Abstract
Glycine betaine (GB), which occurs freely in the environment and is an intermediate in the catabolism of choline and carnitine, can serve as a sole source of carbon or nitrogen in Pseudomonas aeruginosa. Twelve mutants defective in growth on GB as the sole carbon source were identified through a genetic screen of a nonredundant PA14 transposon mutant library. Further growth experiments showed that strains with mutations in two genes, gbcA (PA5410) and gbcB (PA5411), were capable of growth on dimethylglycine (DMG), a catabolic product of GB, but not on GB itself. Subsequent nuclear magnetic resonance (NMR) experiments with 1,2-13C-labeled choline indicated that these genes are necessary for conversion of GB to DMG. Similar experiments showed that strains with mutations in the dgcAB (PA5398-PA5399) genes, which exhibit homology to genes that encode other enzymes with demethylase activity, are required for the conversion of DMG to sarcosine. Mutant analyses and 13C NMR studies also confirmed that the soxBDAG genes, predicted to encode a sarcosine oxidase, are required for sarcosine catabolism. Our screen also identified a predicted AraC family transcriptional regulator, encoded by gbdR (PA5380), that is required for growth on GB and DMG and for the induction of gbcA, gbcB, and dgcAB in response to GB or DMG. Mutants defective in the previously described gbt gene (PA3082) grew on GB with kinetics similar to those of the wild type in both the PAO1 and PA14 strain backgrounds. These studies provided important insight into both the mechanism and the regulation of the catabolism of GB in P. aeruginosa.
A number of microbes, including Pseudomonas aeruginosa, can utilize glycine betaine (GB) as a sole carbon, nitrogen, and energy source (17, 35, 41). GB, an important osmoprotectant for many bacteria (6), is available to organisms in a variety of environments (5, 14, 34, 41). Free GB can be released by roots (9), microbes (14, 15), or decaying animal (20) and plant (10) matter. Alternatively, GB can be derived from choline or carnitine (4, 5, 12, 15, 20, 36). Choline and carnitine can be found in many eukaryote-associated environments, and bacteria, including P. aeruginosa, can use phospholipases and choline phosphatases to release choline from phosphatidylcholine (30, 38). In P. aeruginosa, choline is oxidized to GB by a two-step process catalyzed by BetA and BetB (29, 36), while carnitine is predicted to be reduced and deacetylated by uncharacterized enzymes, ultimately yielding GB (16).
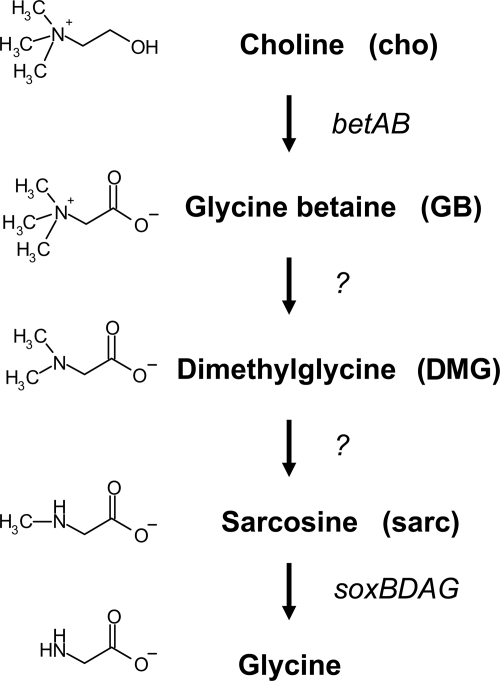
The aerobic catabolism of GB in bacteria is best understood in Sinohizobium (35), Corynebacterium (8, 37), and Arthrobacter species (24). The data from these studies suggest that GB catabolism occurs via serial demethylation that forms dimethylglycine (DMG), then sarcosine (also called monomethylglycine), and finally glycine (Fig. 1). Thin-layer chromatographic analyses indicated that in P. aeruginosa DMG and sarcosine are also intermediates formed during GB catabolism (11). Furthermore, in the same study, a proteomics analysis of P. aeruginosa cultures grown on GB showed that higher levels of the putative sarcosine oxidase subunits are present during growth on GB (11).
FIG. 1.
Predicted pathway for choline catabolism in P. aeruginosa (5, 11).
Some enzymes involved in GB and DMG catabolism have been characterized in different bacterial species. For example, the demethylation of GB to form DMG has been hypothesized to occur via a betaine homocysteine methyltransferase in Sinorhizobium meliloti and Pseudomonas denitrificans based on measurements of enzyme activity in cell extracts (35, 43). In Arthrobacter globiformis, DMG demethylation to sarcosine is catalyzed by a DMG oxidase with similarity to eukaryotic DMG dehydrogenases (24). In Corynebacterium (8, 37) and Arthrobacter (24), a heterotetrameric sarcosine oxidase converts sarcosine into glycine. However, the complete gene set required for conversion of GB to glycine has not been described in any bacterium, nor is there any information about how the genes are regulated at the molecular level.
Using a screen of a nonredundant P. aeruginosa strain PA14 transposon mutant library for mutants unable to grow on GB, we identified genes required for growth on GB and DMG that share no obvious homology to other genes known to be involved in the catabolism of these two compounds (21). Growth experiments, nuclear magnetic resonance (NMR) studies, and reverse transcription-PCR (RT-PCR) analyses led to identification of a gene cluster encoding a putative GB demethylase. In addition, we identified an operon encoding a putative DMG demethylase that is more similar to a bacterial N-methylproline demethylase than to the only verified bacterial DMG demethylase from Arthrobacter (24). We identified a transcription factor which regulates these putative catabolic genes in response to GB and DMG and thus described the first transcriptional regulator of GB catabolism in bacteria. Finally, we confirmed the role of the soxBDAG genes in the conversion of sarcosine to glycine in P. aeruginosa.
MATERIALS AND METHODS
Strains, media, and growth conditions.
P. aeruginosa PA14 and PAO1 wild-type strains, transposon mutants, and deletion strains, as well as Escherichia coli strains (Table 1) were maintained on LB medium. For experiments with single carbon sources, morpholinepropanesulfonic acid (MOPS) minimal medium (25) containing a specified carbon source at a concentration of 20 mM was used unless otherwise noted. When necessary, gentamicin was added to a final concentration of 15 μg/ml for E. coli in liquid, to a final concentration of 20 μg/ml for E. coli on plates, and to a final concentration of 60 μg/ml for P. aeruginosa.
TABLE 1.
