Abstract
Cells of the gliding bacterium Flavobacterium johnsoniae move rapidly over surfaces by an unknown mechanism. Transposon insertions in sprB resulted in cells that were defective in gliding. SprB is a highly repetitive 669-kDa cell surface protein, and antibodies against SprB inhibited the motility of wild-type cells. Polystyrene microspheres coated with antibodies against SprB attached to and were rapidly propelled along the cell surface, suggesting that SprB is one of the outermost components of the motility machinery. The movement of SprB along the cell surface supports a model of gliding motility in which motors anchored to the cell wall rapidly propel cell surface adhesins.
Cells of Flavobacterium johnsoniae and of many related bacteria in the phylum Bacteroidetes crawl over surfaces at a rate of approximately 2 μm/s in a process called gliding motility. F. johnsoniae cells glide on agar, glass, polystyrene, Teflon, and many other surfaces (25, 31). Cells suspended in liquid bind and propel added particles, such as polystyrene spheres (34). The mechanism of this form of cell movement remains unknown despite decades of research (8, 24, 34, 37). Genes and proteins required for F. johnsoniae motility have been identified (1, 5, 6, 16-18, 26, 27). GldA, GldF, and GldG appear to form an ATP-binding cassette transporter that is required for gliding (1, 16). Eight other Gld proteins (encoded by gldB, -D, -H, -I, -J, -K, -L, and -M) are also required for movement (5, 6, 17, 18, 26, 27). Disruption of the gene encoding any of these 11 proteins results in a complete loss of motility. The mutants form nonspreading colonies, and individual cells exhibit no movement on agar, glass, and other surfaces tested. Genetic analyses have suggested that most of the genes that are absolutely required for gliding have been identified (5). Many of these genes are unique to members of the phylum Bacteroidetes. Genome analyses have suggested that gliding of F. johnsoniae and related bacteria is not related to other well-studied forms of bacterial movement, such as bacterial flagellar motility, type IV pilus-mediated twitching motility, myxobacterial gliding motility, and mycoplasma gliding motility (5, 25, 40).
The F. johnsoniae Gld proteins localize to the cell envelope, but surprisingly, none of them appear to be exposed on the cell surface (5, 6, 16-18, 26, 27). Cell surface proteins play critical roles in most other forms of bacterial motility (3), and it is difficult to explain F. johnsoniae gliding without including such proteins. We hypothesized that the cell surface components of the machinery have escaped detection because they are redundant and their absence does not result in a complete loss of motility. In this paper we describe isolation and analysis of transposon-induced mutants with partial defects in gliding. Many of these mutants had insertions in a gene encoding a large cell surface protein that may be a moving component of the gliding motility machinery.
MATERIALS AND METHODS
Bacterial and bacteriophage strains, plasmids, and growth conditions.
F. johnsoniae strain FJ1 (= ATCC 17061) was the wild-type strain used in this study (26). Thirty-seven spontaneous and chemically induced motile nonspreading mutants of F. johnsoniae were obtained from J. Pate and are designated UW102-1, -2, -3, -18, -24, -37, -43, -45, -46, -50, -51, -67, -73, -88, -91, -93, -106, -128, -133, -135, -136, -142, -143, -148, -149, -150, -155, -156, -157, -158, -168, -171, -172, -176, -298, -344, and -345 (9, 39). Other motility mutants used in this study were CJ101-288 (1), CJ282 (17), CJ569 (18), CJ974 (32), CJ1043 (27), CJ1300 (5), CJ1304 (5), FJ113 (5), FJ118 (31), UW102-41 (26), UW102-48 (6), UW102-57 (5), and UW102-77 (16). F. johnsoniae strains were grown in Casitone-yeast extract (CYE) medium at 30°C, as previously described (28). To observe colony spreading, F. johnsoniae was grown on PY2 agar medium (1) at 25°C. Motility medium (MM) was used to observe movement of individual cells in wet mounts (22). Antibiotics were used at the following concentrations when needed: ampicillin, 100 μg/ml; cefoxitin, 100 μg/ml; chloramphenicol, 30 μg/ml; erythromycin, 100 μg/ml; kanamycin, 35 μg/ml; and tetracycline, 20 μg/ml.
Transposon mutagenesis and identification of sites of insertion.
Tn4351, HimarEm1, and HimarEm2 were introduced into wild-type F. johnsoniae by conjugation, and mutants were selected by plating cells on PY2 agar containing erythromycin as previously described (5, 18). Mutants that formed nonspreading colonies but retained some ability to move on glass in wet mounts were chosen for further study. Most of the transposon-induced mutants were derived from F. johnsoniae FJ1; the only exceptions were CJ987 and CJ1270, which were derived from F. johnsoniae MM101 (26). Chromosomal DNA was isolated from mutants, and the sites of transposon insertions were determined as previously described (5, 16, 19). For highly repetitive regions of sprB that were prone to rearrangement by recombination, fragments were cloned into pBC SK+ (Stratagene, La Jolla, CA) and propagated in recombination-deficient Escherichia coli Stbl2 or Stbl4 (Invitrogen, Carlsbad, CA). DNA sequencing was performed by the University of Chicago Cancer Research Center DNA Sequencing Facility. Sequences were analyzed with MacVector and AssemblyLign software (Accelrys, San Diego, CA), and comparisons to database sequences were made using the BLAST algorithm (2). Secondary structure was predicted using the Chou-Fasman and Robson-Garnier analyses (11, 12).
