Abstract
Copper (Cu) is a required micronutrient, but it is highly toxic at high concentrations. Therefore, the levels of Cu must be tightly regulated in all living cells. The phagosome of Mycobacterium tuberculosis has been shown to have variable levels of Cu. Previously, we showed that M. tuberculosis contains a copper-sensitive operon, cso, that is induced during early infection in mice. In this study, we showed that ctpV, a gene in the cso operon, is a copper-responsive gene and most likely encodes an efflux pump for Cu. Furthermore, the transcription of key genes in the cso operon is induced by Cu ions and not by other ions, such as Ni and Zn ions. To elucidate copper-responsive genes other than those in the cso operon, we utilized DNA microarrays to profile mycobacterial responses to physiological levels of Cu. A transcriptome analysis identified a novel set of 30 copper-responsive genes in M. tuberculosis, one-half of which were induced only when toxic levels of Cu were added. Interestingly, several transcriptional regulators, including the furA gene, were induced during toxic Cu exposure, indicating that there was a generalized response to oxidative stressors rather than a Cu-specific response. In general, the Cu-induced transcriptome generated should help elucidate the role of the Cu response in maintaining M. tuberculosis survival during infection and could provide novel targets for controlling this virulent pathogen.
Mycobacterium tuberculosis is the causative agent of the human disease tuberculosis, which infects approximately one-third of the world's population and causes almost 2 million deaths per year (37). In general, M. tuberculosis is spread via aerosolization of the bacteria from an infected host to a naive individual. Once the bacilli are inhaled, they localize to the alveoli of the lungs, where they are phagocytosed by alveolar macrophages. While most bacteria are rapidly killed after phagocytosis, virulent M. tuberculosis strains interfere with host signaling and prevent maturation of the phagosome (29). However, the mycobacterial phagosome is the source of many stresses, such as low levels of nutrients, iron, and oxygen and high levels of reactive nitrogen intermediates (24). Over its lengthy evolution as an obligate human pathogen, M. tuberculosis has developed mechanisms to effectively deal with all of these stresses. For instance, to prevent iron (Fe) starvation, M. tuberculosis encodes iron-scavenging mechanisms, such as siderophores, and regulates their expression via IdeR, an iron-binding transcriptional repressor (22). Characterization of the environment to which the bacilli are exposed within the phagosome and determination of how the bacteria are able to respond to this environment are keys to understanding how M. tuberculosis is such a successful pathogen. Here, we investigated an important aspect of the mycobacterial response to environmental levels of copper.
To characterize the response of M. tuberculosis to the host environment, we previously utilized microarrays to compare the transcriptome of M. tuberculosis in a murine model to the transcriptome of M. tuberculosis growing in culture (31). A 34-kb region of the genome was identified as a region that is predominantly upregulated in mouse lungs compared to in vitro culture. This region was designated the in vivo-expressed genomic island (iVEGI) (31). Several genes in the iVEGI have been shown to be relevant to the pathogenesis of M. tuberculosis. For instance, the iVEGI genes mprA and mprB encode a two-component signal transduction system required for infection (40). Within the iVEGI, the Rv0967 gene was predicted to encode a transcriptional regulator. Using biochemical and structural experiments, it was shown that the Rv0967 gene encodes a copper-binding transcriptional repressor protein (14). Because the repressor regulates its own four-gene operon in a copper-dependent manner, this operon was designated cso (copper sensitive operon), and the Rv0967 transcriptional regulator was designated CsoR (cso repressor). The induction of cso genes during murine tuberculosis suggested that the copper concentration might fluctuate within the phagosome and that the ability to respond to this fluctuation could be important for mycobacterial virulence. In fact, a previous study of cultured macrophages showed that the level of intraphagosomal Cu increases from 25 to 500 μM after phagocytosis of M. tuberculosis, but no such increase occurs after phagocytosis of less virulent species of Mycobacterium (36).
To our knowledge, Cu homeostasis in M. tuberculosis has not been studied previously. However, bacterial mechanisms of copper homeostasis have been established in two model systems: the K-12 laboratory strain of Escherichia coli and Enterococcus hirae, a gram-positive, extracellular member of the intestinal flora (13, 26, 39). The mechanisms of copper homeostasis identified in these systems include copper transporters (e.g., CopA [20]), the detoxifying enzyme multicopper oxidase (CueO [7]), copper-sensing regulatory proteins (e.g., CueR and CopY [18, 28]), and copper chaperones (e.g., CopZ [5]). Based on previous studies, the most important aspect of copper homeostasis is considered to be copper exporters because of their ability to pump excess Cu outside the cell before it can damage intracellular components (32). When free inside the cell, copper can facilitate the generation of toxic reactive oxygen species, and Cu(I) has been shown to damage proteins, particularly the proteins containing thiol groups (19). Furthermore, excess Cu nonspecifically binds to proteins that require other cofactors (e.g., iron) for activity (32). In this study, we further characterized the cso genes and obtained evidence that CtpV is a copper export transporter. Further, we elucidated the whole-genome transcriptional response of M. tuberculosis to variable levels of Cu by identifying both novel and known metalloregulatory proteins triggered by Cu stimulation. Based on our results, we began to sketch the components utilized by M. tuberculosis to survive the fluctuation in Cu levels inside the phagosome.