Strains and plasmids
| Strain, genotype, or plasmid | Proposed gene designationa | Strain | Description |
|---|---|---|---|
| P. aeruginosa strains | |||
| PA14 | DH122 | P. aeruginosa wild typeb | |
| ΔPA5380 | ΔgbdR | DH466 | In-frame PA5380 deletion in PA14 |
| ΔPA5410-PA5411 | ΔgbcA-gbcB | DH802 | Deletion of PA5410 and PA5411 in PA14 |
| ΔPA5398 | ΔdgcA | DH848 | In-frame PA5398 deletion in PA14 |
| ΔPA5380 | ΔgbdR-pGbdR | DH478 | DH466 with pGbdR, Gmr |
| ΔPA5410-PA5411 | ΔgbcAB-pGbcAB | DH804 | DH802 with pGbcAB, Gmr |
| betA::TnM | DH846 | Mutant 40757c | |
| PA5380::TnM | DH662 | Mutant 24457c | |
| PA5396::TnM | DH624 | Mutant 40467c | |
| PA5397::TnM | DH628 | Mutant 35214c | |
| PA5398::TnM | DH636 | Mutant 48591c | |
| PA5399::TnM | DH679 | Mutant 32077c | |
| PA5410::TnM | DH641 | Mutant 53996c | |
| PA5411::TnM | DH667 | Mutant 26808c | |
| soxA::TnM | DH647 | Mutant 56207c | |
| ΔPA5380 | ΔgbdR | DH543 | In-frame PA5380 deletion in PAO1 |
| ΔPA5410-PA5411 | ΔgbcA-gbcB | DH841 | Deletion of PA5410 and PA5411 in PAO1 |
| PA3082::TnM | DH969 | Mutant 4673c | |
| PA3082::TnM | DH970 | Mutant 30085c | |
| PA3082::TnM | DH971 | Mutant 41423c | |
| PA3082::ISlacZ/hah | DH501 | Mutant 8922 in P. aeruginosa PAO1d | |
| ΔPA3082 | DH972 | In-frame PA3082 deletion in PA14 | |
| E. coli strains | |||
| S17/l-pir | DH522 | ||
| Ec-PA5380KO | DH540 | DH522 with pPA5380KO, Gmr | |
| Ec-PA5410-5411KO | DH791 | DH522 with pPA5410-5411KO, Gmr | |
| Ec-PA5398KO | DH843 | DH522 with pPA5398KO, Gmr | |
| Ec-PA3082KO | DH973 | DH522 with pPA3082KO, Gmr | |
| Plasmids | |||
| pUCP22 | Gmr, contains P. aeruginosa stabilization fragment | ||
| pEX18-Gm | Gmr, integrating vector in P. aeruginosa | ||
| pGbdR | PA5380 coding region in frame with the N terminus of lacZa in pUCP22 | ||
| pGbcAB | PA5410-PA5411 genomic region in pUCP22 | ||
| pPA5380KO | PA5380 deletion construct in pEX18-Gm | ||
| pPA5410-5411KO | PA5410-PA5411 deletion construct in pEX18-Gm | ||
| pPA5398KO | PA5398 deletion construct in pEX18-Gm | ||
| pPA3082KO | PA3082 deletion construct in pMQ30 |
Proposed designations for genes based on the data presented in this paper.
See reference 27.
See the website of F. M. Ausubel for more information (http://ausubellab.mgh.harvard.edu/cgi-bin/pa14/home.cgi).
PAO1 transposon mutants were obtained from the University of Washington Genome Center.
Genetic screen.
The PA14 nonredundant transposon mutant library was replicated as described by Liberati et al. (21). Using frozen stocks, strains were replicated onto LB agar plates with a 48-pin replicator and incubated overnight at 37°C. Mutants were then transferred onto MOPS medium plates containing GB and incubated for 24 h at 37°C. Strains that were defective in growth on MOPS medium containing GB were recovered from the LB agar master plates and arrayed manually on 48-spot LB agar plates. After overnight growth, mutants were transferred to MOPS agar plates with either 20 mM glucose, 20 mM pyruvate, 20 mM choline, 20 mM GB, or 20 mM DMG and incubated at 37°C for 24 h. Transposon insertion sites were confirmed using gene-specific primers and the PMFLGM.GB-4a primer described by Liberati et al. (21).
Growth assays.
Growth assays were conducted in 96-well polystyrene plates using a Spectromax plate reader (Molecular Devices). To obtain carbon source growth curves, cells were grown overnight in MOPS medium with 20 mM pyruvate and 5 mM glucose. The overnight cultures were pelleted, washed, and inoculated into MOPS medium with the appropriate carbon source to an initial A600 of 0.05. Plates were shaken at 37°C, and growth was measured by determining the optical density (A600) every hour. To test whether different compounds could serve as sole sources of nitrogen, growth experiments were performed in a similar manner, except that M63 medium containing 0.2% glucose but lacking ammonium chloride (3) and amended with 20 mM choline, 20 mM lysine, 20 mM GB, 20 mM DMG, or 20 mM sarcosine was used. Mutants defective in PA3082 were grown on the media described above, as well as in the minimal medium described by Serra et al. (33), and compared to the wild type.
Construction of deletion strains and complementation constructs.
All gene numbers in this paper are the numbers for the PAO1 genome. The genes described here had at least 99% identity at both the nucleotide and amino acid levels when PAO1 and PA14 sequences were compared. In addition, the genomic region from PA5380 to PA5411 is organized in the same manner in both genomes.
Deletion constructs for PA5380, PA5410-PA5411, PA3082, and PA5398 were obtained using the pEX18-Gm plasmid, and the mutations in P. aeruginosa were obtained by recombination as described previously (32). Briefly, pEX18-Gm constructs were transformed into E. coli S17/λpir. E. coli was mated with the recipient P. aeruginosa strain, and single-crossover mutants were selected by growth on gentamicin. After selection for strains in which double-crossover events had occurred by growth on LB medium plates containing 5% sucrose with no NaCl, in-frame deletion mutants were identified by PCR. The primers used for construction of the deletion constructs are listed in Table S1 in the supplemental material. The nucleotides corresponding to the region from amino acid 8 to amino acid 361 were deleted to construct the PA5380 deletion strain, and the region from amino acid 12 to amino acid 657 was deleted to construct the PA5398 deletion strain. The PA5410-PA5411 deletion eliminated the start codons, as they are divergently transcribed, and the majority of the coding regions of both genes. The PA3082 deletion eliminated the region from amino acid 7 to amino acid 648. Complementing plasmids were constructed using the pUCP22 vector (42). The PA5410-PA5411 region was amplified using primers 5410-5411For_resc (5′-CACAGGGGATTGTTTTCCAC-3′) and 5410-5411Rev_resc (5′-GAGTACCCGTGCTTCGACA-3′), cloned into PCR2.1 (Invitrogen), excised from PCR2.1 using EcoRI, and ligated into pUCP22 treated with EcoRI and shrimp alkaline phosphatase (Roche). For PA5380, the gene was amplified using primers PA5380_coverstart_EcoRI (5′-ATAGGAATTCTACACCCATGACCACGTACG-3′) and PA5380_C-term_Rev (5′-GTTAAAGCTTGATCCGCACGCTGGCGAAGGTCGACTCG-3′), cloned into PCR2.1, excised from PCR2.1 with SacI and EcoRV, and ligated into pUCP22 cut with SacI and SmaI, forming an in-frame fusion with the first seven amino acids of the N terminus of the β-galactosidase alpha fragment.
RNA isolation, RT-PCR, and real time RT-PCR.
For gene induction experiments, cells were grown overnight in MOPS medium with 20 mM pyruvate and 5 mM glucose. Cells were harvested by centrifugation, resuspended in MOPS medium with 20 mM pyruvate and the inducing carbon source at a concentration of 10 mM, and grown for 2 h at 37°C. RNA was isolated from ∼107 cells using an RNeasy kit (Qiagen). Pyruvate was used as a growth substrate for a strain that was not capable of growth on GB without detectable catabolite repression (11; data not shown). RNA was treated with 2 U of RQ1 DNase (Promega) for 1 h at 37°C, followed by a second purification using the RNeasy kit. A final DNase step using a DNA-free kit (Ambion) was performed. The resulting RNA was subjected to PCR to verify the absence of contaminating DNA before it was quantified using a Nanodrop spectrophotometer. cDNA was synthesized using SuperScript III (Invitrogen), 250 ng of starting RNA, and a 5′-NSNSNSNSNS-3′ primer instead of random hexamers. The primers used are shown in Table S1 in the supplemental material. The PCR regimen was 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 55°C for 40 s, and 72°C for 30 s. Quantitative real-time RT-PCR was performed with SYBR green and AmpliTaq Gold DNA polymerase used according to the manufacturer's instructions (Applied Biosystems). The amplification conditions with an Applied Biosystems 7500 instrument were 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 30 s. Only one PCR product was obtained for all primers and all samples based on melting curve analysis.