Cloning of sprB.
F. johnsoniae FJ162 has a HimarEm2 insertion 279 bp upstream of the sprB start codon. This insertion did not alter the expression of sprB and was therefore upstream of the sprB promoter region. The region flanking this insertion was cloned as a ClaI fragment into pBC SK+ in E. coli Stbl4 to generate pSN54. The 21.5-kb XhoI fragment of this plasmid spanning sprB and a portion of the transposon including the kanamycin resistance gene was ligated into the SalI site of the Flavobacterium-E. coli shuttle vector pCP29 to generate pSN60. pSN60 carries the entire sprB gene and includes 279 bp of DNA upstream of the sprB start codon and 439 bp downstream of the stop codon. pSN60 replicates with a copy number of approximately 2 to 4 in F. johnsoniae.
Protein expression and antibody production.
A 12,173-bp HindIII fragment was cut from the HimarEm2 sprB mutant Fj104 (Fig. 1). This fragment contained 9,236 bp of sprB upstream of the transposon insertion and 2,937 bp of the transposon, including the kanamycin resistance gene, which allowed selection. The fragment was ligated into pMalC2 (New England Biolabs, Ipswich, MA), creating pSN91. pSN91 was introduced into E. coli Rosetta 2(DE3) (Novagen, Madison, WI), which expresses seven rare tRNAs required for efficient expression of SprB. To isolate recombinant SprB, cells were grown to mid-log phase at 37°C in rich medium with glucose (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl, 2 g/liter glucose), induced by addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and incubated for an additional 48 h at 25°C. Cells were disrupted using a French press, and inclusion bodies containing recombinant SprB were isolated by centrifugation at 6,415 × g for 30 min. Inclusion bodies were suspended in 20 mM Tris-HCl (pH 7.4) containing 200 mM NaCl, 1 mM EDTA, and 200 μg/ml lysozyme and incubated for 15 min at 24°C. Inclusions were collected by centrifugation at 20,000 × g for 15 min and washed by resuspending them in 10 mM HEPES containing 2% Triton X-100 by sonication a total of three times. The inclusions were boiled in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) loading buffer, and SprB was purified on 5% acrylamide gels by SDS-PAGE. Recombinant SprB was visualized by CuCl2 staining (21), the band was cut from the gel and destained in 0.25 M Tris (pH 9.0), 0.25 M EDTA, and the protein was electroeluted at 60 mA for 5 h into 25 mM Tris, 192 mM glycine, 0.1% SDS using a model 422 Electro-Eluter (Bio-Rad). Polyclonal antibodies against recombinant SprB were produced and affinity purified using the recombinant protein by Proteintech Group, Inc. (Chicago, IL).
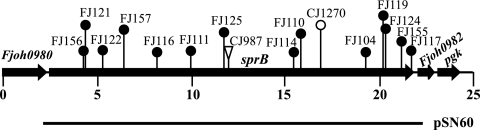
FIG. 1.
Map of the sprB region of F. johnsoniae. The numbers below the map indicate positions (in kilobase pairs) in the sequence. The sites of Tn4351, HimarEm1, and HimarEm2 insertions are indicated by a triangle, an open circle, and filled circles, respectively. The region of DNA carried by complementing plasmid pSN60 is indicated below the map.
Microscopic observations of cell movement.
The movement of wild-type and mutant cells of F. johnsoniae over agar and glass was examined by phase-contrast microscopy at 25°C. Cells were grown in MM at 25°C without shaking as previously described (22). To analyze movement on agar, cells were spotted onto MM containing 1% agar on a glass slide and covered with an oxygen-permeable Teflon membrane (Yellow Springs Instrument Co., Yellow Springs, OH), and motility was examined by phase-contrast microscopy. To observe movement over glass, cells in MM were introduced into a Palmer counting cell (Wildlife Supply Company, Saginaw, MI), covered with an glass coverslip (18 by 18 mm), and incubated for 2 min. Unbound cells were removed by wicking the fluid from beneath the coverslip with a Kimwipe, followed by washing with MM, a total of three times. Cells were incubated in fresh MM for 2 min following the final wash, and then motility was observed. Images were recorded using a Photometrics CoolSNAPcf2 camera mounted on an Olympus BH-2 phase-contrast microscope and were analyzed using MetaMorph software (Molecular Devices, Downingtown, PA). Tracks illustrating the movements of cells were obtained by superimposing individual digital video frames using the Logical AND operation of MetaMorph.
Inhibition of cell movement by antisera against SprB.
To determine the effect of antiserum on movement over agar, wild-type cells in MM at a concentration of approximately 5 × 108 cells per ml were mixed with an equal volume of anti-SprB serum or preimmune serum, and 2-μl samples were spotted on MM agar-coated slides. The spots were allowed to dry for 30 min at 25°C and were then covered with an oxygen-permeable Teflon membrane (Yellow Springs Instrument Co.). After incubation for an additional 45 min at 25°C, images were recorded. To determine the effect of added antiserum on movement of cells over glass, 100 μl of cells in MM was introduced into a Palmer counting cell and incubated for 2 min at 25°C to allow cells to attach to the cover glass. The medium and unattached cells were wicked away with a Kimwipe, and 50 μl of MM and 50 μl of either preimmune serum or antiserum against SprB were added. Cells were incubated for 2 min at 25°C, and the medium containing unbound antiserum was removed and replaced with 75 μl of MM. Cells were then incubated for an additional 2 min at 25°C before observation.
