Abstract
Of the nine genes comprising the l-rhamnose operon of Rhizobium leguminosarum, rhaU has not been assigned a function. The construction of a ΔrhaU strain revealed a growth phenotype that was slower than that of the wild-type strain, although the ultimate cell yields were equivalent. The transport of l-rhamnose into the cell and the rate of its phosphorylation were unaffected by the mutation. RhaU exhibits weak sequence similarity to the formerly hypothetical protein YiiL of Escherichia coli that has recently been characterized as an l-rhamnose mutarotase. To characterize RhaU further, a His-tagged variant of the protein was prepared and subjected to mass spectrometry analysis, confirming the subunit size and demonstrating its dimeric structure. After crystallization, the structure was refined to a 1.6-Å resolution to reveal a dimer in the asymmetric unit with a very similar structure to that of YiiL. Soaking a RhaU crystal with l-rhamnose resulted in the appearance of β-l-rhamnose in the active site.
Rhizobium leguminosarum bv. trifolii is a gram-negative soil bacterium that can form symbiotic associations with various species of clover. The plant provides the bacteria with energy for growth, and in return the bacteria provide the plant with fixed nitrogen from nitrogen-fixing nodules. Competition for nodule occupancy exists among strains of Rhizobium within the rhizosphere (10, 40), and strains of R. leguminosarum unable to catabolize l-rhamnose are compromised in their ability to compete for nodule occupancy (24).
l-Rhamnose is a 6-deoxyhexose monosaccharide found in the mucilage of a number of legume plants and is a constituent of pectin in the form of rhamnogalacturonan within the cell walls of dicotyledonous plants (17, 20). The 11-kb l-rhamnose locus in R. leguminosarum comprises l-rhamnose transport and catabolism genes (24, 28) organized in two divergently transcribed operons controlled by a negative regulator, rhaR (28). One transcript contains rhaD and rhaI, encoding a dehydrogenase/aldolase and an isomerase, respectively, while the other consists of rhaRSTPQUK (28). rhaS, rhaT, rhaP, and rhaQ encode the components of an ABC transporter, including a periplasmic sugar binding protein, an ABC ATPase, and two permeases, respectively (3). Within the l-rhamnose catabolic pathway, the enzymatic action of the kinase, encoded by rhaK, has been shown to precede the action of the dehydrogenase and the isomerase encoded by rhaD and rhaI, respectively (28). Moreover, the biochemical activity of the kinase appears to be necessary for l-rhamnose transport (29).
Among the nine genes in the operon, only rhaU does not have an assigned role, and database searches revealed a number of similar open reading frames, none of which had an assigned function. However, YiiL, originally annotated as a hypothetical protein from the Escherichia coli genome, has recently been shown to be an l-rhamnose mutarotase, providing a strong clue to the identity of RhaU. Mutarotases facilitate the interconversion of α and β anomers where the stereochemically less-favored anomer is required for a subsequent step in a catabolic sequence. Three such examples have so far been identified, including l-rhamnose mutarotase (YiiL in E. coli) (30, 31), galactose mutarotase (GalM) (5, 6, 8, 9, 14, 36, 37, 38, 39), and fucose mutarotase (FucU) (16, 30). In this report, we characterize rhaU from R. leguminosarum and provide evidence that is consistent with the hypothesis that RhaU is an l-rhamnose mutarotase.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used and generated in this work are listed in Table 1. R. leguminosarum strain Rlt100 (original designation W14-2 [4]) and strains derived from this wild-type strain were routinely grown at 30°C on TY as a complex medium (7) and VMM (42), a defined medium, as previously described (28). Carbon sources were filter sterilized and added to defined media to a final concentration of 15 mM. When required, antibiotics were added to solid or liquid media at the following concentrations: tetracycline, either 10 μg ml−1 or 5 μg ml−1; neomycin, 200 μg ml−1; streptomycin, 200 μg ml−1; gentamicin, either 15 μg ml−1 or 30 μg ml−1; ampicillin, 100 μg ml−1; and kanamycin, 50 μg ml−1. Bacterial growth was monitored spectrophotometrically at 600 nm.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotypea | Reference or source |
|---|---|---|
| Strains | ||
| R. leguminosarum | ||
| Rlt100 | W14-2, R. leguminosarum bv. trifolii, Smr wild type | 4 |
| Rlt100/pMR110 | Wild type carrying rhaK coding sequence in pRK7813 | 29 |
| Rlt105 | Rlt100 rhaD1::Tn5B20 | 24 |
| Rlt144 | Rlt100 rhaK50::Tn5B20 | 28 |
| Rlt144/pMR110 | Rlt100 rhaK50::Tn5B20 carrying rhaK coding sequence in pRK7813 | 29 |
| Rlt146 | Rlt100 rhaK52::Tn5B20 | 28 |
| Rlt211 | Rlt105 rhaK58::pKNOCK-Tc | 28 |
| Rlt218 | Rlt100 rhaU::pJQ200 SK+ single crossover | This work |
| Rlt218/pMR110 | Rlt100 rhaU::pJQ200 SK+ pMR174 single crossover carrying rhaK coding sequence in pRK7813 | This work |
| Rlt243 | Rlt100 ΔrhaU | This work |
| Rlt243/pMR110 | Rlt100 ΔrhaU carrying rhaK coding sequence in pRK7813 | This work |
| Rlt243/pMR183 | Rlt100 ΔrhaU carrying rhaU coding sequence in pRK7813 | This work |
| Rlt243/pW3C1 | Rlt100 ΔrhaU cosmid from Rlt100 wild-type cosmid, containing entire l-rhamnose locus | This work |
| E. coli | ||
| MT616 | MT607/pRK600 | 24 |
| DH5α | endA hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA)U169 φ80dlacZΔM15 | BRL |
| Plasmids | ||
| pBlueScript II SK | Cloning vector, ColE1 oriV Apr | Stratagene |
| pRK600 | pRK2013 npt::Tn9 Cmr Nm-Kms | 24 |
| pRK7813 | RK2 ori, pUC9 polylinker, lambda cos site, Tcr | 15 |
| pRSETA | N-terminal His6 tag, T7 promoter, Apr | Invitrogen |
| pW3C1 | pRK7813 cosmid from Rlt100 wild-type cosmid bank, containing entire l-rhamnose locus | 24 |
| pJQ200 SK+ | Suicide vector for homogenotization; P15a ori mob sacB, Gmr | 27 |
| pMR110 | rhaK coding sequence in pRK7813 | 29 |
| pMR113 | rhaU coding sequence in pRK7813 | This work |
| pMR133 | rhaU coding sequence in pRSETA | This work |
| pMR174 | ΔrhaU flanking sequence in pJQ200 SK+ | This work |
Ap, ampicillin; Cm, chloramphenicol; Gm, gentamicin; Km, kanamycin; Nm, neomycin; Sm, streptomycin; Tc, tetracycline.