13C NMR analysis.
For NMR analysis, cells were grown overnight in MOPS medium with 20 mM pyruvate and 5 mM choline. These cells were resuspended in fresh MOPS medium with 7 mM [1,2-13C]choline (Cambridge Isotope Laboratories) and grown for the specified period of time (3 or 9 h). Cell pellets were extracted twice with 80% ethanol at room temperature. The extracts were lyophilized and resuspended in 200 μl of 50 mM phosphate buffer (pH 7.25) that included 20% D2O in 5-mm NMR tubes. 13C NMR spectra were acquired with a Varian Unity 500 spectrometer at 125.69 MHz using 60° pulses and on average 20,000 to 40,000 free induction decays. Identification of the peaks for choline and its metabolites was based on a comparison of the experimental spectra with a database of chemical shifts compiled from NMR analyses of authentic standards. The two observable peaks for choline corresponding to the C-O carbon and the C-N carbon are at 68.33 and 56.63 ppm, respectively. The C-N peaks for GB, DMG, and sarcosine are at 67.00, 60.74, and 51.75 ppm, respectively.
RESULTS
Genetic screen of the PA14 transposon mutant library for mutants defective in growth on GB.
To identify the genes required for growth on GB, the P. aeruginosa PA14 nonredundant library, which contains 5,459 mutants with transposon insertions in 4,596 predicted genes (21), was screened to find mutants that lacked the ability to grow in minimal medium with GB as the sole carbon source.
Because the initial screen identified auxotrophs in addition to mutants defective in GB catabolism, we performed secondary screens to identify mutants capable of growth on minimal media containing glucose or pyruvate but not on medium with GB as the sole carbon source. Mutants that could not grow on pyruvate and glucose were omitted from the subsequent analyses. The mutant strains with growth defects on GB were also analyzed to determine their growth on choline, GB, and DMG in order to sort mutants into classes based on their apparent catabolic defects. Growth on sarcosine or glycine was not tested because P. aeruginosa grows poorly on these carbon sources on agar plates. The 12 mutants with growth defects on GB could be separated into two classes. Class I mutant strains were unable to grow on choline or GB but could grow on DMG (2 genes), and class II mutant strains were unable to grow on choline, GB, or DMG (10 genes). In all cases, the location of the transposon insertion was confirmed by PCR using gene-specific primers in combination with a primer located in the transposon.
The class I strains, which were defective in growth on choline and GB but not on DMG, included two mutant strains, each defective in one of two adjacent, divergently transcribed genes, PA14_71410 (corresponding to PA5410) and PA14_71420 (corresponding to PA5411).
The 10 mutants in class II were unable to grow on choline, GB, or DMG, suggesting that they were defective at or downstream of the conversion of DMG to sarcosine. As predicted, mutants with transposon insertions in the putative sarcosine catabolic genes (soxABDG) and a putative glycine catabolic gene (glyA1) were members of this class. These data provide functional evidence that supports the hypothesis that the sox genes in P. aeruginosa, designated based on their strong homology to the sarcosine oxidase system in Corynebacterium (8), are required for sarcosine catabolism in P. aeruginosa (11). In addition, we identified mutants with transposon insertions in a predicted operon, PA14_71240-PA14_71280 (corresponding to PA5396-PA5399). Finally, a mutant defective in a gene encoding an AraC family transcription factor, PA14_71070 (corresponding to PA5380), was found to be incapable of growth on any of the GB-related substrates (choline, GB, or DMG).
Analysis of the role of PA5410 and PA5411 in GB catabolism.
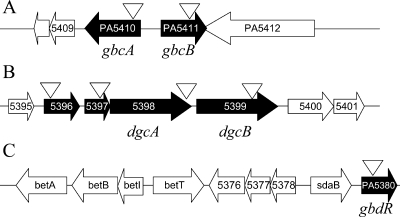
Two transposon mutant strains were categorized as class I GB catabolic mutants based on their inability to grow on choline or GB, but they were able to grow on DMG (data not shown). This growth phenotype suggested a role for the gene products in the catabolism of GB to DMG. To obtain insight into the roles of PA5410 and PA5411, we performed BLASTP analysis (1). The PA5410 gene encodes a protein with homology to the large subunit of numerous predicted and validated bacterial hydroxylating dioxygenases. The PA5411 gene encodes a probable flavin adenine dinucleotide-binding ferrodoxin reductase based on similarity to a number of predicted gram-negative proteins and COG analysis, although the closest homologue with a known function is PaaK in Pseudomonas fluorescens (46% similarity and 29% identity; accession number ABF82243). PA5410 and PA5411 are adjacent and divergently transcribed (Fig. 2A), a common arrangement of the dioxygenase gene and associated ferrodoxin reductase gene for many of the predicted homologues (http://cmr.tigr.org/).
FIG. 2.
Genomic arrangement of the genes involved in GB catabolism. Genes identified in the screen are indicated by filled arrows, and the transposon insertion sites are indicated by open triangles. (A) Genomic region surrounding PA5410 and PA5411. (B) Genomic region including the PA5396-PA5399 putative operon. (C) Genes in the vicinity of PA5380.
Transposon insertion into either PA5410 or PA5411 resulted in an inability of the strains to grow on choline or GB as a sole carbon source. To confirm that this growth defect was due to transposon insertion into these open reading frames (ORFs), we generated a deletion of both reading frames and the intervening intragenic sequence in P. aeruginosa PA14. Consistent with the phenotypes of the transposon mutants, the P. aeruginosa ΔPA5410-PA5411 strain was defective in growth on choline or GB but could grow on DMG (Table 2). When the ΔPA5410-PA5411 strain was grown in minimal medium with glucose or pyruvate as the sole carbon source, its growth rate was indistinguishable from that of PA14 (Table 2 and data not shown). Complementation with the PA5410 and PA5411 genes on a plasmid (pGbcAB) under control of their native promoters partially restored the ability to grow on choline and GB (Table 2). To test whether these genes were necessary for growth using choline or GB as a sole source of nitrogen, the ΔPA5410-PA5411 mutant was grown in minimal medium with glucose as the carbon source and lysine, choline, GB, DMG, or sarcosine as the only source of nitrogen. These analyses showed that the PA5410 and PA5411 genes are also necessary for the utilization of choline and GB as sole nitrogen sources but are not required for the utilization DMG, sarcosine, or lysine (data not shown). Deletion of PA5410 and PA5411 in P. aeruginosa strain PAO1 also abrogated the ability to grow on GB as a sole source of carbon (data not shown).
TABLE 2.
Doubling times of wild-type and mutant strains grown with glucose, choline, GB, or DMG as the sole source of carbon
| Strain or genotype | Proposed designationa | Plasmid | Doubling time (h) with:
|
|||
|---|---|---|---|---|---|---|
| Glucose | Choline | GB | DMG | |||
| PA14 | 0.91 (0.01)b | 2.72 (0.01) | 2.57 (0.12) | 2.07 (0.09) | ||
| PA14 | pUCP22 | 1.19 (0.13) | 2.99 (0.29) | 4.75 (0.35) | 5.13 (0.31) | |
| ΔPA5410-PA5411 | ΔgbcA-gbcB | pUCP22 | 1.02 (0.01) | NGc | NG | 6.51 (0.38) |
| ΔPA5410-PA5411 | ΔgbcA-gbcB | pGbcAB | 1.29 (0.01) | 3.52 (0.03) | 7.49 (0.20) | 4.64 (0.32) |
| ΔPA5398 | ΔdgcA | 1.26 (0.02) | NG | NG | NG | |
| PA5399:TnM | dgcB::TnM | 1.06 (0.07) | NG | NG | NG | |
| ΔPA5380 | ΔgbdR | pUCP22 | 1.44 (0.45) | NG | NG | NG |
| ΔPA5380 | ΔgbdR | pGbdR | 1.10 (0.09) | 2.81 (0.83) | 7.36 (1.59) | 7.53 (0.21) |
Proposed designations based on data presented in this paper.