MATERIALS AND METHODS
Strains, media, and growth conditions.
All experiments were performed with M. tuberculosis strain H37Rv grown with shaking at 37°C. Cultures of bacteria were grown in liquid Middlebrook 7H9 medium (Remel, Lenexa, KS) supplemented with 10% albumin-dextrose-catalase (ADC) and 0.05% Tween 80 until the optical density at 600 nm (OD600) was 1.0. These rich medium cultures were used to start cultures in Sauton's minimal medium, which is copper free. The Middlebrook 7H9 broth cultures were pelleted by centrifugation and washed twice in Sauton's medium before they were diluted 10-fold in Sauton's broth to obtain an OD600 of 0.10. All metal-free cultures were grown in acid-washed glassware (1 N nitric acid). Cultures used for microarrays and quantitative real-time PCR (qRT-PCR) were then grown to an OD600 of 0.6, after which defined amounts of a metal(s) were added. After 3 h of exposure, cultures were pelleted by centrifugation at 4°C and immediately stored at −80°C prior to RNA extraction as described below. Cultures used to determine growth curves were likewise inoculated to obtain an OD600 of 0.10, and defined amounts of metals were added. These cultures were grown for 14 days, and the numbers of CFU were determined by plating on Middlebrook 7H10 medium in the presence of ADC, followed by incubation at 37°C for 4 weeks. At least two biological replicates of each experiment were performed.
RNA extraction and cDNA synthesis and labeling.
RNA was extracted by resuspending mycobacterial cell pellets in 1 ml Trizol (Invitrogen, Carlsbad, CA) per 5-ml culture. The suspensions were placed in 2-ml screw-cap tubes containing 1 g zirconia/silica beads (Biospec Products, Inc., Bartlesville, OK). The tubes were pulsed twice for 45 s with a mini-Bead-Beater-8 (Biospec Products, Inc.). Following processing with the Bead-Beater, RNA extraction was performed as recommended by the manufacturer (Invitrogen), with modifications described previously (38). Extracted RNA was treated with DNase I (Ambion, Austin, TX) until no DNA was detected using PCR primers for the 16S rRNA gene (see Table S1 in the supplemental material). For qRT-PCR, cDNA was synthesized from 1 μg of total RNA using SuperScript III (Invitrogen) as directed by the manufacturer in the presence of 250 ng of mycobacterial genome-directed primers (30). For microarrays, double-stranded cDNA (ds-cDNA) was synthesized from 10 μg of total RNA using an Invitrogen SuperScript ds-cDNA synthesis kit as directed by the manufacturer in the presence of 250 ng genome-directed primers (30). The ds-cDNA was cleaned and labeled using the NimbleGen gene expression analysis protocol (NimbleGen Systems, Inc., Madison, WI). Briefly, ds-cDNA was incubated with 10 ng RNase A (Novagen) at 37°C for 10 min and cleaned using phenol-chloroform extraction, followed by ethanol precipitation. For Cy3 labeling, 1 μg ds-cDNA was incubated for 10 min at 98°C with 1 OD600 unit of Cy3-9mer Wobble primer (TriLink Biotechnologies, San Diego, CA). Then 8 mmol of deoxynucleoside triphosphates and 100 U of the Klenow fragment (New England Biolabs, Ipswich, MA) were added, which was followed by incubation at 37°C for 2 h. The reaction was stopped by adding 0.1 volume of 0.5 M EDTA, and the labeled ds-cDNA was cleaned by isopropanol precipitation.
DNA microarrays.
Microarray chips were purchased from NimbleGen Systems, Inc., and they contained 19 replicates of each 60-mer oligonucleotide probe designed for the 3,989 open reading frames in the genome of M. tuberculosis H37Rv (3). Further, the whole genome was represented five times on each chip (i.e., five technical replicates/chip) so that there were a total of 95 probes per gene. Hybridization of 3 μg of ds-cDNA with NimbleGen hybridization buffer and Nimblegen hybridization component A was performed in hybridization chambers (TeleChem International, Inc., Sunnyvale, CA) overnight at 42°C. Following hybridization, washing was performed using NimbleGen wash solutions I, II, and III as recommended by the manufacturer. Slides were scanned using an Axon GenePix 4000B scanner (Molecular Devices Corporation, Sunnyvale, CA), and fluorescence intensity levels were determined using NimbleScan (NimbleGen) and normalized to a mean value of 1,000. Significantly changed genes were identified using a flexible empirical Bayes model (specifically, the LNN model in the EBArrays package in R [http://www.bioconductor.org]). A cutoff value of 0.50 for the probability of differential expression was used to determine significantly changed genes (12). To determine functional groups of genes enriched in the microarray data set, identified genes were assigned TIGR roles (http://cmr.tigr.org). Statistical enrichment of each role category within the microarray genes compared with the whole H37Rv genome was performed using a standard hypergeometric distribution function in Microsoft Excel. Complete microarray data are shown in Table S4 in the supplemental material and are available online at http://www.ahabs.wisc.edu/Faculty/talaat-a/lab/data.php.
qRT-PCR.