DNA manipulations, sequencing, and sequence analysis.
Standard techniques were used for plasmid isolation, restriction enzyme digestion, ligation, transformation, and agarose gel electrophoresis (32, 33). Genomic DNA was isolated using a modified version of the protocol outlined by Meade et al. (21), as previously described (24). Sequencing was carried out by the University of Calgary Core DNA Services. Sequence data were analyzed using DNASIS (Hitachi Software Engineering Co., San Bruno, CA). Database searches were done using the BLASTX program (2).
Overexpression and purification of RhaU.
To construct a gene encoding a His6-tagged RhaU, the coding region was amplified and cloned into pRSETA (34) using BamHI and EcoRI restriction sites introduced within the primers such that rhaU was under the control of the T7 promoter and in frame with the N-terminal His6 tag. The primers used in the PCR amplification were 5′-ATATGGATCCGGAGATATGACATTGGAAAAACACGC and 5′-ATATGAATTCTCATGGCATATGGAAGAGG (bold type indicates restriction sites). The sequence of the resulting construct, pMR133, was confirmed. For protein expression, pMR133 was transformed into E. coli BL21(DE3) (22), grown to mid-log phase (optical density at 600, 0.5), and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 37°C for 4 h. For selenium methionine (SeM) labeling, cultures were grown in M9 minimal medium supplemented with SeM at the time of induction. Cells were harvested, resuspended in buffer A (20 mM Tris [pH 7.8], 300 mM NaCl, 5 mM β-mercaptoethanol) supplemented with 10 mM imidazole, and lysed by two passages through a French pressure cell (16,000 lb/in2). Cell extracts were passed through a nickel affinity column to bind the His6-RhaU, and after the extracts were washed with buffer A plus 20 mM imidazole for 60 min, RhaU was eluted with 250 mM imidazole in buffer A. Protein samples were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (19) and transferred to a nitrocellulose membrane for Western blot analysis by using a His6 monoclonal antibody as a primary antibody and a goat anti-mouse secondary antibody conjugated with horseradish peroxidase antibody. Horseradish peroxidase was detected colorimetrically with an Opti-4CN substrate detection kit (Bio-Rad Laboratories).
Generation of the ΔrhaU strain.
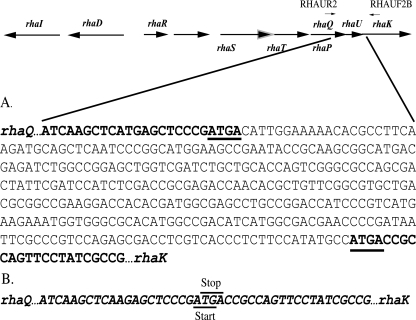
The ΔrhaU strain was constructed using the overlap extension PCR method (32), with pW3C1 as a template. The primers used were 5′-AAGGATCCGGTCAGGGCTATGTCGTC and 5′-AACTGCAGGCAGCCGAGAGAGGTCAA, with the mutagenic primers 5′-TGGCGGTCATCGGGAGCTCATGAGCTTG and 5′-TGAGCTCCCGATGACCGCCAGTTCCTATC (bold type indicates BamHI and PstI sites). The amplification product was cloned as a BamHI/PstI fragment into pJQ200SK (27) to generate pMR174. The gene replacement of rhaU in Rlt100 with pMR174 (ΔrhaU) was carried out as previously described (27) to generate strain Rlt243. The deletion of rhaU from strain Rlt243 was verified by sequencing (Fig. 1).
FIG. 1.
Sequence of the wild type (Rlt100 rhaQUK) (A) and a rhaU deletion strain (Rlt243) (B) generated by site-specific mutagenesis by overlap extension PCR. The sequences corresponding to rhaQ and rhaK are bold and italicized. The rhaQ termination site and the rhaK initiation site that overlap the rhaU initiation and termination sites, respectively, are underlined in panels A and B. A schematic representation of the l-rhamnose catabolic operon is shown above panel A, with the corresponding flanking primers, RHAUR2 and RHAUF2B, used for sequencing.
Transport assays.
The uptake of l-rhamnose was carried out as described previously (28) using [3H]l-rhamnose (5 Ci/mmol) (American Radiolabeled Chemicals, Ltd.). Transport assays were initiated by the addition of [3H]l-rhamnose to a final concentration of 2 μM (125,000 dpm), and aliquots of 0.5 ml were removed, rapidly filtered through a Millipore 0.45-μm HV filter, and washed with 5 ml of the defined salts medium. The 3H on the filters was quantified by liquid scintillation counting.