Binding and movement of protein G-coated polystyrene spheres.
Purified anti-SprB (1 μl of a 1:10 dilution of 300 mg/liter stock), 0.5-μm-diameter protein G-coated polystyrene spheres (1 μl of a 0.1% stock preparation; Spherotech Inc., Libertyville, IL), and bovine serum albumin (BSA) (1 μl of a 1% solution) were added to 7 μl of cells (approximately 5 × 108 cells per ml) in MM. The cells were spotted on a glass slide and covered with a glass coverslip, and images were recorded and analyzed using MetaMorph software.
Detection and localization of SprB.
Antisera to SprB were used to detect SprB in cell extracts. Overnight cultures of F. johnsoniae were grown in MM at 25°C without shaking. Cells were pelleted by centrifugation at 4,000 × g for 15 min and suspended in 20 mM sodium phosphate (pH 7.4). Cells were disrupted with a French press and fractionated into soluble and membrane fractions as described previously (16), except that Halt protease inhibitor (Pierce, Rockford, IL) was added to cell extracts. SprB in spent culture medium was concentrated by precipitation with trichloroacetic acid. Briefly, 1 volume of 100% (wt/vol) trichloroacetic acid was added to 4 volumes of spent medium, the mixture was incubated 10 min at 4°C, and the precipitate was collected by centrifugation at 18,400 × g for 5 min, washed twice with cold acetone, dried, and suspended in SDS-PAGE loading buffer. Proteins were separated on 3 to 8% Criterion XT Tris-acetate acrylamide gels (Bio-Rad, Hercules, CA), and Western blot analyses were performed as described previously (18), except that antigens were detected using Opti-4CN (Bio-Rad, Hercules, CA).
Localization of SprB by immunoelectron microscopy.
Cells were grown in MM in static cultures at 25°C to a density of approximately 5 × 108 cells per ml. Cells were settled onto Formvar-coated 400-mesh Ni grids which had been exposed to UV light for 30 s using an Ultra-Lum UV cross-linker to reduce the hydrophobicity. Cells were allowed to adsorb to the grids for 5 min and were fixed for 15 min at 25°C in 1% formaldehyde. Fixed cells were washed three times with 25 mM sodium phosphate (pH 7.5)-100 mM NaCl (PBS), and the grids were blocked with 100 μl PBS containing 1% BSA for 30 min. Samples were washed three times with PBS and exposed to affinity-purified anti-SprB polyclonal antiserum (1:100 dilution) in PBS with 1% BSA at 25°C for 1 h. Samples were washed three times in PBS and incubated with gold-labeled goat anti-rabbit immunoglobulin G (1:10 dilution; Sigma Chemical Co., St. Louis, MO) in PBS with 1% BSA for 1 h at 25°C. Cells were washed three times with PBS, dried, and examined using a Hitachi H-600 transmission electron microscope at 75 kV.
Measurement of bacteriophage sensitivity.
Sensitivity to F. johnsoniae bacteriophages φCj1, φCj13, φCj23, φCj28, φCj29, φCj42, φCj48, and φCj54 (9, 35, 39) was determined essentially as previously described by spotting 3 μl of phage lysate (109 PFU/ml) onto lawns of cells in CYE overlay agar (18), except that the cells used to initiate the lawns were obtained from static overnight cultures in MM. The plates were incubated for 24 h at 25°C to observe lysis.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been deposited in the GenBank database under accession no. EF111026.
RESULTS AND DISCUSSION
To identify redundant cell surface components of the motility machinery, Tn4351, HimarEm1, and HimarEm2 induced mutants with partial motility defects were isolated as described previously (5, 16-18). These mutants formed nonspreading colonies on agar that were indistinguishable from those of completely nonmotile gld mutants, but individual cells exhibited some ability to glide on glass in wet mounts. The sites of transposon insertions were determined for 37 randomly selected motile nonspreading mutants as previously described (5, 16, 19, 33). Six mutants had insertions in sprA (31), and two had insertions in secDF (32). Of the remaining 29 mutants, 16 had insertions in a 19.5-kbp gene that we designated sprB (Fig. 1).
The sprB mutants were defective in gliding on agar and formed completely nonspreading colonies, in contrast to the thin spreading colonies produced by wild-type cells (Fig. 2). Cells of sprB mutants were also partially defective in movement on glass (Fig. 3). Unlike cells with mutations in sprA, which displayed a defect in initial binding to glass (31), sprB mutants attached to glass as well as wild-type cells (Fig. 3 and data not shown). Introduction of pSN60, which carries wild-type sprB, into the transposon-induced mutants restored the ability to form spreading colonies (Fig. 2) and resulted in cells that moved as rapidly as wild-type cells on glass (Fig. 3). In addition, pSN60 complemented 4 of 37 spontaneous or chemically induced “motile nonspreading” mutants (UW102-24, UW102-45, UW102-128, and UW102-298) isolated by Pate and colleagues (9, 39), suggesting that these mutants also have mutations in sprB. In addition to the defects in motility described above, sprB mutants also displayed partial resistance to some bacteriophages (φCj1, φCj13, φCj23, and φCj29) that infect wild-type cells (Fig. 4). SprB may be a receptor for these bacteriophages.