A SYBR green-based qRT-PCR protocol was used for confirmation of the microarray results. SYBR green qRT-PCR was performed using 100 ng cDNA as the template in a reaction with iTaq SYBR green Supermix with ROX (Bio-Rad Laboratories, Hercules, CA) in the presence of gene-specific primers (see Table S1 in the supplemental material) at a concentration of 200 nM. For experiments to determine metal ion specificity and induction of cso, a TaqMan-based qRT-PCR protocol was used. TaqMan qRT-PCR was performed using 100 ng cDNA as the template in a reaction with Platinum quantitative PCR SuperMix-UDG with ROX (Invitrogen) in the presence of gene-specific primers at a concentration of 200 nM and 6-carboxyfluorescein-labeled TaqMan probes (Biosearch Technologies, Novato, CA) (see Table S1 in the supplemental material). For both SYBR green and TaqMan qRT-PCR, the cycle conditions were 50°C for 2 min, 95°C for 3 min, and 40 cycles of 95°C for 15 s and 60°C for 30 s. Reactions were performed in triplicate with an AB7300 with fluorescence read at the 60°C step. The threshold cycle values were normalized to levels of 16S rRNA transcripts and then expressed as changes compared to the metal-free sample (ΔΔCT method) (30).
RESULTS AND DISCUSSION
Impact of Cu on the growth of M. tuberculosis.
Previously, our analysis suggested that M. tuberculosis has the ability to respond to copper fluctuations in its environment (14), and a hard X-ray microprobe study showed that the physiological levels of copper within the mycobacterial phagosome were between 25 and 500 μM (36). To examine the relationship between intraphagosomal Cu levels and mycobacterial survival, we tested the ability of M. tuberculosis to grow in the presence of physiologically relevant concentrations of Cu. Cultures of virulent M. tuberculosis strain H37Rv were grown in copper-free medium or in medium supplemented with 5, 50, or 500 μM CuCl2, and all cultures were monitored for 14 days. Growth curves showed that with 0, 5, and 50 μM Cu, the bacilli displayed normal growth and doubling times (Fig. 1). However, there was a dramatic bactericidal effect at a Cu concentration of 500 μM, resulting in a 100-fold decrease in the number of CFU compared with the initial inoculum after 14 days of growth. Clearly, high physiological levels of Cu can have bactericidal affects on the mycobacterial bacilli. However, at lower physiological levels (e.g., 50 μM), the cells grow normally, suggesting that Cu homeostasis mechanisms are encoded in the genome of M. tuberculosis.
FIG. 1.
Growth of M. tuberculosis in the presence of different levels of Cu. Cultures were inoculated to obtain an OD600 of 0.1 and then grown for 14 days (∼14 doubling times) in the presence of 0, 5, 50, or 500 μM CuCl2. Colony counts were determined by plating on Middlebrook 7H10 medium supplemented with ADC. The results for a representative sample of two biological replicates are shown. The error bars indicate standard deviations of the mean colony counts.
Specificity of the induction of the cso operon.
Previously, the transcriptional profile of the Cu metalloregulatory gene, csoR, and the cation transporter gene, ctpV, were analyzed using SYBR green qRT-PCR (14). To confirm the differential induction of both csoR and ctpV using a more sensitive protocol, we examined the induction of the cso genes when M. tuberculosis cultures were exposed to different levels of copper using a TaqMan-based protocol. The transcriptional levels obtained (Fig. 2A) showed that there was strong induction of the csoR and ctpV genes in response to increasing levels of Cu, a finding consistent with CsoR's ability to bind the cso promoter region and block transcription only in the absence of Cu ions. Notably, the highest level of induction occurred with 500 μM Cu. Additionally, we tested the specificity of the Cu induction by using the same TaqMan protocol to estimate the level of induction of the cso genes when M. tuberculosis cultures were exposed to other divalent cations, such as silver (Ag), nickel (Ni), and zinc (Zn). We focused on these metal ions based on the sequence analysis (see below) of the protein encoded by ctpV. As expected, the transcripts of both the csoR and ctpV genes were highest when Cu ions were added to the medium and were induced to a lesser extent when Ag was added (Fig. 2b). However, the physiological relevance of Ag-stimulated induction is unknown, as Ag serves no biological purpose in the human body and is probably not transported within macrophages. In contrast, addition of the biologically active metals Ni and Zn did not induce any of the transcripts examined, indicating the specificity of the cso induction by Cu ions.
FIG. 2.
Metal induction of genes in the cso operon. Cultures were grown to an OD600 of 0.6 in metal-free (Sauton's) medium and exposed to the metal ions indicated for 3 h prior to RNA extraction and TaqMan qRT-PCR analysis. Expression values were normalized to 16S rRNA values and are expressed as changes relative to the culture with no metal added. (A) qRT-PCR showing transcriptional induction of csoR and ctpV as the Cu concentration increases. (B) Induction of the cso operon in the presence of Cu and other divalent cations (Ag, Ni, and Zn). Concentrations were chosen based on the upper limit of toxicity for mycobacteria (14).
Sequence analysis of the cso genes.
The Cu-specific regulation of cso via CsoR suggested that the other genes in the operon might have a function related to the intracellular Cu concentration. Two of the genes in the cso operon, the Rv0968 and Rv0970 genes, have no known function, although Rv0970 is predicted to encode an integral membrane protein. However, the remaining downstream gene, Rv0969 (ctpV), is predicted to encode a copper-translocating P-type ATPase based on significant similarity to experimentally characterized Cu transporters. such as CopA in E. coli and CopA in E. hirae (62 and 66% amino acid similarity, respectively) (17, 20).