The values in parentheses are standard deviations.
NG, no detectable growth.
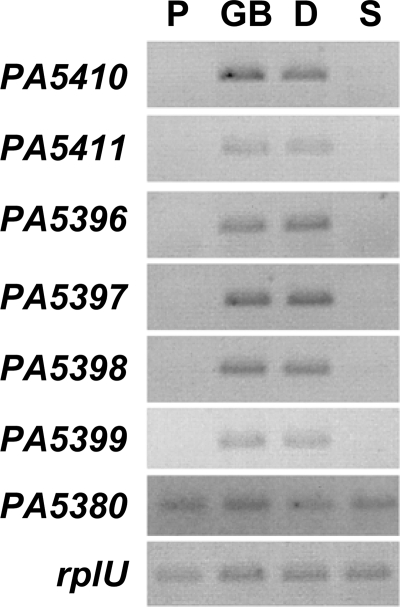
To determine if PA5410 and PA5411 were induced during growth on GB or related compounds, cultures were grown in minimal medium with pyruvate and then transferred to medium containing pyruvate, GB, DMG, or sarcosine for 2 h. While PA5410 and PA5411 transcripts were not detected in medium with pyruvate or sarcosine, high levels of these transcripts were present in medium containing GB or DMG (Fig. 3). This finding is supported by the results of a quantitative transcript analysis which showed that PA5410 was present at levels that were 262- ± 19-fold higher in the presence of GB than in the presence of pyruvate and that PA5411 was present at levels that were 41.5- ± 8.8-fold higher in the presence of GB than in the presence of pyruvate. The rplU transcript, which encodes a ribosomal subunit and has been shown to remain unchanged relative to the total RNA under a variety of conditions, was used as a control transcript in this and other analyses and to normalize the quantitative real-time PCR signals (18).
FIG. 3.
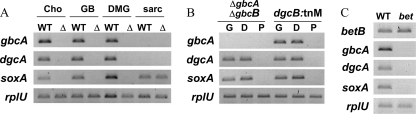
Semiquantitative RT-PCR analysis of P. aeruginosa cells after 2 h of induction with pyruvate (P), GB, DMG (D), or sarcosine (S). PA5410 (gbcA), PA5411 (gbcB), PA5398 (dgcA), PA5399 (dgcB), and PA5380 (gbdR) transcript levels are shown. rplU was used as a control transcript.
The inability of the PA5410 and PA5411 transposon mutants and the ΔPA5410-PA5411 mutant strain to grow on choline or GB and the absence of defects in growth on DMG suggested that these genes encode proteins involved in the conversion of GB to DMG. Based on our findings, these genes were designated gbcA and gbcB (glycine betaine catabolism A and B), respectively.
Analysis of the role of the PA5396-PA5399 putative operon in DMG catabolism.
Four mutant strains with transposon insertions in each gene in the PA5396-PA5399 putative operon (Fig. 2B) were unable to grow on choline, GB, or DMG (Table 2 and data not shown). BLASTP analysis predicted that PA5396 encodes a putative Zn-dependent dipeptidase with high similarity to a predicted aminohydrolase in P. aeruginosa that has been crystallized (accession no. 2I5G_A). PA5397 encodes a small protein with an unknown function. PA5398 encodes a predicted flavin mononucleotide oxidoreductase that is 60% similar and 43% identical (over 635 amino acids) to a putative N-methylproline demethylase from Mesorhizobium loti (accession no. NP_108417). PA5399 encodes a probable membrane-spanning ferrodoxin Fe-S oxidoreductase with high levels of similarity to a large number of predicted membrane-spanning ferrodoxins in gram-negative bacteria.
Transposon insertion into PA5396, PA5397, PA5398, and PA5399, which are predicted to form an operon, resulted in an inability of P. aeruginosa to grow on choline, GB, or DMG as a sole carbon source (Table 2 and data not shown). The similarity of the PA5398 protein to enzymes with demethylation activity suggested that this gene may be directly involved in the demethylation of DMG to sarcosine. To test this hypothesis, we generated a mutant with an in-frame deletion in the PA5398 gene. As predicted, the P. aeruginosa ΔPA5398 mutant was defective in growth on choline, GB, and DMG (Table 2). When the ΔPA5398 mutant was grown in minimal medium with glucose or pyruvate, the growth rate was indistinguishable from that of wild-type strain PA14 (Table 2 and data not shown). To further test the hypothesis that PA5398 is involved in DMG catalysis, the PA5398 and PA5399 transposon mutants and the ΔPA5398 mutant were tested for growth on choline, GB, DMG, and sarcosine as sole sources of nitrogen. Because these mutant strains were able to use sarcosine and lysine but not choline, GB, or DMG (data not shown), we predicted that PA5398 and PA5399 encode proteins critical for the conversion of DMG to sarcosine, which involves a demethylation step. It is not known whether the growth defects on DMG exhibited by the PA5396::TnM and PA5397::TnM mutants, which have disruptions in the two genes upstream of PA5398-PA5399 (Fig. 2), are due to polar effects on PA5398 or if these genes also play a role in DMG catabolism (TnM is the MAR2×T7 transposon insertion).
We predicted that, like transcription of gbcA and gbcB, transcription of PA5398-PA5399 would be induced by growth on GB and DMG but not by growth on sarcosine. The transcript levels for all of the genes in this predicted operon were strongly increased in medium containing GB and DMG, but the transcripts were not detectable in medium with pyruvate or sarcosine (Fig. 3), consistent with the predicted function of the enzymes. As determined by real-time PCR, the transcript levels for PA5398 and PA5399 were 194- ± 58-fold and 181- ± 44-fold higher, respectively, in the presence of GB than in the presence of pyruvate.
Based on the growth phenotypes of the ΔPA5398 mutant and the transposon mutants with mutations in the PA5396-PA5399 genes, we designated the PA5398 and PA5399 genes dgcAB (dimethylglycine catabolism).
Analysis of the accumulation of catabolic intermediates in mutants defective in gbcA and gbcB, dgcB, and soxA.