In vitro RhaK assays.
Cultures were grown on minimal medium to mid-log phase, harvested by centrifugation, resuspended, and lysed by two passages through a French pressure cell. After the removal of cell debris, the cell extracts were mixed with 50 mM HEPES buffer (pH 7.6), 5 mM MgCl2, 10 mM ATP, and 2 mM [3H]l-rhamnose to a volume of 100 μl, from which aliquots were removed and boiled for 10 min. Samples were spotted on Whatman DE 81 cellulose chromatography paper that had been previously washed sequentially with 1 M formic acid and water to remove any impurities and then dried. After descending chromatography was done with 50 mM formic acid (pH 3.2), the paper was cut into 1-in.-square pieces and assayed using a liquid scintillation counter.
Crystallization and structure determination.
The His tag-labeled RhaU was crystallized at room temperature by using the vapor diffusion hanging-drop method at a protein concentration of about 6 mg/ml over a reservoir containing 2 M sodium formate, 0.1 M sodium citrate (pH 5.6), and 20% glycerol. The crystallization buffer was supplemented with 10 mM l-rhamnose for a 1-min soaking. Crystals were trigonal, space group P3212, with two subunits in the crystal asymmetric unit. Diffraction data were obtained from cryocooled crystals, giving the following unit cell parameters: a = 69.2 Å, b = 69.2 Å, c = 101.1 Å, α and β = 90.0°, and γ = 120.0°. A multiwavelength anomalous diffraction experiment was initially performed with a SeM derivative crystal. The heavy metal substructure was finally solved by the single-wavelength anomalous diffraction method using only the remote wavelength data set due to problems in data collection. The substructure of 15 selenium sites, solved by using SHELXD (41) through the graphic interface HKL2MAP (25), showed a correlation between observed and calculated E values of 44.28 (all data) and 22.41 (weak data). Phases were calculated and refined with SHELXE (35) by using all 15 selenium sites and using the slightly higher resolution peak data set. Two SHELXE jobs were started for the two possible enantiomorphs. The contrast and connectivity figures of merit for the correct solution were 0.680 and 0.937, respectively, and the pseudo-free correlation coefficient was 74%, well above the figures of merit for the wrong hand. Automatic model building performed with ARP/wARP (26) traced almost 100% of the asymmetric unit. Model rebuilding was carried out with Coot software (11), and refinement was effected using the REFMAC program (23). Refinement statistics are given in Table 2. All figures were prepared using the PyMOL molecular graphics system (W. L. DeLano; http://www.pymol.org).
TABLE 2.
Data collection, phasing, and structural refinement statistics for an RhaU SeM derivative and an l-rhamnose-soaked crystal
| Statistic or parametera | Result forb:
|
|
|---|---|---|
| Remote-wavelength derivative | Soaked crystal | |
| Data collection statistics for unit cell parameters | ||
| Space group | P3212 | P3212 |
| a (Å)30-1.6 (1.66-1.60) | 69.2 | 68.7 |
| b (Å) | 69.2 | 68.7 |
| c (Å) | 101.1 | 100.7 |
| α, β, γ (degree) | 90, 90, 120 | 90, 90, 120 |
| Wavelength | 0.9079 | 0.9330 |
| Resolution (Å) | 30-1.6 (1.66-1.60) | 30-1.85 (1.92-1.85) |
| No. of unique reflections | 35,978 (3,574) | 23,317 (2,169) |
| Completeness (%) | 97.1 (97.0) | 98.3 (92.5) |
| Rsym (%) | 9.7 (57.6) | 9.1 (30.7) |
| <I/σI> | 11.0 (3.2) | 8.0 (3.7) |
| Redundancy | 5.7 (5.6) | 4.2 (2.8) |
| Model refinement statistics for: | ||
| Resolution | 20-1.6 (1.64-1.60) | 29-2.00 (2.05-2.00) |
| No. of reflections | 33,997 (2,483) | 17,475 (1,281) |
| No. of free reflections | 1,786 (118) | 942 (64) |
| Rcryst (%) | 15.2 (21.3) | 14.4 (14.8) |
| Rfree (%) | 18.0 (27.2) | 19.1 (23.6) |
| No. of residues | 1,428 | 216 |
| No. of water molecules | 220 | 204 |
| Avg B factor (Å2) | ||
| Protein | 12.7 | 19.9 |
| Water | 27.4 | 33.0 |
| All atoms | 17.3 | 21.9 |
| RMSD | ||
| Bond length (Å) | 0.018 | 0.013 |
| Bond angle (degree) | 1.69 | 1.43 |
Rsym = Σhkl Σj|Ihkl,j−<Ihkl>|/Σhkl<Ihkl>, where j extends to all the observed hkl symmetry-related reflections. Rcryst = Σ||Fobs| − |Fcalc||/Σ|Fobs|. Rfree is as for Rcryst but calculated for a test set comprising reflections not used for the refinement.
Values in parentheses correspond with the highest-resolution shell.
Protein structure accession numbers.
Structure factors and coordinates have been submitted to the Protein Data Bank under accession numbers 2QLW (native) and 2QLX (with l-rhamnose).
RESULTS
Generation of the ΔrhaU strain Rlt243.
Previous random transposon mutagenesis of the R. leguminosarum l-rhamnose catabolic locus yielded a total of 56 independent mutants, none of which provided a lesion within rhaU (28). In order to determine if the product of rhaU was required for growth on or the transport of l-rhamnose, site-specific mutagenesis by overlap extension PCR (32) was used to generate the rhaU deletion strain, Rlt243. Nucleotide sequencing of the rhaQ-rhaK region in Rlt243 confirmed that no inadvertent mutations had been introduced and that the predicted upstream rhaQ termination site and predicted downstream rhaK start site were not interrupted (Fig. 1).