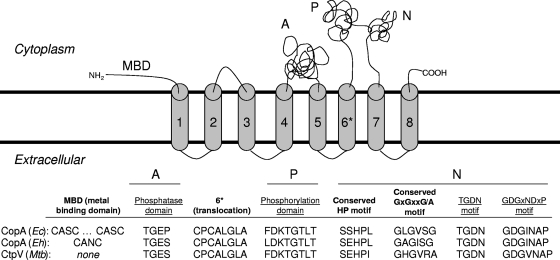
More in-depth sequence analysis revealed that, like other metal translocation P-type ATPases, CtpV is predicted to have eight transmembrane helices with three major cytoplasmic domains (11). Within these domains are conserved motifs important for ATPase activity and substrate translocation (Fig. 3). Helix 6 contains the residues thought to confer metal specificity in the family of metal translocation P-type ATPases, and CtpV contains the CPCALGLA motif found in most other Cu transporters (26). CtpV does not, however, contain the cytoplasm-facing Cu binding sites common in metal translocation P-type ATPases. These metal binding domains have been proposed to aid in the transfer of Cu ions from Cu chaperones to the transporter (10), but deletion of them in two separate human Cu transporters, the Wilson protein and the Menkes protein, did not affect Cu translocation activity (4, 35). The significance of the apparent loss of the metal binding domain motif in CtpV is unknown. The directionality of ion transport (import versus export) cannot be determined based on sequence data alone. However, the induction of ctpV with increasing concentrations of Cu and particularly its high level of induction with 500 μM Cu suggested that it may function as a Cu export pump.
FIG. 3.
Predicted structure and alignment of CtpV with experimentally characterized Cu transporters in E. coli (Ec) and E. hirae (Eh). The A, P, and N domains and the underlined motifs are common to P-type ATPases (34). The motifs in bold type are specific to metal transport P-type ATPases (27). Helix 6 motif is associated with metal specificity (33). MBD, metal binding domain; Mtb, M. tuberculosis.
Identification of copper-responsive genes in M. tuberculosis.
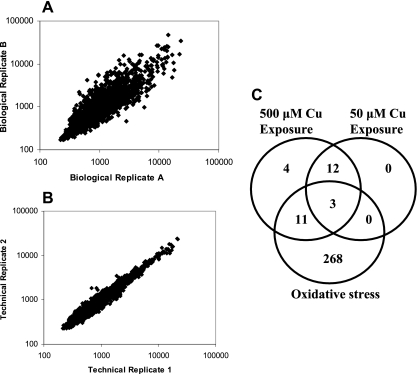
Although CtpV is the only putative ion transporter encoded in the iVEGI and thus was the initial focus of our investigation, there is potential redundancy of proteins involved in Cu homeostasis in proteins encoded by the M. tuberculosis genome. In fact, the genome of M. tuberculosis encodes an unusually high number of predicted metal transport P-type ATPases (n = 11) (www.patbase.kvl.dk). Also, in addition to Cu homeostasis genes, high levels of Cu are expected to activate general stress response genes. To elucidate the mycobacterial response to Cu stress and to identify Cu-responsive genes other than the genes in the cso operon, we compared the transcriptome of M. tuberculosis grown in Cu-free medium to the transcriptome of bacilli grown in medium containing 50 or 500 μM CuCl2 for 3 h. In our preliminary experiments we also included cultures supplemented with 5 μM CuCl2, but we found that the differences between the transcriptional responses to these conditions and the responses observed in medium without Cu were minimal and that the results were unreliable. Because it was difficult to verify that the medium used was completely copper free, it was possible that the differences between the results obtained with 0 μM Cu and the results obtained with 5 μM Cu were negligible. Two biological replicates of the experiment were performed, in which 95 probes per gene were utilized to estimate the transcriptional level of each gene in each replicate. Overall, there was an excellent level of correlation between copies of the genome on each chip (technical replicates) (Fig. 4A) (R = 1.0) and a high level of correlation between separate chip hybridizations (biological replicates) (Fig. 4B) (R = 0.6 to 0.8).
FIG. 4.
Microarray analysis of the Cu-responsive transcriptome in M. tuberculosis. (A) Reproducibility of technical replicates in NimbleGen-based arrays following hybridization with Cu-treated samples. The correlation (r > 0.95) between signals of two copies of the genome on a chip hybridized with a sample containing no Cu is shown. (B) Correlation (r > 0.67) of signals between biological replicates. The results for two biological replicates for samples containing no Cu are shown. (C) Venn diagram showing common and unique sets of Cu-responsive genes. The overlap with a previously identified oxidative stress data set (24) is shown for a comparison to genes induced with both low and high levels of Cu or with only high levels of Cu. As expected, most of the overlap occurs at the toxic concentration of Cu (500 μM).