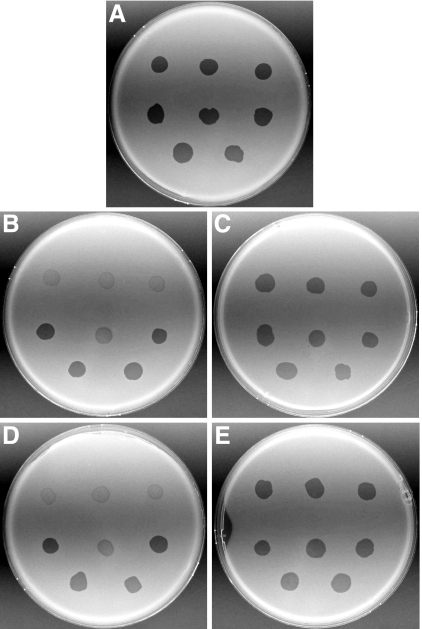
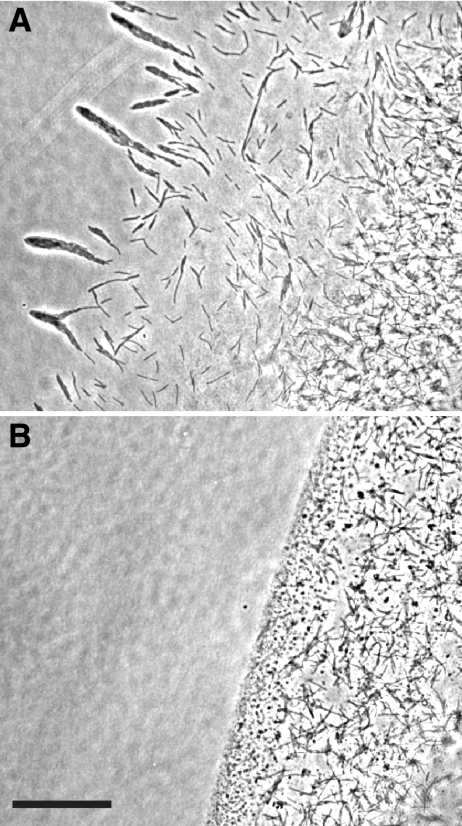
FIG. 2.
Photomicrographs of F. johnsoniae colonies. Colonies of wild-type F. johnsoniae FJ1 (A), sprB mutant FJ156 (B), FJ156 complemented with pSN60 (C), sprB mutant FJ117 (D), and FJ117 complemented with pSN60 (E) were grown for 48 h at 25°C on PY2 agar medium. Photomicrographs were taken with a Photometrics CoolSNAPcf2 camera mounted on an Olympus IMT-2 phase-contrast microscope. Bar in panel E = 0.5 mm (applies to all panels).
FIG. 3.
Effect of mutations in sprB on gliding of cells on glass. Cells attached to a glass coverslip were observed by phase-contrast microscopy, and digital images of cells of the wild-type strain (A), sprB mutant FJ156 (B), sprB mutant FJ117 (C), FJ156 complemented with pSN60 (D), and FJ117 complemented with pSN60 (E) were obtained at time zero. Tracks illustrating the movements of the cells shown in panels A to E over a 106-s period were obtained by superimposing individual digital video frames of wild-type strain FJ1 (F), sprB mutant FJ156 (G), sprB mutant FJ117 (H), FJ156 complemented with pSN60 (I), and FJ117 complemented with pSN60 (J). Images were recorded using a Photometrics CoolSNAPcf2 camera mounted on an Olympus BH-2 phase-contrast microscope. Bar in panel E = 75 μm (applies to all panels).
FIG. 4.
Effect of mutations in sprB on bacteriophage resistance. Bacteriophages (3 μl of lysate containing approximately 109 PFU/ml) were spotted onto lawns of cells in CYE overlay agar. The plates were incubated at 25°C for 24 h to observe lysis. Bacteriophages were spotted in the following order from left to right: top row, φCj1, φCj13, and φCj23; middle row, φCj28, φCj29, and φCj42; bottom row, φCj48 and φCj54. (A) Wild type F. johnsoniae FJ1. (B) sprB mutant FJ156. (C) FJ156 complemented with pSN60, which carries sprB. (D) sprB mutant FJ117. (E) FJ117 complemented with pSN60.
sprB encodes a primary product consisting of 6,497 amino acids with a predicted 24-amino-acid cleavable signal peptide. The processed protein is predicted to have a molecular mass of 669 kDa and an extensive β-sheet structure. The sequences of both the gene and the protein are highly repetitive. A 567-amino-acid sequence starting at proline 1230 is present in three tandemly repeated copies that exhibit 92% amino acid identity, and a 594-amino-acid sequence starting at lysine 3444 is present in three tandem copies that exhibit 99% amino acid identity. Short repeated sequences are also abundant within the large repeats and throughout most of the remainder of the protein sequence. The sequence of SprB is similar to the sequences of large hypothetical proteins from other members of the phylum Bacteroidetes. This protein also exhibits lower but potentially significant similarity to mouse mucin 19 (BLASTP e value, 5e-113; 21% amino acid identity over 5,111 amino acids) (10), the Pseudomonas fluorescens adhesin LapA (BLASTP e value, 1e-88; 21% amino acid identity over 4,987 amino acids) (14), and the Synechococcus sp. strain WH 8102 motility protein SwmB (BLASTP e value, 2e-26; 20% amino acid identity over 4,442 amino acids) (29). These proteins are all large, repetitive, cell surface or extracellular proteins. The F. johnsoniae genome contains multiple paralogs of sprB. The protein encoded by the most similar of these paralogs exhibits 28% identity with SprB over 2,205 amino acids. The presence of paralogs may explain why mutations in sprB do not completely eliminate motility. Strains with mutations in sprB and in one of the related genes have much more severe motility defects than strains with either single mutation, suggesting that the protein products have partially redundant roles in motility (S. Pochiraju and M. J. McBride, unpublished).