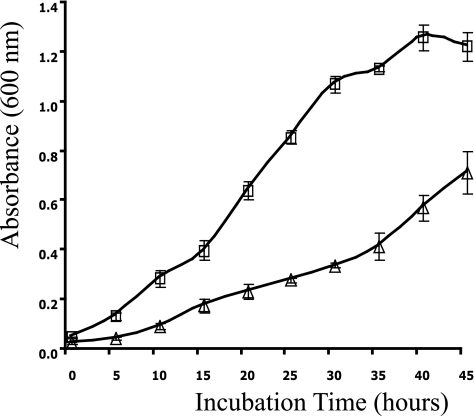
Rlt243 exhibits a slow-growth phenotype on l-rhamnose.
Sequence similarity (41% identity) suggested that RhaU was related to a group of hypothetical proteins that included YiiL of E. coli (28), which was recently characterized as an l-rhamnose mutarotase, catalyzing the α- to β-anomeric conversion of l-rhamnose (30, 31). Based on the phenotype produced by yiiL in E. coli, it was expected that a rhaU-containing mutant would grow normally at high concentrations of l-rhamnose (0.2%) and more slowly at low concentrations (0.03%), but Rlt243 exhibited a slow-growth phenotype even at high l-rhamnose concentrations (Table 3). The mean growth rates for Rlt243 and Rlt100 were 0.0563 and 0.109 generations per hour, respectively (Fig. 2). Despite the low growth rate, the ultimate cell yields of Rlt243 and wild-type Rlt100 were indistinguishable, indicating that carbon utilization and final growth potential were not affected. To ensure that the slow-growth phenotype of Rlt243 was unique to l-rhamnose catabolism and not the result of a pleiotropic effect causing a generalized slow-growth potential, Rlt243 and Rlt100 were grown on a complex medium (TY) and VMM-glucose, with no discernible differences in growth rate or yield. Furthermore, identical slow-growth phenotypes were observed after every round of repeated streaking on VMM-rhamnose, confirming that the phenotype was not a result of the up-regulation of some uncharacterized l-rhamnose catabolic genes or the presence of a second site mutation. Plasmid pW3C1 bearing wild-type rhaU complemented the slow-growth phenotype, confirming that the slow-growth phenotype in Rlt243 was a result of ΔrhaU (Table 3).
TABLE 3.
Strain growth on glucose, l-rhamnose, and glycerol-rhamnose
| Strain | Genotype | Growth ona:
|
||
|---|---|---|---|---|
| Glycerol | Glycerol- rhamnose | Rhamnose | ||
| Rlt100 | Wild type | + | + | + |
| Rlt105 | rhaDI | + | − | − |
| Rlt144 | rhaK | + | + | − |
| Rlt144/pMR110 | rhaK rhaK+ | + | + | + |
| Rlt211 | rhaDI rhaK | + | + | − |
| Rlt243 | ΔrhaU | + | + | ± |
| Rlt243/pMR110 | Δrha rhaK+ | + | + | + |
| Rlt243/pW3C1 | ΔrhaU rha locus+ | + | + | + |
+, similar to wild-type growth; −, no growth; ±, slower growth compared to that of Rlt100. A 15 mM concentration of each carbon source was added to defined medium (VMM).
FIG. 2.
Growth curves of R. leguminosarum wild-type (Rlt100) and rhaU deletion (Rlt243) strains based on absorbance measured at a wavelength of 600 nm. Strains were grown on minimal medium supplemented with 15 mM l-rhamnose as the sole carbon source. Squares, Rlt100; triangles, Rlt243.
Placement of RhaU in a biochemical pathway.
RhaU would most reasonably be situated in a metabolic pathway to catalyze anomerization after transport into the cell prior to phosphorylation. A biochemical lesion in a catabolic pathway following the synthesis of a phosphorylated sugar intermediate can give rise to a conditional phenotype if the bacterium is grown on a medium that contains a noninducing carbon source that can be catabolized (1). For example, the conditional phenotype of the rhaD- and rhaI-containing Rlt105 was relieved by the introduction of a rhaK mutation in Rlt211, demonstrating that the action of RhaK occurs prior to RhaD and RhaI functions in R. leguminosarum (28). Rlt243 grew at wild-type levels on glycerol-rhamnose plates, implying that RhaU enzymatic action occurs before l-rhamnose phosphorylation (Table 3). However, a rhaU-containing strain may simply have been less sensitive to the accumulation of phosphorylated l-rhamnose intermediates. To test this possibility, RhaK levels were increased by introducing a rhaK-bearing plasmid into the rhaU deletion strain. The fact that even this RhaK-overproducing strain, Rlt243/pMR110, did not exhibit a conditional phenotype is consistent with RhaU working prior to RhaK in the catabolic pathway. Interestingly, the overexpressed RhaK had the unexpected effect of eliminating the slow-growth phenotype on VMM-l-rhamnose (Table 3).
rhaU is not necessary for the uptake or phosphorylation of l-rhamnose.
Since the slow-growth phenotype of the rhaU deletion strain Rlt243 was complemented by plasmid-borne wild-type rhaK, RhaK-dependent l-rhamnose phosphorylation and transport levels were investigated. Transport rates in Rlt100 (wild type), Rlt243 (ΔrhaU), and Rlt243/pWC3C1 (ΔrhaU complemented with rhaU) were similar (Table 4), suggesting that the uptake of l-rhamnose was normal in the absence of RhaU and that extra copies of RhaU did not enhance uptake. In contrast, transport in the rhaK mutant strain, Rlt144, was undetectable but could be rescued by the plasmid-borne rhaK in Rlt144/pMR110 (data not shown). Rhamnose phosphorylation levels were unaffected by the ΔrhaU mutation in Rlt243 compared to that of wild-type Rlt100 (Table 5), and the RhaK-bearing plasmid pMR110 did not significantly enhance the amount of phosphorylation (Table 5) or increase l-rhamnose transport (Table 4).