Genes were considered significantly different in the Cu-supplemented and Cu-free conditions only if there was a probability of differential expression of >0.50 based on a Bayesian statistical model and there was an absolute change of >1.5-fold (12). Based on these criteria, 30 genes were identified as genes that were differentially regulated in Cu-containing cultures and Cu-free cultures (Table 1). As expected, the four genes constituting the known Cu-responsive operon, cso, were induced in cultures exposed to Cu at both concentrations tested. To further confirm the validity of the microarray data, 11 genes were chosen for qRT-PCR confirmation, and the confirmation rate was 91% in the direction of induction or repression of transcripts (see Table S2 in the supplemental material). The change obtained using qRT-PCR was often much greater than the microarray value, indicating the increased sensitivity of qRT-PCR compared with the whole-genome technique. Of the 30 genes identified as copper-responsive genes, 15 were identified as genes that are responsive to 50 μM Cu, while all 30 were identified as genes that are responsive to 500 μM Cu. Because exposure to 50 μM Cu is growth permissive, while exposure to 500 μM Cu is bactericidal (Fig. 1), it is expected that the 15 genes in the data set obtained with 50 μM Cu represent the core Cu response genes, while the 15 additional genes may be induced mainly as a result of Cu-based toxicity. In fact, 11 of the 15 genes identified with 500 μM Cu (67%) were identified using oxidative stress in a previous microarray study of cultures exposed to H2O2 (24). On the other hand, only 3 of the 15 genes (cadI, Rv2963, and Rv3463) from the samples exposed to 50 μM Cu (20%) were associated with the oxidative stress response (Fig. 4C).
TABLE 1.
Genes identified as genes that are significantly up- or downregulated in medium containing 50 or 500 μM CuCl2 compared with Cu-free medium
| Category and genea | Change (fold) with 50 μM Cu | Change (fold) with 500 μM Cu | Putative function | Oxidative stress |
|---|---|---|---|---|
| Regulators | ||||
| Rv0967 (csoR) | 4.3 | 30.8 | Copper-sensitive repressorb | |
| Rv1909c (furA) | 3.8 | Ferric uptake regulation protein | Yes | |
| Rv1994c | 4.0 | Probable MerR transcriptional regulatory proteinb | Yes | |
| Rv2642 | 3.9 | Probable ArsR family transcriptional regulatory protein | ||
| Transporters | ||||
| Rv0969 (ctpV) | 1.7 | 2.9 | Probable metal cation transporter P-type ATPase | |
| Rv0849 | 4.4 | Probable permease | Yes | |
| Rv2398c (cysW) | 3.8 | Probable sulfate transport ABC transporter | ||
| Rv2963 | 2.6 | 15.7 | Possible permeaseb | Yes |
| Membrane protein | ||||
| Rv0970 | 3.2 | Probable integral membrane protein | ||
| Transposases | ||||
| Rv1765A | −3.3 | −2.8 | Putative transposase (fragment) | |
| Rv0850 | 6.4 | Putative transposase (fragment) | Yes | |
| Enzymes | ||||
| Rv0815c (cysA2) | 3.4 | Probable thiosulfate sulfurtransferase | ||
| Rv0848 (cysM3) | 20.1 | Probable cysteine synthase A | Yes | |
| Rv0851c | 2.9 | Probable short-chain dehydrogenase | ||
| Rv0988 | −6.5 | −3.2 | Predicted secreted hydrolaseb | |
| Rv1471 (trxB) | 3.2 | Probable thioredoxin | Yes | |
| Rv1908c (katG) | 5.3 | Catalase peroxidase | Yes | |
| Rv2199c | −7.7 | −3.1 | Probable cytochrome c oxidase polypeptide 4b | |
| Rv2641 (cadI) | 4.1 | 71.6 | Predicted glyoxylase I enzymeb | Yes |
| Rv2962c | 5.0 | Probable glycosyl transferase | ||
| Rv3117 (cysA3) | 3.3 | Probable thiosulfate sulfur transferase | ||
| Chaperone | ||||
| Rv0350 (dnaK) | 3.0 | Molecular chaperone | ||
| Hypothetical | ||||
| Rv0057 | −4.9 | −3.9 | Hypothetical protein | |
| Rv0140 | 7.3 | Hypothetical protein | Yes | |
| Rv0190 | 8.5 | Hypothetical protein | Yes | |
| Rv0430 | −3.3 | Hypothetical protein | Yes | |
| Rv0500B | −4.6 | −3.1 | Hypothetical protein | |
| Rv0968 | 1.6 | 7.7 | Hypothetical protein | |
| Rv2466c | 5.5 | Hypothetical protein | Yes | |
| Rv3463 | 1.9 | 3.2 | Hypothetical protein | Yes |
Genes were identified as genes that were previously associated with oxidative stress based on identification in the hydrogen peroxide stress study of Schnappinger et al. (24).
Annotation based on BLASTP searches (http://www.ncbi.nlm.nih.gov/BLAST/).
To identify common metal-responsive genes in M. tuberculosis, we also compared the Cu-induced transcriptome to the sets of genes previously identified by other workers as genes that are responsive to Fe or Zn (15, 24). Interestingly, no genes overlapped with the Zn-regulated data set, and only one Fe-responsive gene, katG, encoding a catalase-peroxidase enzyme involved in oxidative stress response, was found in the Cu transcriptome. Additionally, the furA gene, encoding the regulator of katG (23), was highly (3.8-fold) induced in samples containing a high concentration of Cu. Overall, our comparative analysis of the Cu-induced transcriptome revealed a strong correlation with genes induced by oxidative stress responses, especially when toxic levels of Cu were used. In contrast, a unique profile for the Cu transcriptome was obtained when M. tuberculosis exposed to Cu was compared to M. tuberculosis exposed to other metal ions. Below we discuss activated groups of Cu-responsive genes to elucidate the role of Cu in M. tuberculosis basic biology.