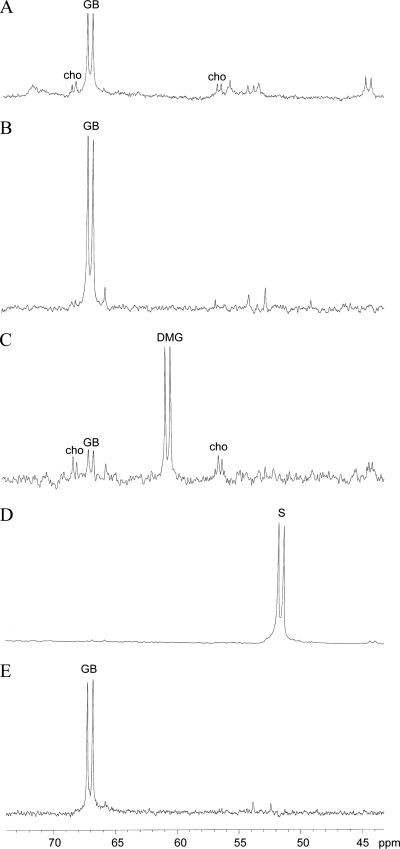
To obtain additional evidence that gbcA and gbcB are involved in conversion of GB to DMG and that dgcA and dgcB are involved in conversion of DMG to sarcosine, cells were fed 1,2-13C-labeled choline, and the accumulation of catabolic intermediates was analyzed by NMR. This isotope enrichment of choline was used because, as shown in Fig. 4, the presence of 13C labels on adjacent carbons results in spectra in which choline and its metabolites appear as characteristic doublets due to the spin-spin coupling between the adjacent 13C nuclei. As shown in Fig. 4A, in wild-type cells choline had been converted primarily to GB after 3 h. After 9 h, only small amounts of GB remained, and no choline or other GB catabolic intermediates were observed, indicating that the choline had been largely mineralized or assimilated into cellular carbon (data not shown). By contrast, when the ΔgbcA-gbcB cells were provided with [1,2-13C]choline as the sole carbon source, only GB was detected at 9 h (Fig. 4B). When dgcB::TnM cells were provided with [1,2-13C]choline as the sole carbon source, some choline and GB were detected, but the majority of the substrate that accumulated was DMG (Fig. 4C). Consistent with the genetic and bioinformatics evidence that soxBDAG genes encode an enzyme that catalyzes the conversion of sarcosine to glycine (8), only sarcosine was detected at 9 h in extracts from soxA::TnM cells provided with [1,2-13C]choline as the sole carbon source (Fig. 4D). These findings are in agreement with our genetic predictions that gbcA and gbcB are required for conversion of GB to DMG, that dgcB is involved in conversion of DMG to sarcosine, and that soxA is required for conversion of sarcosine to glycine in P. aeruginosa.
FIG. 4.
13C NMR spectra of P. aeruginosa cell extracts after incubation with [1,2-13C]choline as the sole carbon source. (A) Wild type at 3 h. (B) ΔgbcA-gbcB mutant at 9 h. (C) ΔdgcB::TnM mutant at 9 h. (D) soxA::TnM mutant at 9 h. (E) ΔPA5380 (ΔgbdR) mutant at 9 h. The peaks are labeled. cho, choline; S, sarcosine.
PA5380 encodes a transcription factor that is required for growth on choline, GB, and DMG.
PA5380, one of the genes disrupted by a transposon in our class II, which was comprised of strains unable to grow on choline, GB, or DMG, encodes a predicted AraC/XylS family transcription factor.
Transposon insertion 159 bp downstream of the PA5380 start codon resulted in a strain unable to utilize choline, GB, and DMG as sole carbon sources (Fig. 2C and data not shown). To confirm this phenotype, we generated an in-frame deletion of PA5380 in P. aeruginosa. P. aeruginosa strain PA14 mutants that lack the PA5380 gene are defective in growth on choline, GB, and DMG as sole carbon or nitrogen sources (Table 2 and data not shown). Complementation with PA5380 on a plasmid (pGbdR) partially restored the ability to grow on all of these compounds as carbon and nitrogen sources. When the P. aeruginosa PA14 ΔPA5380 mutant was grown in minimal medium with glucose, the growth rate was similar to that of the wild-type reference strains (Table 2).
Based on the inability of the ΔPA5380 strain to grow on GB or DMG, we predicted that the PA5380 gene is a transcriptional activator of genes involved in GB and DMG catabolism. To test this hypothesis, the transcript levels of the dgcA and gbcA genes were analyzed in the wild-type and ΔPA5380 mutant backgrounds in media with choline, GB, DMG, or sarcosine (Fig. 5A). The soxA gene involved in sarcosine catabolism was also analyzed, and the rplU transcript was used as the control transcript. As shown in Fig. 5A, PA5380 is required for induction of gbcA, dgcA, and soxA in the presence of choline, GB, and DMG (13). The presence of sarcosine led to increased soxA transcript levels in both the wild type and the ΔPA5380 mutant. In choline-containing medium, mutants defective in PA5380 were able to induce betB, a gene in the betABI choline catabolic operon that is derepressed in the presence of choline (13, 36), with kinetics indistinguishable from those of the wild type (data not shown), suggesting that catabolism of choline to GB (Fig. 1) is not regulated by PA5380. Transcription of the PA5380 gene does not appear to be regulated by GB or DMG, as the PA5380 transcript was detected in cells grown on pyruvate, GB, DMG, and sarcosine (Fig. 3) and did not change significantly under these conditions as measured using quantitative real-time RT-PCR (data not shown). Deletion of PA5380 in P. aeruginosa strain PAO1 also eliminated the ability to grow on choline, GB, or DMG (data not shown), and, similar to observations for strain PA14, deletion of PA5380 prevented the accumulation of gbcA and dgcA transcripts in media with GB or DMG as the sole carbon source (data not shown).
FIG. 5.
Semiquantitative RT-PCR analysis of P. aeruginosa RNA after 2 h of induction by different compounds. The primer sets used in the PCR are indicated on the left. (A) Wild-type (WT) and ΔgbdR (Δ) cells induced with different carbon sources, including choline (cho), GB, DMG, and sarcosine (sarc). (B) ΔgbcA-gbcB and dgcB::TnM cells induced with different carbon sources, including GB (G), DMG (D), and pyruvate (P). (C) Wild-type and betA::TnM cells induced with choline.
To obtain insight into whether GB or DMG was capable of inducing PA5380-controlled genes, the induction of gbcA, dgcA, and soxA in the ΔgbcA-gbcB and ΔdgcA mutants was analyzed (Fig. 5B). When grown with GB, the mutant lacking gbcA and gbcB accumulated GB but not DMG (Fig. 4). If DMG was the sole inducer of PA5380-dependent transcription, GB would not induce GB or DMG catabolic genes in the ΔgbcA-gbcB strain. However, as shown in Fig. 5B, GB induced dgcA and soxA in response to both GB and DMG in the ΔgbcA-gbcB strain. In a similar experiment, it was observed that catabolism of DMG is not required for induction of GB catabolic genes by DMG. The dgcB::TnM mutant strain could still induce the gbcA, dgcA, and soxA genes in response to either GB or DMG. None of the three genes (gbcA, dgcA, or soxA) was induced in medium with pyruvate (Fig. 5B). Together, these results suggest that both GB and DMG induce PA5380-dependent transcriptional activation.
To determine if choline could induce PA5380-dependent transcription of gbcA, dgcA, or soxA, we tested whether choline could stimulate expression of the transcripts in a betA::TnM transposon mutant, which is unable to convert choline to GB (19, 29) (Fig. 5C). As shown in Fig. 5C, choline induced betB in both the wild type and the betA mutant background, but choline did not induce gbcA, dgcA, or soxA in the betA mutant despite the fact that PA5380 was present, indicating that these genes are not induced in response to choline. These data support the hypothesis that GB and DMG induce the gbc, dgc, and sox genes.
Based on these induction experiments and the finding that sarcosine does not induce the gbc and dgc transcripts in the wild type (Fig. 3), we suggest that PA5380-dependent gene activation is induced by both GB and DMG but not by choline or sarcosine. In addition, when the ΔPA5380 strain was provided with [1,2-13C]choline as the sole carbon source, GB was the sole catabolite that accumulated after 9 h (Fig. 4E), supporting the role of PA5380 in transcriptional regulation of GB catabolic genes. Because our data suggest that PA5380 has a direct role in regulation of the GB catabolic pathway, PA5380 was designated gbdR (glycine betaine- and dimethylglycine-responsive regulator).