TABLE 4.
Transport of [3H]l-rhamnose
| Strain | Relevant genotype | [3H]l-rhamnose transported (nmol/gfw/min)a |
|---|---|---|
| Rlt100 | Wild type | 30.7 ± 1 |
| Rlt100/pMR110 | Wild type rhaK+ | 24.7 ± 6.6 |
| Rlt243 | ΔrhaU | 36.9 ± 6.4 |
| Rlt243/pMR110 | ΔrhaU rhaK+ | 28.3 ± 6.7 |
| Rlt243/pW3C1 | ΔrhaU rha+ | 28.5 ± 4.5 |
| Rlt144 | rhaK | <0.05 |
Strains were grown as broth cultures in minimal medium (VMM) supplemented with 15 mM l-rhamnose-glycerol. Initial transport rates were determined using tritiated l-rhamnose. The data are presented as the means ± standard deviations (n = 3). gfw, grams (fresh weight).
TABLE 5.
Rate of l-rhamnose phosphorylationa
| Strain | Relevant genotype | Activity (μmol/min/mg) |
|---|---|---|
| Rlt100 | Wild type | 426 |
| Rlt100/pMR110 | Wild type rhaK+ | 300 |
| Rlt146 | rhaK52 | 12 |
| Rlt243 | ΔrhaU | 382 |
| Rlt243/pMR110 | ΔrhaU rhaK+ | 461 |
| Rlt243/pW3C1 | ΔrhaU rha+ | 479 |
Strains were grown in minimal medium (VMM) supplemented with 15 mM l-rhamnose-glycerol; induced cell extract was isolated and processed as described in Materials and Methods. Values are presented as one representative set of data. Data are the average of two independent replicates.
RhaU overexpression and purification.
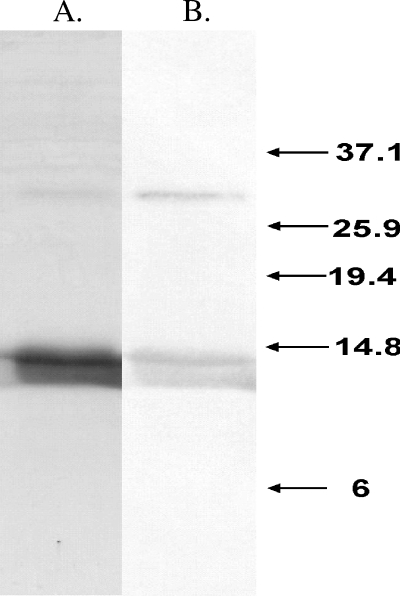
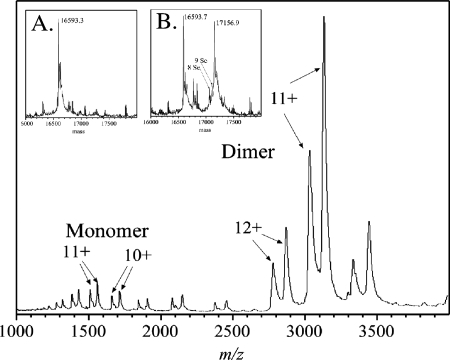
To investigate further the role that RhaU has in l-rhamnose catabolism in R. leguminosarum, a biochemical approach was initiated. Since the nearest homologue to rhaU was annotated as a hypothetical open reading frame, the first step was to demonstrate that rhaU encoded an expressible protein. An in-frame, His6-tagged version of rhaU under the control of a T7 promoter was constructed in the plasmid pMR133, and the protein was purified on a nickel affinity column containing a mixture of three proteins, all of which cross-reacted with monoclonal antibodies to the RGS-His6 tag in a Western blot (Fig. 3). The lower two bands migrated with apparent masses of 12,000 and 14,000 Da, while the smaller upper band appeared to be a dimer with a mass of approximately 28,000 Da. Because the sizes of the lower two bands were smaller than the predicted 16,595 Da, the protein was analyzed by mass spectrometry, confirming a deconvoluted monomer size of 16,593 Da and providing no evidence of a second protein (Fig. 4, inset A). The mass spectrum of RhaU also revealed that RhaU existed predominantly as a dimer and that increasing ionization voltage caused its dissociation to a monomer (Fig. 4). While it is possible that the dimer is due to different charge states of the protein due to partial decarboxylations of the glutamic acid residues, we do not have a definitive explanation for the existence of the two bands of protein on the SDS gel.
FIG. 3.
A 15% SDS-polyacrylamide gel stained with Coomassie blue (A) and an associated Western blot (B) of purified His-tagged RhaU. The numbers indicate in kDa the locations of Invitrogen benchmark prestained ladder proteins run as size markers.
FIG. 4.
Electrospray mass spectrometry analysis of RhaU protein prepared from medium containing methionine or SeM. After purification, the proteins were dialyzed into 5 mM ammonium acetate, diluted to approximately 10 μM, and sprayed directly into a 16-kV time-of-flight instrument at the Department of Physics and Astronomy, University of Manitoba (18). Part of the spectrum from the SeM protein was obtained at a 110-V spray voltage, where most of the ions are from the folded species of protein. Each of the two ion envelopes shows pairs of ions from monomers at 1,500 m/z, as shown in panel B, and from dimers at 3,000 m/z. Inset A, deconvolutions of spectra obtained at a 250-V spray voltage for the methionine-containing protein; inset B, the SeM-containing protein showing ions with 10, 9, and 8 SeM substitutions. Unlabeled ions are from residual buffer salts. At this voltage, most of the ions were unfolded monomers.
