archaeon, Pyrococcus furiosus. J. Biol. Chem. 28012351-12358. [DOI] [PubMed] [Google Scholar]
- 9.Fujikane, R., H. Shinagawa, and Y. Ishino. 2006. The archaeal Hjm helicase has recQ-like functions, and may be involved in repair of stalled replication fork. Genes Cells 1199-110. [DOI] [PubMed] [Google Scholar]
- 10.Garcia, P. L., Y. Liu, J. Jiricny, S. C. West, and P. Janscak. 2004. Human RECQ5β, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 232882-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guy, C. P., and E. L. Bolt. 2005. Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands. Nucleic Acids Res. 333678-3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guy, C. P., S. Haldenby, A. Brindley, D. A. Walsh, G. S. Briggs, M. J. Warren, T. Allers, and E. L. Bolt. 2006. Interactions of RadB, a DNA repair protein in Archaea, with DNA and ATP. J. Mol. Biol. 35846-56. [DOI] [PubMed] [Google Scholar]
- 13.Ishino, Y., T. Nishino, and K. Morikawa. 2006. Mechanisms of maintaining genetic stability by homologous recombination. Chem. Rev. 106324-339. [DOI] [PubMed] [Google Scholar]
- 14.Kadyrov, F. A., and J. W. Drake. 2004. UvsX recombinase and Dda helicase rescue stalled bacteriophage T4 DNA replication forks in vitro. J. Biol. Chem. 27935735-35740. [DOI] [PubMed] [Google Scholar]
- 15.Kanagaraj, R., N. Saydam, P. L. Garcia, L. Zheng, and P. Janscak. 2006. Human RECQ5β helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 345217-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komori, K., T. Miyata, J. DiRuggiero, R. Holley-Shanks, I. Hayashi, I. K. O. Cann, K. Mayanagi, H. Shinagawa, and Y. Ishino. 2000. Both RadA and RadB are involved in Homologous recombination in Pyrococcus furiosus. J. Biol. Chem. 27533782-33790. [DOI] [PubMed] [Google Scholar]
- 17.Komori, K., S. Sakae, R. Fujikane, K. Morikawa, H. Shinagawa, and Y. Ishino. 2000. Biochemical characterization of the Hjc Holliday junction resolvase of Pyrococcus furiosus. Nucleic Acids Res. 284544-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindahl, T., and R. D. Wood. 1999. Quality control by DNA repair. Science 2861897-1905. [DOI] [PubMed] [Google Scholar]
- 19.Machwe, A., L. Xiao, J. Groden, and D. K. Orren. 2006. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry 4513939-13946. [DOI] [PubMed] [Google Scholar]
- 20.Mashimo, K., M. Kawata, and K. Yamamoto. 2003. Roles of the RecJ and RecQ proteins in spontaneous formation of deletion mutations in the Escherichia coli K12 endogenous tonB gene. Mutagenesis 18355-363. [DOI] [PubMed] [Google Scholar]
- 21.McGlynn, P., and R. G. Lloyd. 2002. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 3859-870. [DOI] [PubMed] [Google Scholar]
- 22.McGlynn, P., and R. G. Lloyd. 2000. Modulation of RNA polymerase by (p) ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 10135-45. [DOI] [PubMed] [Google Scholar]
- 23.McGlynn, P., and R. G. Lloyd. 2002. Genome stability and the processing of damaged replication forks by RecG. Trends Genet. 18413-419. [DOI] [PubMed] [Google Scholar]
- 24.McGlynn, P., and R. G. Lloyd. 2001. Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Natl. Acad. Sci. USA 988227-8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGlynn, P., R. G. Lloyd, and K. J. Marians. 2001. Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc. Natl. Acad. Sci. USA 988235-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ralf, C., I. D. Hickson, and L. Wu. 2006. The Bloom's syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 28122839-22846. [DOI] [PubMed] [Google Scholar]
- 27.Rothstein, R., B. Michel, and S. Gangloff. 2000. Replication fork pausing and recombination or “gimme a break”. Genes Dev. 141-10. [PubMed] [Google Scholar]
- 28.Seigneur, M., V. Bidnenko, S. D. Ehrlich, and B. Michel. 1998. RuvAB acts at arrested replication forks. Cell 95419-430. [DOI] [PubMed] [Google Scholar]
- 29.Sharma, S., J. A. Sommers, S. Choudhary, J. K. Faulkner, S. Cui, L. Andreoli, L. Muzzolini, A. Vindigni, and R. M. Brosh, Jr. 2005. Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J. Biol. Chem. 28028072-28084. [DOI] [PubMed] [Google Scholar]
- 30.Sharples, G. J. 2001. The X philes: structure-specific endonucleases that resolve Holliday junctions. Mol. Microbiol. 39823-834. [DOI] [PubMed] [Google Scholar]
- 31.Shen, Y., K. Musti, M. Hiramoto, H. Kikuchi, Y. Kawarabayashi, and I. Matsui. 2001. Invariant Asp-1122 and Asp-1124 are essential residues for polymerization catalysis of family D DNA polymerase from Pyrococcus horikoshii. J. Biol. Chem. 27627376-27383. [DOI] [PubMed] [Google Scholar]
- 32.Singleton, M. R., S. Scaife, and D. B. Wigley. 2001. Structural analysis of DNA replication fork reversal by RecG. Cell 10779-89. [DOI] [PubMed] [Google Scholar]
- 33.Sogo, J. M., M. Lopes, and M. Foiani. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297599-602. [DOI] [PubMed] [Google Scholar]
- 34.Viguera, E., P. Hernandez, D. B. Krimer, R. Lurz, and J. B. Schvartzman. 2000. Visualisation of plasmid replication intermediates containing reversed forks. Nucleic Acids Res. 28498-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wirtenberger, M., B. Frank, K. Hemminki, R. Klaes, R. K. Schmutzler, B. Wappenschmidt, A. Meindl, M. Kiechle, N. Arnold, B. H. F. Weber, D. Niederacher, C. R. Bartram, and B. Burwinkel. 2006. Interaction of Werner and Bloom syndrome genes with p53 in familial breast cancer. Carcinogenesis 271655-1660. [DOI] [PubMed] [Google Scholar]
- 36.Zerbib, D., C. Mezard, H. George, and S. C. West. 1998. Coordinated actions of RuvABC in Holliday junction processing. J. Mol. Biol. 281621-630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.