Analysis of the role of PA3082 in GB catabolism in PAO1 and PA14.
The PA3082 gene, as reported by Serra et al. (33), was predicted to encode a homocysteine-dependent GB methyltransferase that was required for growth of P. aeruginosa strain PRS on GB. To determine if mutants defective in PA3082 exhibited defects in growth on GB, the growth on GB of three PA14 transposon mutants with insertions in PA14-24290 (corresponding to PA3082) in the PA14 nonredundant library and one PAO1 transposon mutant with an insertion in PA3082 (Table 1) was examined. In addition, an in-frame deletion in PA3082 was generated in P. aeruginosa strain PA14. Growth analyses were performed using both MOPS minimal medium and the minimal medium described by Serra et al. (33). The growth studies showed that all five strains with independent defects in PA3082 grew in media with choline, GB, or DMG as the sole source of carbon with kinetics similar to those of wild-type strains (data not shown). When data were normalized using the rplU control transcript, semiquantitative analysis of the PA3082 transcript showed that there was no difference in its levels between cultures grown using GB or pyruvate as the carbon source (data not shown).
DISCUSSION
We screened the nonredundant PA14 transposon mutant library for mutants defective in growth on GB to identify genes involved in the GB catabolic pathway in P. aeruginosa. We identified a putative GB demethylase, encoded by gbcA and gbcB, based on phenotypic data that indicated that the ΔgbcA-gbcB mutant could not grow on GB but could use DMG as a carbon and nitrogen source (Table 2 and data not shown). Furthermore, when the the ΔgbcA-gbcB mutant was fed choline, GB accumulated in the cells (Fig. 4). The gbcA and gbcB transcript levels increased in response to GB and DMG in a GbdR-dependent manner. The transcript accumulation was mirrored in a proteomics analysis, in which the GbcB protein was shown to be more abundant in P. aeruginosa grown in the presence of GB as the sole carbon source (11). We also identified a putative DMG demethylase, encoded by the dgcAB genes, which is necessary for conversion of DMG to sarcosine. Experiments with 13C-labeled choline confirmed that DMG accumulated in the dgcB::TnM mutant strain. The dgcAB genes were also induced by GB and DMG in a GbdR-dependent manner. In addition, we identified a transcriptional regulator, GbdR, which is required for induction of the gbc and dgc genes in the presence of either GB or DMG and thus is required for growth on these substrates. Finally, our mutagenesis and NMR data verified the role of the predicted sarcosine oxidase (soxBDAG) genes in the catabolism of sarcosine to glycine in P. aeruginosa. In addition to the data presented here obtained using P. aeruginosa strain PA14, our studies also showed that gbdR is required for growth on choline, GB, or DMG and that gbcAB is required for growth on choline and GB but not for growth on DMG in P. aeruginosa strain PAO1.
While the enzymatic activities of the products of the gbcA and gbcB genes have not been examined yet biochemically, in silico analyses predicted that GbcAB may remove a methyl group from GB via a dioxygenase mechanism that yields DMG and perhaps formaldehyde, a known product of DMG oxidases from other bacteria (11, 24). Thus, these enzymes may represent a new type of GB catabolism that is distinct from the homocysteine-dependent pathway that has been described for other bacteria (2, 5).
The dgcAB genes are predicted to encode proteins necessary for the conversion of DMG to sarcosine based on the inability of four independent mutants with transposon insertions in the PA5396-PA5399 predicted operon (Fig. 2) and an in-frame dgcA deletion mutant to grow on DMG and based on the accumulation of 13C-labeled DMG in P. aeruginosa ΔdgcA cultures. The inability of multiple dgcA rescue constructs to restore growth in the ΔdgcA strain (data not shown) may suggest that the stoichiometry of the complex is critical for its function or that the processing or assembly of a functional complex can occur only when the genes are cotranscribed. While characterization of the biochemical activities of the dgcA and dgcB gene products remains a subject for future research, these genes have homology to an oxidoreductase gene in M. loti (NP_108417) involved in N-methylproline demethylation which also involves cleavage of a C—N bond (7).
These studies also validated the role of the soxAD genes in catabolism of choline, GB, DMG, and sarcosine. Furthermore, the transcript analysis data shown in Fig. 5 suggest that there may be two mechanisms for transcriptional induction of the sox genes, one that is GbdR dependent and one that is GbdR independent. As shown in Fig. 5A, the soxA gene was induced in the presence of sarcosine in the wild-type strain and the ΔgbdR mutant, suggesting that there is a GbdR-independent mechanism for soxA induction in the presence of sarcosine. In the ΔgbcA-gbcB and ΔdgcA strains, in which GbdR is present, GB and DMG could still induce soxA despite the fact that sarcosine was not produced (Fig. 4). Furthermore, the ΔgbdR strain grew more slowly with sarcosine as the sole nitrogen source than either the wild type or the complemented ΔgbdR derivative (data not shown), suggesting that GbdR may participate in regulation of sarcosine utilization but is not necessary.
GbdR, which was found to be essential for transcriptional activation of the gbc and dgc genes, is similar to the ArgR transcription factor (59% similarity and 43% identity over 322 amino acids), which binds arginine and functions as an activator of arginine catabolic gene transcription in P. aeruginosa (26). We predict that, like ArgR, GbdR activates transcription at target loci only when it has bound GB or DMG. Transcript levels of gbdR did not change significantly during growth in GB or DMG as determined by semiquantitative (Fig. 3) or quantitative real-time RT-PCR methods (data not shown), suggesting that GbdR does not regulate its own transcription under these conditions.
We examined the genomic context of gbdR in P. aeruginosa and its closest homologs in a number of bacteria (http://cmr.tigr.org/). The genomic neighbors of gbdR in a variety of bacteria suggest that GbdR homologs might also regulate GB catabolism or uptake in other organisms. In P. aeruginosa, gbdR is in a region rich in genes involved in choline and GB catabolism (Fig. 2) (PA5372 to PA5421) and is adjacent to a group of genes involved in choline catabolism, including betABI (PA5372-PA5374), betT (PA5375), transporter genes (PA5376-PA5378), and sdaB (PA5379) (Fig. 2c). BetAB are required for oxidation of choline to GB, while BetI is a repressor of the betABI operon. PA5376-PA5378 is hypothesized to encode an inducible GB transporter, and SdaB is a predicted serine deaminase that converts serine to pyruvate. Similar genomic contexts have been observed for the gbdR homologues in Pseudomonas entomophila, Pseudomonas putida, Pseudomonas fluorescens, and Pseudomonas syringae. In Ralstonia eutropha, Burkholderia mallei, Burkholderia pseudomallei, Burkholderia thailandensis, Burkholderia cenocepacia, and Burkholderia xenovorans, the gbdR homologue is also proximal to the PA5376-PA5378 putative operon. In Rhizobium etli, the gbdR homologue (RHE PF00390) is adjacent to gbcA and gbcB homologues.