Antisera raised against recombinant SprB were used to detect SprB in intact cells of F. johnsoniae by immunoelectron microscopy. SprB was found to be unevenly distributed on the surface of cells (Fig. 5). In many cases bands of SprB appeared to be wrapped around the outside of the cell. This distribution of SprB on the cell surface is reminiscent of the organization of GldJ in the periplasm (6). GldJ and the other periplasmic Gld proteins may form a scaffold on which the cell surface SprB is mounted. Patches of cell surface filaments on gliding F. johnsoniae cells have recently been reported (22). The composition of these filaments is not known, but the similar distribution of filaments and of SprB on the cell surface suggests that they may be related.
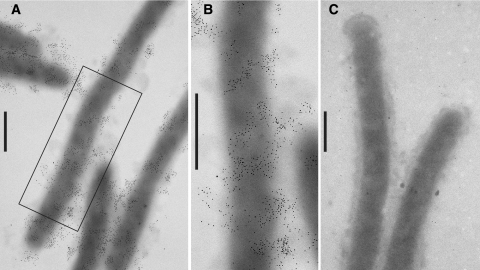
FIG. 5.
Localization of SprB by immunoelectron microscopy. Cells were fixed with 1% formaldehyde for 15 min, and SprB was detected by transmission electron microscopy using affinity-purified antiserum and gold-labeled secondary antibodies. Bars = 0.5 μm. (A) Cells of wild-type F. johnsoniae FJ1. (B) Higher magnification of a wild-type cell in panel A. (C) Cells of sprB mutant FJ156.
Western blot analysis revealed an extremely large protein corresponding to SprB that was present in wild-type cells but not in cells of the sprB mutant FJ156 (Fig. 6A, lanes 2 and 3). Numerous shorter fragments of SprB were also observed and may be functionally important. Most of the SprB protein was cell associated, and only small amounts were found in the spent medium (Fig. 6B, lane 5). SprB in cell extracts was soluble and was not pelleted by centrifugation at 223,000 × g for 60 min, suggesting that it was not an integral membrane protein. SprB may interact with other components of the motility apparatus, and cell disruption may cause the machinery to dissociate, releasing soluble SprB. Although the largest form of SprB was easily detected when cells were disrupted with a French press, it was less apparent when intact cells were solubilized with SDS without mechanical disruption (Fig. 6A, lane 1). Full-length SprB may have been entangled with some other large component, such as the peptidoglycan layer, which likely remained intact following solubilization with SDS and could have impeded entry of the protein into a polyacrylamide gel. The C-terminal 34 amino acids of SprB appear to be needed for function. The FJ117 mutant has a transposon insertion that results in the production of truncated SprB missing this region (Fig. 1). Cells of FJ117 exhibit the same motility defects as other sprB mutants (Fig. 2 and 3) despite production of nearly full-length protein (Fig. 6A, lane 4). Strains with mutations in other genes involved in gliding were examined for the presence of the SprB protein. SprB was present in cells of each of the gld mutants, but cells with a mutation in secDF (32) had no detectable SprB (Fig. 7). SecDF may play a role in secretion of large cell surface proteins, such as SprB, and the mislocalized SprB in secDF mutants may be subject to degradation.
FIG. 6.
Immunodetection and localization of SprB. (A) Total cell extracts examined for SprB by SDS-PAGE and Western blot analysis. The extract in lane 1 was prepared by boiling cells in SDS-PAGE loading buffer, and the extracts in lanes 2 to 6 were prepared by disruption of cells by passage through a French pressure cell, followed by boiling of the extract in SDS loading buffer. Lanes 1 and 2, wild-type F. johnsoniae FJ1; lane 3, sprB mutant FJ156; lane 4, sprB mutant FJ117; lane 5, FJ156 complemented with pSN60; lane 6, FJ117 complemented with pSN60. The arrow indicates the position of the highest-molecular-weight form of SprB. Fifty micrograms of protein was loaded in each lane. (B) Detection of SprB in cell fractions by Western blot analysis. Lane 1, whole-cell extract of wild-type strain FJ1; lane 2, whole-cell extract of sprB mutant FJ156; lane 3, soluble fraction of FJ1; lane 4, insoluble (membrane and particulate) fraction of FJ1; lane 5, concentrated spent growth medium of FJ1. The arrow indicates the position of the highest-molecular-weight form of SprB. Equal amounts, corresponding to 30 μg of cell protein of starting material, were loaded in the lanes.
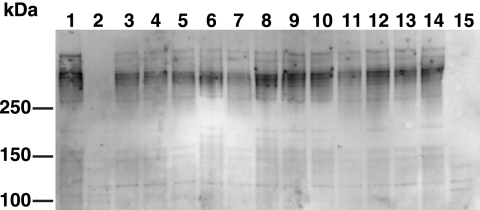
FIG. 7.