Crystal structure of RhaU.
Attempts to solve the structure of RhaU by molecular replacement using the YiiL structure were not successful. However, a solution was found using multiple-wavelength anomalous scattering from a crystal containing SeM. Mass spectrometry analysis of the His tag-labeled protein revealed approximately 70% replacement of sulfur by selenium (Fig. 4, inset B), but this was clearly sufficient selenium for phase determination. The mass of the Se-labeled RhaU was 17,157 Da compared to a predicted mass of 17,158 Da (Fig. 4). Only two sorts of dimers are evident. One contains exclusively methionine and must have been synthesized prior to the shift to SeM medium; the second type contains only SeM and must have been synthesized after the medium change. Some of the latter have less than the 10 expected substitutions, which is reasonable if there was residual methionine in the culture medium. From the relative peak areas, the relative incorporation of SeM for the total protein preparation was about 60%. The electron density map defines main chain and side chain atoms of 216 amino acids in two subunits and 220 water molecules. The maps show clear continuity in both subunits over the complete length from Met1 to Pro106. Of the predicted 38 residues in the His tag sequence at the N terminus, only Gly1 and Asp0 were clearly visible and the remaining 36 residues were presumably disordered. The model has crystallographic agreement Rcryst and Rfree factors of 15.2 and 18.0% for 33,997 reflections in the resolution shell between 1.6 and 20 Å. The average root mean square deviation (RMSD) after the superimposition of the two subunits is 0.26 Å for the Cα atoms and 0.41 Å for all atoms. Ramachandran plots confirm that all residues lie within the energetically favorable regions in both subunits. Based on coordination geometry and electron density, a single metal ion assigned as Mg was associated with C=O of Ala83, the C=O of Met86, and Oγ of Thr88 on the surfaces of both subunits, something that was not observed in YiiL. This is 10 Å from the l-rhamnose binding site and is not affected by l-rhamnose binding.
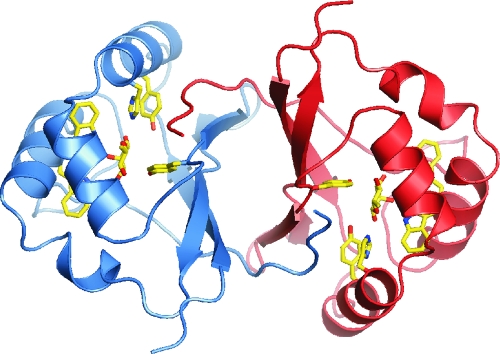
Despite the lack of success in using the YiiL structure as a probe for a molecular replacement structural solution, the structures of YiiL and RhaU are very similar, with an RMSD after the superimposition of 1.01 and 0.98 Å for the Cα atoms in subunits A and B, respectively, when the subunits were superimposed separately. The RMSD increased to 1.56 Å when the subunits were superimposed simultaneously, indicative of a slightly different orientation of the two subunits in the dimer between the two enzymes. The only significant deviation of the main chains lies between residues 60 and 62 at the transition between β-strand 3 and a short α-helical section where the RMSD increases to >4 Å. Like YiiL, a single subunit is comprised of four long β strands in an antiparallel organization that form the interface with the second subunit in the dimer and three α-helical sections that form a cage with the β sheets to house the active site (Fig. 5).
FIG. 5.
View of RhaU to illustrate the relationship of the subunits, the location of rhamnose binding sites in the dimer, and the orientation of the key binding residues. Subunit A is blue, and subunit B is red.
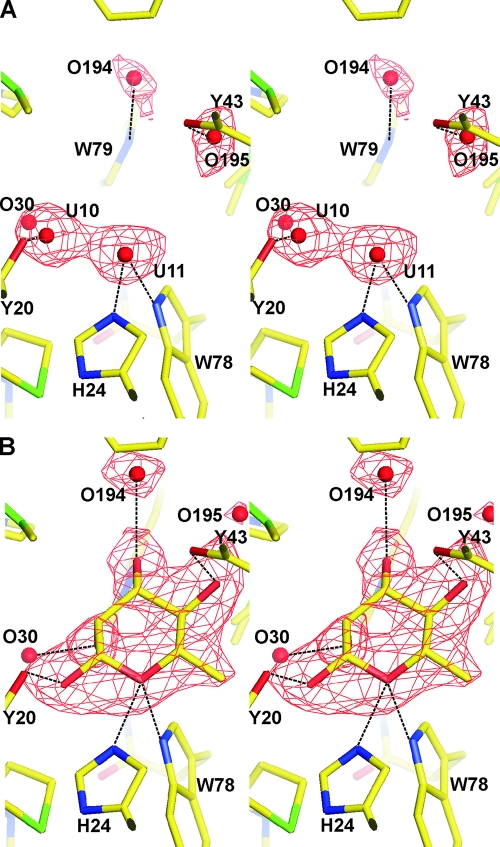
Unlike YiiL, RhaU crystallizes without l-rhamnose bound in the active site, thereby providing an opportunity to study structural changes arising from substrate binding. Soaking RhaU crystals with l-rhamnose results in the appearance of new electron density in the central cavity adjacent to His24 that can be modeled satisfactorily as β-l-rhamnose (Fig. 6B). In subunit B, where the l-rhamnose bound with near 100% occupancy, each of its oxygen atoms forms a hydrogen bond with the protein, including O(H)-1 with Tyr20, OH-2 through water molecules O30 and O94 to the main chain C=O of Trp79 and Asn92, OH-3 with Trp79 and through water molecule O194 with Gln96, OH-4 with Tyr43, and the ring oxygen O-5 with the imidazole of His24 and the indole of Trp78. The C-6 methyl group falls in a relatively hydrophobic pocket composed of Ile27 (3.9 Å), Leu35 (4.12 Å), Leu31 (4.25 Å), and Phe103 (4.44 Å). Omit maps reveal that two water molecules from the native structure (Fig. 6A) are replaced by l-rhamnose, including U-10 by OH-1 and U-11 by ring O-5 (where U indicates a water molecule present when rhamnose was unbound). In addition, water molecule O194 is displaced by 2.3 Å by OH-3, and water molecule O195 is displaced 2.5 Å by OH-4. The only portion of the protein that changes as a result of l-rhamnose binding is the indole ring of Trp79, which is shifted almost 1 Å in order for it to interact with OH-3 of the l-rhamnose. In subunit A, there is an estimated 60% occupancy of l-rhamnose, with a corresponding overlap of l-rhamnose and native water molecules.