The PA3082 gene was previously reported to be required for GB catabolism in P. aeruginosa strain PRS (33). Our growth analyses with multiple validated transposon mutants and an in-frame deletion mutant showed that PA3082 is not required for GB catabolism in P. aeruginosa strains PAO1 and PA14. While this finding may reflect a strain-dependent difference in regulation of GB catabolism, we have some concerns about identification of this gene as a gbt (GB methyltransferase) sequence. Serra et al. (33) reported homology to sequences deposited under accession numbers AF293354 (the sequence for a Streptomyces collinus polyketide cluster), X91736 (the sequence for a Chlamydomonas reinhardtii ADP-glucose pyrophosphorylase), and AB024601 (which contains the sequences for three P. aeruginosa genes, including dapD, which encodes a tetrahydrodipicolinate succinylase; PA3660, which encodes a probable sodium/hydrogen antiporter; and glnD, which is predicted to encode a uridylyltransferase [PA3658]). Our analyses did not detect any ORFs in the PA3082 region with these homologies (33). To determine if other P. aeruginosa ORFs elsewhere in the genome had similarity to the sequences deposited under accession numbers AF293354, AB024601, and X91736, BLASTP and TBLASTN analyses were performed, and there were no matches with an E value less than 0.1 except for the accession number AB024601 sequence, which is a P. aeruginosa sequence that had perfect identity with itself. None of these sequences had any detectable similarity to any ORFs in the PA3082 region. Based on our growth data and the lack of similarity of any ORF in the PA3082 region to a sequence encoding a protein with any methyltransferase capability, we propose that the PA3082 region does not contain a gene related to GB catabolism in PAO1 and PA14, although our analyses do not exclude the possibility that PA3082 has a regulatory role during GB catabolism in P. aeruginosa strain PRS.
GB catabolic pathway in P. aeruginosa.
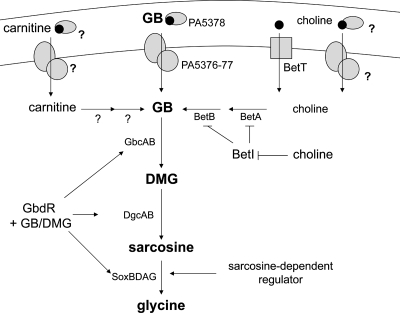
We illustrated our model of GB catabolism in P. aeruginosa based on the data presented here and the model of Diab et al. (Fig. 6) (11). P. aeruginosa can access GB as a free molecule or form GB from the precursors choline and carnitine (16, 40). Choline and GB can be taken into P. aeruginosa cells by a constitutive transporter, as well as by high- and low-affinity inducible transport systems (22, 31), one of which is predicted to be the transporter encoded by PA5376-PA5378 (11). Once in the cytoplasm, choline binds to BetI, resulting in release of transcriptional repression at the betIBA promoter (28). We have shown that transcription of the bet operon in response to choline does not require GbdR. BetA and BetB catalyze the formation of GB from choline (35, 39). Thus, P. aeruginosa can generate GB, a potent osmoprotectant and an inducer of choline-related virulence factors (30), by catabolizing choline to GB without regulation by GbdR. We hypothesize that once free GB is in the cytoplasm, whether due to catabolism of choline or carnitine or due to GB uptake from the periplasm, the GB binds to GbdR. GbdR, when bound to GB, activates the transcription of gbcA, gbcB, dgcAB, and soxBDAG. Since DMG is also predicted to activate GbdR-dependent transcription, the production of DMG may further stimulate DMG and sarcosine catabolic genes. Once DMG is catabolized to sarcosine, an uncharacterized sarcosine-responsive transcription factor may contribute to induction of the soxBDAG operon (Fig. 5C).
FIG. 6.
Model of P. aeruginosa GB catabolism modified from the model of Diab et al. (11), with additions based on the data presented in this paper. BetA is the choline oxidase. BetB is betaine aldehyde dehydrogenase. BetI is the transcriptional repressor of the betABI locus. GbcA and GbcB are the predicted GB demethylase. DgcAB are the predicted DMG demethylase. SoxBDAG are the sarcosine oxidase complex members. Question marks indicate steps that have not been thoroughly evaluated in P. aeruginosa.
Catabolism of GB is also under the control of two other regulatory systems in bacteria, catabolite repression (11) and osmotic regulation (35). In P. aeruginosa, glucose and succinate have both been shown to repress the production of the predicted GB catabolic protein GbcB and the DMG catabolism-associated protein encoded by PA5396, which is predicted to be part of the dgcAB operon (11). We have observed the same effect at the level of transcription for these genes (data not shown). The gbdR transcript was detected even under catabolite-repressing conditions; thus, if GbdR is involved in the regulation of GB catabolism during catabolite repression, the regulation may be in the form of translational regulation, posttranslational modification, or occlusion of the GbdR binding site at one or more promoters. Osmotic stress can also repress bacterial GB catabolism (35), although this has not been demonstrated in P. aeruginosa. This regulatory network would ensure the stability of the intracellular GB pool during a prolonged period of osmotic stress. The role of GbdR and its regulation under osmotic stress conditions are not yet known.
Because P. aeruginosa can derive GB from catabolism of choline and carnitine, molecules that are abundant in host-associated environments, the catabolism of GB may be important during interactions with eukaryotic hosts. In addition, GB can induce expression of known and predicted virulence-related proteins (23, 30), which may impact P. aeruginosa survival and/or pathogenesis in the host. By controlling the catabolism of GB, GbdR may also play an important role during P. aeruginosa interactions with eukaryotes.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant P20-RR018787 from the IDeA Program of the National Center for Research Resources (to D.A.H.) and by a Ruth Kirchstein NRSA institutional fellowship awarded to the Department of Microbiology and Immunology, Dartmouth Medical School (grant T32 AI07519 supporting M.J.W.).
Footnotes
Published ahead of print on 19 October 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barra, L., C. Fontenelle, G. Ermel, A. Trautwetter, G. C. Walker, and C. Blanco. 2006. Interrelations between glycine betaine catabolism and methionine biosynthesis in Sinorhizobium meliloti strain 102F34. J. Bacteriol. 1887195-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazire, A., A. Dheilly, F. Diab, D. Morin, M. Jebbar, D. Haras, and A. Dufour. 2005. Osmotic stress and phosphate limitation alter production of cell-to-cell signal molecules and rhamnolipid biosurfactant by Pseudomonas aeruginosa. FEMS Microbiol. Lett. 253125-131. [DOI] [PubMed] [Google Scholar]
- 4.Boch, J., B. Kempf, and E. Bremer. 1994. Osmoregulation in Bacillus subtilis: synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J. Bacteriol. 1765364-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boncompagni, E., M. Osteras, M. C. Poggi, and D. le Rudulier. 1999. Occurrence of choline and glycine betaine uptake and metabolism in the family Rhizobiaceae and their roles in osmoprotection. Appl. Environ. Microbiol. 652072-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, A. D. 1976. Microbial water stress. Bacteriol. Rev. 40803-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnet, M. W., A. Goldmann, B. Message, R. Drong, A. El Amrani, O. Loreau, J. Slightom, and D. Tepfer. 2000. The stachydrine catabolism region in Sinorhizobium meliloti encodes a multi-enzyme complex similar to the xenobiotic degrading systems in other bacteria. Gene 244151-161. [DOI] [PubMed] [Google Scholar]
- 8.Chlumsky, L. J., L. Zhang, and M. S. Jorns. 1995. Sequence analysis of sarcosine oxidase and nearby genes reveals homologies with key enzymes of folate one-carbon metabolism. J. Biol. Chem. 27018252-18259. [DOI] [PubMed] [Google Scholar]