Effect of mutations in motility genes on SprB levels. Cells were lysed by passage through a French press, and proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. SprB was detected using antisera against the recombinant protein. Lane 1, wild-type F. johnsoniae FJ1; lane 2, sprB mutant FJ156; lane 3, gldA mutant CJ101-288; lane 4, gldB mutant CJ569; lane 5, gldD mutant CJ282; lane 6, gldF mutant UW102-77; lane 7, gldH mutant CJ1043; lane 8, gldI mutant UW102-41; lane 9, gldJ mutant UW102-48; lane 10, gldK mutant UW102-57; lane 11, gldL mutant CJ1300; lane 12, gldM mutant FJ113; lane 13, gldN mutant CJ1304; lane 14, sprA mutant FJ118; lane 15, secDF mutant CJ974. Thirty micrograms of protein was loaded in each lane.
Antibodies against SprB were used to probe the role of SprB in motility. Addition of antiserum to wild-type cells inhibited movement on agar and glass (Fig. 8; see Movies S1, S2, and S3 in the supplemental material). As expected, antisera against SprB had no effect on the residual motility of cells of the SprB mutant FJ156 on glass (data not shown). Protein G-coated polystyrene spheres were used to monitor the movement of SprB on the cell surface. Protein G served two purposes; it bound specifically to antibodies, and it interfered with the nonspecific binding of polystyrene spheres to F. johnsoniae cells. BSA (0.1%) was added to further limit nonspecific binding. Under these conditions protein G-coated spheres failed to interact with wild-type cells (Fig. 9; see Movie S4 in the supplemental material). Addition of antibodies against SprB to protein G-coated spheres resulted in attachment of the spheres to cells and rapid propulsion along the cell surface (Fig. 9; see Movies S5 and S6 in the supplemental material). The spheres were often observed to move continuously along the length of the cell, around the pole, and back along the opposite side of the cell. They also often crossed the width of the cell, and occasionally their direction of movement was reversed. The paths followed by spheres were somewhat irregular rather than rigidly linear or helical. Often, multiple spheres were attached to a single cell, as shown in Fig. 9E. In such cases the spheres could travel independently, or they could follow identical paths. Spheres moved at a rate of approximately 2 μm per s, which is similar to the rate of cell movement. Spheres carrying antibodies against SprB failed to bind to cells of the sprB mutants FJ156 and FJ117 (Fig. 9; see Movie S7 in the supplemental material), and spheres carrying antibodies against other proteins, such as GldJ and SprA, did not bind to wild-type cells. The sphere movements on wild-type cells are similar to those described by previous investigators who used unlabeled polystyrene spheres adsorbed to cells of F. johnsoniae or related bacteria (20, 34, 36). The key differences are that the protein G and BSA prevented nonspecific binding and that the antibodies provided a specific interaction with a known protein on the cell surface (SprB). This makes it less likely that spheres are passed along the cell surface from adsorption site to adsorption site and suggests that SprB itself is propelled rapidly along the cell surface. Antibodies do not provide a permanent interaction, so it is still possible that antibody-coated spheres could be transferred from one molecule of SprB to another. However, it would be difficult to explain the observed rapid and smooth movement of spheres over long distances by such intermolecular transfer. It is also possible that spheres are propelled by some other cell surface component and that SprB, bound to the spheres, is carried along. This also seems unlikely since we did not observe such movement of protein G-coated spheres in the absence of antibodies against SprB.
FIG. 8.
Effect of antisera against SprB on cell movement over agar. Cells of wild-type F. johnsoniae FJ1 in MM at a density of approximately 5 × 108 cells per ml were mixed with an equal volume of preimmune serum (A) or anti-SprB (B). Aliquots (2 μl) were spotted on MM agar-coated slides and incubated for 75 min at 25°C, and images were recorded using a Photometrics CoolSNAPcf2 camera mounted on an Olympus BH-2 phase-contrast microscope. Bar = 50 μm.
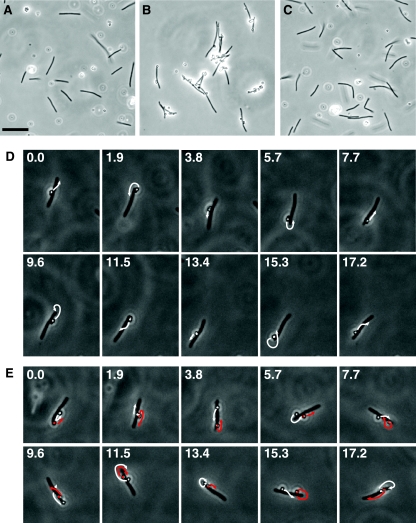
FIG. 9.
Polystyrene spheres coated with anti-SprB move rapidly along the cell surface. Protein G-coated 0.5-μm polystyrene spheres with or without anti-SprB antibodies were added to cells of F. johnsoniae, and images were recorded using a Photometrics CoolSNAPcf2 camera mounted on an Olympus BH-2 phase-contrast microscope. (A) Cells of wild-type F. johnsoniae FJ1 and polystyrene spheres without anti-SprB antibodies. (B) Cells of wild-type F. johnsoniae FJ1 and polystyrene spheres with anti-SprB antibodies. (C) Cells of sprB mutant FJ156 and polystyrene spheres with anti-SprB antibodies. Bar in panel A = 10 μm (applies to panels A to C). The movements of the cells and spheres in panels A, B, and C are documented in Movies S4, S5, and S7 in the supplemental material, respectively. (D) Consecutive images of a cell with one attached sphere with anti-SprB antibodies. The numbers indicate time (in seconds). The arrows indicate the movement of the sphere during the next 1.9 s. The cell was 5.7 μm long. (E) Consecutive images of a cell with two attached spheres with anti-SprB antibodies. The numbers indicate time (in seconds). One sphere was artificially colored to allow easy identification. The arrows indicate the movement of the spheres during the next 1.9 s. The cell was 5.2 μm long. The cells in panels D and E were in the same field of view, and their movements are documented in Movie S6 in the supplemental material. Final concentrations of purified anti-SprB antibodies were 30 mg/liter for panels B and C and 3 mg/liter for panels D and E.