FIG. 6.
Stereoview omit Fobs-Fcalc electron density maps of the rhamnose binding site on RhaU. The maps were calculated with no water molecules or rhamnose in the binding site. (A) The omit map of the native protein prior to rhamnose binding. The red spheres indicate the water molecules that were ultimately refined into the model. (B) The omit map calculated after rhamnose binding. A model of rhamnose is superimposed on the map in the location where it was subsequently refined. In both panels, the Fobs-Fcalc density shown in red is modeled at a σ of 3.0.
DISCUSSION
The primary phenotype exhibited by the R. leguminosarum ΔrhaU strain is a lower growth rate than that of the wild-type strain but one that ultimately leads to the same growth yield as the wild type's. The phenotype is consistent with the l-rhamnose mutarotase activity ascribed to RhaU, which would be expected to speed up l-rhamnose catabolism but not be essential. In contrast to E. coli, in which the slow-growth phenotype of a yiiL-containing mutant strain was observed only at low l-rhamnose concentrations, the slow-growth phenotype of a rhaU-containing mutant strain was evident even at high l-rhamnose concentrations. The fact that elevated RhaK levels mask the slow-growth phenotype of the R. leguminosarum rhaU mutant at high l-rhamnose concentrations suggests that the discrepancy with the E. coli yiiL mutant may lie in RhaK levels in R. leguminosarum that are lower than the RhaA/RhaB levels in E. coli. Thus, the uncatalyzed α- to β-anomeric conversion is sufficient to support a low rate of growth in R. leguminosarum, and the role of RhaU is to facilitate the anomerization, thereby allowing a higher rate of growth. Indeed, the equilibrium ratio of α and β anomers in l-rhamnose, determined by nuclear magnetic resonance analysis to be 56:44 (31), suggests a relatively low energy barrier for α to β anomerization of approximately 0.15 to 0.2 kcal/mol, and displacement of the α ⇌ β anomerization equilibrium by the phosphorylation of the β anomer or a subsequent metabolite is not unexpected.
Three possibilities exist for the position at which RhaU might function in the l-rhamnose catabolic pathway. The first is in the periplasm prior to transport, but RhaU is phylogenetically unrelated to other periplasmic mutarotases like GalM of Acinetobacter calcoaceticus (9, 12, 13), and RhaU does not contain a leader targeting sequence. The other two options have RhaU functioning either before or after RhaK-dependent phosphorylation, and differentiation is complicated by the requirement of RhaK for both transport and phosphorylation (29). If RhaU functions before phosphorylation, as in the E. coli l-rhamnose catabolism pathway, RhaK will have to function in two steps separated by the RhaU mutarotation step. On the other hand, the placement of RhaU following phosphorylation would allow RhaK to carry out transport and phosphorylation as part of the same step but would require RhaU to isomerize a phosphorylated sugar. Some data can be interpreted as supporting the concept of RhaU functioning after phosphorylation, including the normal transport and accumulation of phosphorylated l-rhamnose in the absence of RhaU. However, the facts that rhaU mutants grow normally on l-rhamnose-glycerol and that unphosphorylated l-rhamnose binds to RhaU strongly suggest that RhaU functions prior to phosphorylation. Furthermore, the ability of elevated RhaK to mask the slow-growth phenotype of rhaU mutants is consistent with the “product pull” or displacement of the α ⇌ β equilibrium toward the β anomer by the phosphorylation step removing the β anomer from the equilibrium. Thus, RhaK must interact in some way with the ABC transporter to activate the transport of l-rhamnose, while approximately 40% of the l-rhamnose (the proportion that is β anomer) can be immediately phosphorylated by RhaK, and the mutarotation of the remaining 55 to 60% that is the α anomer is facilitated by RhaU. Whether RhaU is physically associated with RhaK and the ABC transporter has not been determined.
The structures of RhaU and the bound l-rhamnose provide important clues to how the α ⇌ β anomerization is catalyzed. The electron density of the bound l-rhamnose is well modeled as the β anomer situating OH-1 3.02 Å from Tyr20, with good geometry for hydrogen bond formation. OH-1 is also just 3.07 Å from His24, but the geometry is less ideal for hydrogen bond formation, whereas His24 is ideally situated to form a hydrogen bond with the ring O-5. In contrast, modeling shows that the α conformation would place OH-1 just 3.0 Å from the δ-CH3 of Ile45, and 3.4 Å from Tyr20, with poor geometry for hydrogen bond formation. The initial binding of the α anomer would be facilitated by the interactions of OH-2, OH-3, OH-4, and O-5 with the protein, placing α-OH-1 in an unfavorable position from which its isomerization to the β anomer becomes an energetically favorable process. The interaction with Ile45 will force α-OH-1 to move toward Tyr20, gradually strengthening that interaction until the optimum β conformation is reached. Ring opening, in which O-5 transiently becomes an OH, is facilitated by hydrogen bonding with His24 and Trp78. The hydrogen bond with His24 provides the proton required for transient O-5-to-OH conversion as part of the sugar ring opening, and protonation of the His24 imidazole is favored by the second imidazole hydrogen bond with the main chain C=O of Tyr20, raising the imidazole pKa slightly. The hydrogen bond of O-5 with the indole of Trp78 serves to stabilize and maintain the proper orientation of O-5 and its transient OH-5 form during ring opening and closing. This reaction pathway which moves the OH-1 from a conformation with an unfavorable interaction to one with a stable hydrogen bond will be favored by 2 to 3 kcal/mol, substantially more than the ∼0.2-kcal/mol barrier to anomerization.