- 9.Cuin, T. A., and S. Shabala. 2005. Exogenously supplied compatible solutes rapidly ameliorate NaCl-induced potassium efflux from barley roots. Plant Cell Physiol. 461924-1933. [DOI] [PubMed] [Google Scholar]
- 10.de Rudder, K. E., C. Sohlenkamp, and O. Geiger. 1999. Plant-exuded choline is used for rhizobial membrane lipid biosynthesis by phosphatidylcholine synthase. J. Biol. Chem. 27420011-20016. [DOI] [PubMed] [Google Scholar]
- 11.Diab, F., T. Bernard, A. Bazire, D. Haras, C. Blanco, and M. Jebbar. 2006. Succinate-mediated catabolite repression control on the production of glycine betaine catabolic enzymes in Pseudomonas aeruginosa PAO1 under low and elevated salinities. Microbiology 1521395-1406. [DOI] [PubMed] [Google Scholar]
- 12.Diaz, M. R., and B. F. Taylor. 1996. Metabolism of methylated osmolytes by aerobic bacteria from Mono Lake, a moderately hypersaline, alkaline environment. FEMS Microbiol. Ecol. 19239-247. [Google Scholar]
- 13.Eshoo, M. W. 1988. lac fusion analysis of the bet genes of Escherichia coli: regulation by osmolarity, temperature, oxygen, choline, and glycine betaine. J. Bacteriol. 1705208-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapfhammer, D., E. Karatan, K. J. Pflughoeft, and P. I. Watnick. 2005. Role for glycine betaine transport in Vibrio cholerae osmoadaptation and biofilm formation within microbial communities. Appl. Environ. Microbiol. 713840-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiene, R. P. 1998. Uptake of choline and its conversion to glycine betaine by bacteria in estuarine waters. Appl. Environ. Microbiol. 641045-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleber, H. P. 1997. Bacterial carnitine metabolism. FEMS Microbiol. Lett. 1471-9. [DOI] [PubMed] [Google Scholar]
- 17.Kortstee, G. J. 1970. The aerobic decomposition of choline by microorganisms. I. The ability of aerobic organisms, particularly coryneform bacteria, to utilize choline as the sole carbon and nitrogen source. Arch. Mikrobiol. 71235-244. [PubMed] [Google Scholar]
- 18.Kuchma, S. L., J. P. Connolly, and G. A. O'Toole. 2005. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 1871441-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landfald, B., and A. R. Strom. 1986. Choline-glycine betaine pathway confers a high level of osmotic tolerance in Escherichia coli. J. Bacteriol. 165849-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leblanc, L., K. Gouffi, F. Leroi, A. Hartke, C. Blanco, Y. Auffray, and V. Pichereau. 2001. Uptake of choline from salmon flesh and its conversion to glycine betaine in response to salt stress in Shewanella putrefaciens. Int. J. Food Microbiol. 6593-103. [DOI] [PubMed] [Google Scholar]
- 21.Liberati, N. T., J. M. Urbach, S. Miyata, D. G. Lee, E. Drenkard, G. Wu, J. Villanueva, T. Wei, and F. M. Ausubel. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 1032833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lucchesi, G. I., C. Pallotti, A. T. Lisa, and C. E. Domenech. 1998. Constitutive choline transport in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 162123-126. [DOI] [PubMed] [Google Scholar]
- 23.Massimelli, M. J., P. R. Beassoni, M. A. Forrellad, J. L. Barra, M. N. Garrido, C. E. Domenech, and A. T. Lisa. 2005. Identification, cloning, and expression of Pseudomonas aeruginosa phosphorylcholine phosphatase gene. Curr. Microbiol. 50251-256. [DOI] [PubMed] [Google Scholar]
- 24.Meskys, R., R. J. Harris, V. Casaite, J. Basran, and N. S. Scrutton. 2001. Organization of the genes involved in dimethylglycine and sarcosine degradation in Arthrobacter spp.: implications for glycine betaine catabolism. Eur. J. Biochem. 2683390-3398. [DOI] [PubMed] [Google Scholar]
- 25.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, S. M., C. D. Lu, and A. T. Abdelal. 1997. Purification and characterization of an arginine regulatory protein, ArgR, from Pseudomonas aeruginosa and its interactions with the control regions for the car, argF, and aru operons. J. Bacteriol. 1795309-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 2681899-1902. [DOI] [PubMed] [Google Scholar]
- 28.Rkenes, T. P., T. Lamark, and A. R. Strom. 1996. DNA-binding properties of the BetI repressor protein of Escherichia coli: the inducer choline stimulates BetI-DNA complex formation. J. Bacteriol. 1781663-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sage, A. E., A. I. Vasil, and M. L. Vasil. 1997. Molecular characterization of mutants affected in the osmoprotectant-dependent induction of phospholipase C in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 2343-56. [DOI] [PubMed] [Google Scholar]
- 30.Sage, A. E., and M. L. Vasil. 1997. Osmoprotectant-dependent expression of plcH, encoding the hemolytic phospholipase C, is subject to novel catabolite repression control in Pseudomonas aeruginosa PAO1. J. Bacteriol. 1794874-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salvano, M. A., T. A. Lisa, and C. E. Domenech. 1989. Choline transport in Pseudomonas aeruginosa. Mol. Cell. Biochem. 8581-89. [DOI] [PubMed] [Google Scholar]
- 32.Schweizer, H. D. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15831-834. [PubMed] [Google Scholar]
- 33.Serra, A. L., J. F. Mariscotti, J. L. Barra, G. I. Lucchesi, C. E. Domenech, and A. T. Lisa. 2002. Glycine betaine transmethylase mutant of Pseudomonas aeruginosa. J. Bacteriol. 1844301-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sleator, R. D., G. A. Francis, D. O'Beirne, C. G. M. Gahan, and C. Hill. 2003. Betaine and carnitine uptake systems in Listeria monocytogenes affect growth and survival in foods and during infection. J. Appl. Microbiol. 95839-846. [DOI] [PubMed] [Google Scholar]
- 35.Smith, L. T., J. A. Pocard, T. Bernard, and D. Le Rudulier. 1988. Osmotic control of glycine betaine biosynthesis and degradation in Rhizobium meliloti. J. Bacteriol. 1703142-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Styrvold, O. B., P. Falkenberg, B. Landfald, M. W. Eshoo, T. Bjornsen, and A. R. Strom. 1986. Selection, mapping, and characterization of osmoregulatory mutants of Escherichia coli blocked in the choline-glycine betaine pathway. J. Bacteriol. 165856-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki, H., R. Tamamura, S. Yajima, M. Kanno, and M. Suguro. 2005. Corynebacterium sp. U-96 contains a cluster of genes of enzymes for the catabolism of sarcosine to pyruvate. Biosci. Biotechnol. Biochem. 69952-956. [DOI] [PubMed] [Google Scholar]
- 38.Vasil, M. L., D. P. Krieg, J. S. Kuhns, J. W. Ogle, V. D. Shortridge, R. M. Ostroff, and A. I. Vasil. 1990. Molecular analysis of hemolytic and phospholipase C activities of Pseudomonas cepacia. Infect. Immun. 584020-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velasco-Garcia, R., L. Gonzalez-Segura, and R. A. Munoz-Clares. 2000. Steady-state kinetic mechanism of the NADP+- and NAD+-dependent reactions catalysed by betaine aldehyde dehydrogenase from Pseudomonas aeruginosa. Biochem. J. 352675-683. [PMC free article] [PubMed] [Google Scholar]
- 40.Velasco-Garcia, R., C. Mujica-Jimenez, G. Mendoza-Hernandez, and R. A. Munoz-Clares. 1999. Rapid purification and properties of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa. J. Bacteriol. 1811292-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Welsh, D. T. 2000. Ecological significance of compatible solute accumulation by micro-organisms: from single cells to global climate. FEMS Microbiol. Rev. 24263-290. [DOI] [PubMed] [Google Scholar]
- 42.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 14881-86. [DOI] [PubMed] [Google Scholar]
- 43.White, R. F., L. Kaplan, and J. Birnbaum. 1973. Betaine-homocysteine transmethylase in Pseudomonas denitrificans, a vitamin B12 overproducer. J. Bacteriol. 113218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.