Numerous models have been proposed to explain gliding of F. johnsoniae and related bacteria (4, 8, 20, 24, 25, 34, 36). Our results support the model described by Lapidus and Berg more than 25 years ago (20), which postulates that machinery in the cell envelope propels cell surface proteins along tracks that are anchored to the cell wall. We suggest that Gld proteins in the cell envelope comprise the motors that convert chemical and/or electrochemical energy into movement of large adhesive proteins, such as SprB, along the cell surface. Other models for bacterial gliding, such as pilus extension and retraction (23), polysaccharide secretion (15, 38), coordinated export and import of adhesive macromolecules (25), generation of cell surface waves (8), contraction of periplasmic or cytoplasmic filaments resulting in inchworm-like movement (7), or the functioning of rotary motors (34), are not supported by the apparent continuous movement of SprB protein along the cell surface. The proposed model for Flavobacterium gliding has some similarities to the focal adhesion models described for Myxococcus xanthus adventurous gliding (30) and for gliding of protists, such as members of the genus Plasmodium (13). In each case “motor” proteins are thought to exert a force on cell surface adhesins and on a large cellular framework, such as the cytoskeleton or the peptidoglycan layer. The molecular components that constitute the motility machineries of F. johnsoniae, M. xanthus, and Plasmodium species are completely different and likely evolved independently. For Plasmodium cells, myosin motors are thought to walk along the actin cytoskeleton as they pull on cell surface adhesins. It would be difficult to explain F. johnsoniae motility by a comparable model since the peptidoglycan meshwork in the cell wall would break the connection between the motor and the adhesin at every step. Rather, we suggest that the F. johnsoniae gliding “motor” is embedded in the cytoplasmic membrane and anchored to the cell wall peptidoglycan. Adhesins, or perhaps groups of adhesins mounted on a large assembly, would be pushed along the cell surface and handed off to the next motor complex. Our results also suggest that there is redundancy in the cell surface components of the motility machinery. SprB is required for gliding on agar, and we suggest that some of the other SprB-like proteins of F. johnsoniae function as adhesive components of the motility machinery that allow cell movement over other surfaces. The ability of F. johnsoniae cells to attach different adhesins to their motility machinery may explain how they are able to move over diverse surfaces that have dramatically different chemical and physical properties.
Supplementary Material
Acknowledgments
This research was supported by grants MCB-0130967 and MCB-0641366 from the National Science Foundation.
We thank D. Saffarini for comments on the manuscript and H. Owen for assistance with electron microscopy.
Footnotes
Published ahead of print on 15 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Agarwal, S., D. W. Hunnicutt, and M. J. McBride. 1997. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc. Natl. Acad. Sci. USA 9412139-12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bardy, S. L., S. Y. M. Ng, and K. F. Jarrell. 2003. Prokaryotic motility structures. Microbiology 149295-304. [DOI] [PubMed] [Google Scholar]
- 4.Beatson, P. J., and K. C. Marshall. 1994. A proposed helical mechanism for gliding motility in three gliding bacteria (order Cytophagales). Can. J. Microbiol. 40173-183. [Google Scholar]
- 5.Braun, T. F., M. K. Khubbar, D. A. Saffarini, and M. J. McBride. 2005. Flavobacterium johnsoniae gliding motility genes identified by mariner mutagenesis. J. Bacteriol. 1876943-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, T. F., and M. J. McBride. 2005. Flavobacterium johnsoniae GldJ is a lipoprotein that is required for gliding motility. J. Bacteriol. 1872628-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burchard, R. P. 1982. Evidence for contractile flexing of the gliding bacterium Flexibacter FS-1. Nature 298663-665. [DOI] [PubMed] [Google Scholar]
- 8.Burchard, R. P. 1981. Gliding motility of prokaryotes: ultrastructure, physiology, and genetics. Annu. Rev. Microbiol. 35497-529. [DOI] [PubMed] [Google Scholar]
- 9.Chang, L. Y. E., J. L. Pate, and R. J. Betzig. 1984. Isolation and characterization of nonspreading mutants of the gliding bacterium Cytophaga johnsonae. J. Bacteriol. 15926-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Y., Y. H. Zhao, T. B. Kalaslavadi, E. Hamati, K. Nehrke, A. D. Le, D. K. Ann., and R. Wu. 2004. Genome-wide search and identification of a novel gel-forming mucin MUC19/Muc19 in glandular tissues. Am. J. Respir. Cell Mol. Biol. 30155-165. [DOI] [PubMed] [Google Scholar]
- 11.Chou, P. Y., and G. D. Fasman. 1978. Empirical predictions of protein conformation. Annu. Rev. Biochem. 47251-276. [DOI] [PubMed] [Google Scholar]
- 12.Garnier, J., D. J. Osguthorpe, and B. Robson. 1978. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J. Mol. Biol. 12097-120. [DOI] [PubMed] [Google Scholar]
- 13.Heintzelman, M. B. 2006. Cellular and molecular mechanics of gliding locomotion in eukaryotes. Int. Rev. Cytol. 25179-129. [DOI] [PubMed] [Google Scholar]