Transporters involved in copper response.
By assigning the 30 genes in the Cu transcriptome to functional groups (http://cmr.tigr.org), we were able to determine which biological functions are statistically overrepresented in our list of Cu-responsive genes (see Table S3 in the supplemental material). Only the transport and binding protein category was identified as significantly enriched (P < 0.05) in response to Cu, and four putative transporters were identified as proteins that were significantly upregulated in the presence of 500 μM Cu. Only one of these transporters, CtpV, is predicted to specifically transport Cu, although two permease proteins (Rv0849 and Rv2963) that could potentially serve as nonspecific ion transporters were identified. Unexpectedly, none of the other predicted metal-translocating P-type ATPases were identified as proteins that were induced by Cu, including two ATPases (CtpA and CtpB) with the predicted Cu-specific helix 6 motif. The lack of induction of CtpA and CtpB was confirmed by qRT-PCR (data not shown).
Copper-responsive enzymes.
Many enzymes were identified in the Cu transcriptome, and most of them were induced upon addition of Cu. With 70-fold induction after exposure to Cu, cadI is by far the gene that is induced most in the Cu-responsive transcriptome. Bioinformatic analyses identified CadI as a putative glyoxalase I metalloenzyme required for the conversion of methylglyoxal, a toxic by-product of glycolysis, to lactate. Because methylglyoxalase enzymes generally contain zinc, it is possible that Cu substitutes for the Zn, which would result in an inactive enzyme (25). Inactivity could then lead to upregulation via a feedback mechanism (e.g., a mechanism related to the accumulation of substrate). Thus, we speculate that the extreme induction of cadI with high levels of Cu reveals that cofactor substitution is a potential pathway for Cu toxicity in M. tuberculosis.
Other induced enzymes include proteins with functions likely related to oxidative stress (e.g., catalase-peroxidase KatG) and Cu-induced protein damage (e.g., cysteine synthase and thioredoxin). Only two enzymes were repressed in response to Cu, a putative cytochrome c oxidase subunit 4 (Rv2199c) and a putative secreted hydrolase (Rv0988). Because cytochrome c oxidase requires Cu for activity (16), Cu-stimulated regulation is expected. However, the mechanism for regulation of Rv2199c remains unknown. Surprisingly, the predicted multicopper oxidase enzyme (Rv0846c) was not identified as a Cu-responsive protein based on the statistical criteria used in our microarray study, despite the clear role of multicopper oxidases in both E. coli and Salmonella Cu responses (6, 21). Multicopper oxidase has been proposed to reduce Cu(I) to the less toxic form Cu(II), as well as to act as a storage mechanism for excess Cu. Further investigation of the microarray data indicated that Rv0846c transcripts were induced twofold in the presence of a high copper concentration, but induction did not meet the criteria used for statistical significance. However, the induction of Rv0846c with both 50 and 500 μM Cu was clearly demonstrated using qRT-PCR (see Fig. S1 in the supplemental material). Additionally, the protein chaperone DnaK was identified as a Cu-induced protein in the microarray study. In E. coli, DnaK is required to chaperone the multicopper oxidase CueR to the TatAB secretion system (8), which then exports it into the periplasm. The Cu-responsive induction of DnaK and the presence of a predicted Tat signal sequence in Rv0846 (1) suggest that a similar process may occur in M. tuberculosis. Because M. tuberculosis lacks periplasm, we hypothesize that the main role of Rv0846c could be Cu storage and extracellular export.
Copper-responsive transcriptional regulators.
As expected, the microarray study identified the previously characterized Cu-responsive regulator CsoR as a protein that is upregulated in response to Cu. Three other transcriptional regulators were also identified, including FurA, an iron-binding regulator associated with oxidative stress (23), and two previously uncharacterized transcriptional regulators, Rv1994c and Rv2642. The Rv1994c protein is a member of the MerR family of transcriptional regulators. MerR regulators are typically metal binding proteins, and the MerR protein CueR is vital for Cu homeostasis in E. coli (9, 18). Similarly, Rv2642 is a member of the ArsR family of metalloregulatory proteins (2). Either of these proteins could complement the Cu-regulatory activities of CsoR, and these proteins are interesting candidates for future study.
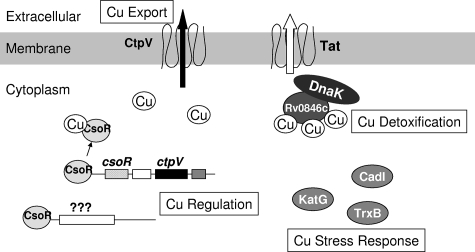
In conclusion, preliminary evidence shows that the Cu level fluctuates in the natural environment of M. tuberculosis, and we showed that physiological Cu levels are toxic to the mycobacterial cell. Furthermore, M. tuberculosis transcriptionally regulates a set of genes to deal with the Cu stress both specifically (e.g., csoR, ctpV, and Rv0846) and nonspecifically via the induction of common stress response genes. The Cu-specific genes were found with both 50 and 500 μM Cu, while the stress response genes were found mainly when the 500 μM Cu data set was examined. Our data indicate that CtpV is the most important copper-regulated cation transporter. Together, these data begin to sketch a model for the interplay of Cu-activated genes within M. tuberculosis (Fig. 5). Currently, experiments are under way to elucidate the specific role(s) played by CtpV in Cu homeostasis and M. tuberculosis virulence, as well as to verify aspects of the proposed model. This analysis should further reveal the role of metal-regulated genes in bacterial pathogenesis in general.
FIG. 5.
Model for Cu response in M. tuberculosis. Based on microarray analysis combined with bioinformatics, four major pathways were identified. (i) For Cu export, the predicted Cu-translocating CtpV protein likely functions as a Cu export pump. Additionally, the Rv0849 and Rv2963 proteins were identified as possible nonspecific metal permeases involved in Cu response. (ii) For Cu regulation, CsoR is the Cu-specific regulator of CtpV and other unknown targets. Possible metalloregulators Rv1994c and Rv2642 are also Cu responsive and may contribute to Cu regulation. (iii) For Cu detoxification, the Cu-induced predicted multicopper oxidase Rv0846c may detoxify Cu(I) in the cell and/or may bind and export Cu ions. Its predicted chaperone (DnaK) and export system (Tat) also show Cu induction. (iv) There are also stress response mechanisms for various aspects of Cu-induced stress, including oxidative stress (e.g., FurA/KatG), protein stress (e.g., TrxB), and cofactor substitution (CadI).
Supplementary Material
Acknowledgments
We acknowledge the technical assistance of Shelly K. Schmoller. In particular, we are indebted to Chia-wei Wu, Bassam Abomoelak, and David Eide for reading the manuscript.
This work was supported in part by grant NIH-R21AI066235, by Animal Formula funds provided to A.M.T., and by NIH training grant T32GM007215 to S.K.W.
Footnotes
Published ahead of print on 8 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bendtsen, J., H. Nielsen, D. Widdick, T. Palmer, and S. Brunak. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Busenlehner, L., M. Pennella, and D. Giedroc. 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27131-143. [DOI] [PubMed] [Google Scholar]
- 3.Camus, J., M. Pryor, C. Médigue, and S. Cole. 2002. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology 1482967-2973. [DOI] [PubMed] [Google Scholar]
- 4.Cater, M., J. Forbes, S. La Fontaine, D. Cox, and J. Mercer. 2004. Intracellular trafficking of the human Wilson protein: the role of the six N-terminal metal-binding sites. Biochem. J. 380805-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cobine, P., W. Wickramasinghe, M. Harrison, T. Weber, M. Solioz, and C. Dameron. 1999. The Enterococcus hirae copper chaperone CopZ delivers copper(I) to the CopY repressor. FEBS Lett. 44527-30. [DOI] [PubMed] [Google Scholar]
- 6.Espariz, M., S. Checa, M. Audero, L. Pontel, and F. Soncini. 2007. Dissecting the Salmonella response to copper. Microbiology 1532989-2997. [DOI] [PubMed] [Google Scholar]
- 7.Grass, G., and C. Rensing. 2001. CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem. Biophys. Res. Commun. 286902-908. [DOI] [PubMed] [Google Scholar]
- 8.Graubner, W., A. Schierhorn, and T. Brüser. 2007. DnaK plays a pivotal role in Tat targeting of CueO and functions beside SlyD as a general Tat signal binding chaperone. J. Biol. Chem. 2827116-7124. [DOI] [PubMed] [Google Scholar]
- 9.Hobman, J., J. Wilkie, and N. Brown. 2005. A design for life: prokaryotic metal-binding MerR family regulators. Biometals 18429-436. [DOI] [PubMed] [Google Scholar]
- 10.Huffman, D., and T. O'Halloran. 2000. Energetics of copper trafficking between the Atx1 metallochaperone and the intracellular copper transporter, Ccc2. J. Biol. Chem. 27518611-18614. [DOI] [PubMed] [Google Scholar]
- 11.Käll, L., A. Krogh, and E. Sonnhammer. 2007. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 35W429-W432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kendziorski, C., M. Newton, H. Lan, and M. Gould. 2003. On parametric empirical Bayes methods for comparing multiple groups using replicated gene expression profiles. Stat. Med. 223899-3914. [DOI] [PubMed] [Google Scholar]
- 13.Kershaw, C., N. Brown, C. Constantinidou, M. Patel, and J. Hobman. 2005. The expression profile of Escherichia coli K-12 in response to minimal, optimal and excess copper concentrations. Microbiology 1511187-1198. [DOI] [PubMed] [Google Scholar]
- 14.Liu, T., A. Ramesh, Z. Ma, S. Ward, L. Zhang, G. George, A. Talaat, J. Sacchettini, and D. Giedroc. 2007. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol. 360-68. [DOI] [PubMed] [Google Scholar]