- 14.Hinsa, S. M., M. Espinosa-Urgel, J. L. Ramos, and G. A. O'Toole. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49905-918. [DOI] [PubMed] [Google Scholar]
- 15.Hoiczyk, E., and W. Baumeister. 1998. The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria. Curr. Biol. 81161-1168. [DOI] [PubMed] [Google Scholar]
- 16.Hunnicutt, D. W., M. J. Kempf, and M. J. McBride. 2002. Mutations in Flavobacterium johnsoniae gldF and gldG disrupt gliding motility and interfere with membrane localization of GldA. J. Bacteriol. 1842370-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunnicutt, D. W., and M. J. McBride. 2001. Cloning and characterization of the Flavobacterium johnsoniae gliding motility genes gldD and gldE. J. Bacteriol. 1834167-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunnicutt, D. W., and M. J. McBride. 2000. Cloning and characterization of the Flavobacterium johnsoniae gliding motility genes, gldB and gldC. J. Bacteriol. 182911-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kempf, M. J., and M. J. McBride. 2000. Transposon insertions in the Flavobacterium johnsoniae ftsX gene disrupt gliding motility and cell division. J. Bacteriol. 1821671-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapidus, I. R., and H. C. Berg. 1982. Gliding motility of Cytophaga sp. strain U67. J. Bacteriol. 151384-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, C., A. Levin, and D. Branton. 1987. Copper staining: a five minute protein stain for sodium dodecyl sulfate-polyacrylamide gels. Anal. Biochem. 166308-312. [DOI] [PubMed] [Google Scholar]
- 22.Liu, J., M. J. McBride, and S. Subramaniam. 2007. Cell-surface filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. J. Bacteriol. 1897503-7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56289-314. [DOI] [PubMed] [Google Scholar]
- 24.McBride, M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 5549-75. [DOI] [PubMed] [Google Scholar]
- 25.McBride, M. J. 2004. Cytophaga-flavobacterium gliding motility. J. Mol. Microbiol. Biotechnol. 763-71. [DOI] [PubMed] [Google Scholar]
- 26.McBride, M. J., and T. F. Braun. 2004. GldI is a lipoprotein that is required for Flavobacterium johnsoniae gliding motility and chitin utilization. J. Bacteriol. 1862295-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBride, M. J., T. F. Braun, and J. L. Brust. 2003. Flavobacterium johnsoniae GldH is a lipoprotein that is required for gliding motility and chitin utilization. J. Bacteriol. 1856648-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride, M. J., and M. J. Kempf. 1996. Development of techniques for the genetic manipulation of the gliding bacterium Cytophaga johnsonae. J. Bacteriol. 178583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarren, J., and B. Brahamsha. 2007. SwmB, a 1.12-megadalton protein that is required for nonflagellar swimming motility in Synechococcus. J. Bacteriol. 1891158-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mignot, T., J. W. Shaevitz, P. L. Hartzell, and D. R. Zusman. 2007. Evidence that focal adhesion complexes power bacterial gliding motility. Science 315853-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson, S. S., P. P. Glocka, S. Agarwal, D. P. Grimm, and M. J. McBride. 2007. Flavobacterium johnsoniae SprA is a cell surface protein involved in gliding motility. J. Bacteriol. 1897145-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson, S. S., and M. J. McBride. 2006. Mutations in Flavobacterium johnsoniae secDF result in defects in gliding motility and chitin utilization. J. Bacteriol. 188348-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pate, J. L., and L.-Y. E. Chang. 1979. Evidence that gliding motility in prokaryotic cells is driven by rotary assemblies in the cell envelopes. Curr. Microbiol. 259-64. [Google Scholar]
- 35.Pate, J. L., S. J. Petzold, and L.-Y. E. Chang. 1979. Phages for the gliding bacterium Cytophaga johnsonae that infect only motile cells. Curr. Microbiol. 2257-262. [Google Scholar]
- 36.Ridgway, H. F., and R. A. Lewin. 1988. Characterization of gliding motility in Flexibacter polymorphus. Cell Motil. Cytoskelet. 1146-63. [DOI] [PubMed] [Google Scholar]
- 37.Stanier, R. Y. 1947. Studies on non-fruiting myxobacteria. I. Cytophaga johnsonae, n. sp., a chitin-decomposing myxobacterium. J. Bacteriol. 53297-315. [PMC free article] [PubMed] [Google Scholar]
- 38.Wolgemuth, C., E. Hoiczyk, D. Kaiser, and G. Oster. 2002. How myxobacteria glide. Curr. Biol. 12369-377. [DOI] [PubMed] [Google Scholar]
- 39.Wolkin, R. H., and J. L. Pate. 1985. Selection for nonadherent or nonhydrophobic mutants co-selects for nonspreading mutants of Cytophaga johnsonae and other gliding bacteria. J. Gen. Microbiol. 131737-750. [Google Scholar]
- 40.Xie, G., D. C. Bruce, J. F. Challacombe, O. Chertkov, J. C. Detter, P. Gilna, C. S. Han, S. Lucas, M. Misra, G. L. Myers, P. Richardson, R. Tapia, N. Thayer, L. S. Thompson, T. S. Brettin, B. Henrissat, D. B. Wilson, and M. J. McBride. 2007. Genome sequence of the cellulolytic gliding bacterium Cytophaga hutchinsonii. Appl. Environ. Microbiol. 733536-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.