- 15.Maciag, A., E. Dainese, G. Rodriguez, A. Milano, R. Provvedi, M. Pasca, I. Smith, G. Palù, G. Riccardi, and R. Manganelli. 2007. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 189730-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michel, H., J. Behr, A. Harrenga, and A. Kannt. 1998. Cytochrome c oxidase: structure and spectroscopy. Annu. Rev. Biophys. Biomol. Struct. 27329-356. [DOI] [PubMed] [Google Scholar]
- 17.Odermatt, A., H. Suter, R. Krapf, and M. Solioz. 1993. Primary structure of two P-type ATPases involved in copper homeostasis in Enterococcus hirae. J. Biol. Chem. 26812775-12779. [PubMed] [Google Scholar]
- 18.Outten, F., C. Outten, J. Hale, and T. O'Halloran. 2000. Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, cueR. J. Biol. Chem. 27531024-31029. [DOI] [PubMed] [Google Scholar]
- 19.Pinto, E., T. C. S. Sigaud-kutner, M. A. S. Leitao, O. K. Okamoto, D. Morse, and P. Colepicolo. 2003. Heavy metal-induced oxidative stress in algae. J. Phycol. 391008-1018. [Google Scholar]
- 20.Rensing, C., B. Fan, R. Sharma, B. Mitra, and B. Rosen. 2000. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc. Natl. Acad. Sci. USA 97652-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rensing, C., and G. Grass. 2003. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27197-213. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez, G., M. Voskuil, B. Gold, G. Schoolnik, and I. Smith. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 703371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sala, C., F. Forti, E. Di Florio, F. Canneva, A. Milano, G. Riccardi, and D. Ghisotti. 2003. Mycobacterium tuberculosis FurA autoregulates its own expression. J. Bacteriol. 1855357-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnappinger, D., S. Ehrt, M. Voskuil, Y. Liu, J. Mangan, I. Monahan, G. Dolganov, B. Efron, P. Butcher, C. Nathan, and G. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sellin, S., L. Eriksson, and B. Mannervik. 1987. Electron paramagnetic resonance study of the active site of copper-substituted human glyoxalase I. Biochemistry 266779-6784. [DOI] [PubMed] [Google Scholar]
- 26.Solioz, M., and J. Stoyanov. 2003. Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 27183-195. [DOI] [PubMed] [Google Scholar]
- 27.Solioz, M., and C. Vulpe. 1996. CPx-type ATPases: a class of P-type ATPases that pump heavy metals. Trends Biochem. Sci. 21237-241. [PubMed] [Google Scholar]
- 28.Strausak, D., and M. Solioz. 1997. CopY is a copper-inducible repressor of the Enterococcus hirae copper ATPases. J. Biol. Chem. 2728932-8936. [DOI] [PubMed] [Google Scholar]
- 29.Sturgill-Koszycki, S., P. Schlesinger, P. Chakraborty, P. Haddix, H. Collins, A. Fok, R. Allen, S. Gluck, J. Heuser, and D. Russell. 1994. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 263678-681. [DOI] [PubMed] [Google Scholar]
- 30.Talaat, A., S. Howard, W. T. Hale, R. Lyons, H. Garner, and S. Johnston. 2002. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 30e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Talaat, A., R. Lyons, S. Howard, and S. Johnston. 2004. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. USA 1014602-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teitzel, G., A. Geddie, S. De Long, M. Kirisits, M. Whiteley, and M. Parsek. 2006. Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J. Bacteriol. 1887242-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tottey, S., P. Rich, S. Rondet, and N. Robinson. 2001. Two Menkes-type ATPases supply copper for photosynthesis in Synechocystis PCC 6803. J. Biol. Chem. 27619999-20004. [DOI] [PubMed] [Google Scholar]
- 34.Toyoshima, C., M. Nakasako, H. Nomura, and H. Ogawa. 2000. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature 405647-655. [DOI] [PubMed] [Google Scholar]
- 35.Voskoboinik, I., D. Strausak, M. Greenough, H. Brooks, M. Petris, S. Smith, J. Mercer, and J. Camakaris. 1999. Functional analysis of the N-terminal CXXC metal-binding motifs in the human Menkes copper-transporting P-type ATPase expressed in cultured mammalian cells. J. Biol. Chem. 27422008-22012. [DOI] [PubMed] [Google Scholar]
- 36.Wagner, D., J. Maser, B. Lai, Z. Cai, C. R. Barry, K. Höner Zu Bentrup, D. Russell, and L. Bermudez. 2005. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J. Immunol. 1741491-1500. [DOI] [PubMed] [Google Scholar]
- 37.World Health Organization. March 2007, posting date. World Health Organization tuberculosis fact sheet. World Health Organization, Geneva, Switzerland. www.who.int/mediacenter/factsheets/fs104/en/.
- 38.Wu, C., S. Schmoller, S. Shin, and A. Talaat. 2007. Defining the stressome of Mycobacterium avium subsp. paratuberculosis in vitro and in naturally infected cows. J. Bacteriol. 1897877-7886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto, K., and A. Ishihama. 2005. Transcriptional response of Escherichia coli to external copper. Mol. Microbiol. 56215-227. [DOI] [PubMed] [Google Scholar]
- 40.Zahrt, T., and V. Deretic. 2001. Mycobacterium tuberculosis signal transduction system required for persistent infections. Proc. Natl. Acad. Sci. USA 9812706-12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.