Acknowledgments
This work was supported by Discovery grants 9600 (to P.C.L.) and 238421 (to I.J.O.) from the Natural Sciences and Engineering Research Council of Canada and by the Canadian Research Chair Program (to P.C.L.). Additional support to I.J.O. from the University of Manitoba URGP program is also acknowledged.
Footnotes
Published ahead of print on 21 December 2007.
REFERENCES
- 1.Adhya, S. L., and J. A. Shapiro. 1969. The galactose operon of E. coli K-12. I. Structural and pleiotropic mutations of the operon. Genetics 62231-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames, G. F. 1986. Bacterial periplasmic transport systems: structure, mechanism, and evolution. Annu. Rev. Biochem. 55397-425. [DOI] [PubMed] [Google Scholar]
- 4.Baldani, J. I., R. W. Weaver, M. F. Hynes, and B. D. Eardly. 1992. Utilization of carbon substrates, electrophoretic enzyme patterns, and symbiotic performance of plasmid-cured clover rhizobia. Appl. Environ. Microbiol. 582308-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beebe, J. A., A. Arabshahi, J. G. Clifton, D. Ringe, G. A. Petsko, and P. A. Frey. 2003. Galactose mutarotase: pH dependence of enzymatic mutarotation. Biochemistry 424414-4420. [DOI] [PubMed] [Google Scholar]
- 6.Beebe, J. A., and P. A. Frey. 1998. Galactose mutarotase: purification, characterization, and investigations of two important histidine residues. Biochemistry 3714989-14997. [DOI] [PubMed] [Google Scholar]
- 7.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84188-198. [DOI] [PubMed] [Google Scholar]
- 8.Bettenbrock, K., and C.-A. Alpert. 1998. The gal genes for the Leloir pathway of Lactobacillus casei 64H. Appl. Environ. Microbiol. 642013-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouffard, G. G., K. E. Rudd, and S. L. Adhya. 1994. Dependence of lactose metabolism upon mutarotase encoded in the gal operon in Escherichia coli. J. Mol. Biol. 244269-278. [DOI] [PubMed] [Google Scholar]
- 10.Dowling, D. N., and W. J. Broughton. 1986. Competition for nodulation of legumes. Annu. Rev. Microbiol. 40131-157. [DOI] [PubMed] [Google Scholar]
- 11.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 602126-2132. [DOI] [PubMed] [Google Scholar]
- 12.Gatz, C., J. Altschmied, and W. Hillen. 1986. Cloning and expression of the Acinetobacter calcoaceticus mutarotase gene in Escherichia coli. J. Bacteriol. 16831-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gatz, C., and W. Hillen. 1986. Acinetobacter calcoaceticus encoded mutarotase: nucleotide sequence analysis of the gene and characterization of its secretion in Escherichia coli. Nucleic Acids Res. 144309-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holden, H. M., I. Rayment, and J. B. Thoden. 2003. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J. Biol. Chem. 27843885-43888. [DOI] [PubMed] [Google Scholar]
- 15.Jones, J. D., and N. Gutterson. 1987. An efficient mobilizable cosmid vector, pRK7813, and its use in a rapid method for marker exchange in Pseudomonas fluorescens strain HV37a. Gene 61299-306. [DOI] [PubMed] [Google Scholar]
- 16.Kim, M. S., J. Shin, W. Lee, H. S. Lee, and B. H. Oh. 2003. Crystal structures of RbsD leading to the identification of cytoplasmic sugar-binding proteins with a novel folding architecture. J. Biol. Chem. 27828173-28180. [DOI] [PubMed] [Google Scholar]
- 17.Knee, E. M., F. C. Gong, M. Gao, M. Teplitski, A. R. Jones, A. Foxworthy, A. J. Mort, and W. D. Bauer. 2001. Root mucilage from pea and its utilization by rhizosphere bacteria as a sole carbon source. Mol. Plant Microbe Interact. 14775-784. [DOI] [PubMed] [Google Scholar]
- 18.Kozlovski, V. I., L. J. Donald, V. Montero-Collado, V. Spicer, A. V. Loboda, I. V. Chernushevich, J. McNabb, W. Ens, and K. G. Standing. 2004. Studies of noncovalent complexes of citrate synthase and catalase on a new TOF mass spectrometer with an ESI source, orthogonal injection, and 16 kV acceleration voltage. Desorption 2004, St. Petersburg, Russia.
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 20.McNeil, M., A. G. Darvill, S. C. Fry, and P. Albersheim. 1984. Structure and function of the primary cell walls of plants. Annu. Rev. Biochem. 53625-663. [DOI] [PubMed] [Google Scholar]
- 21.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260289-298. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53240-255. [DOI] [PubMed] [Google Scholar]
- 24.Oresnik, I. J., L. A. Pacarynuk, S. A. P. O'Brien, C. K. Yost, and M. F. Hynes. 1998. Plasmid-encoded catabolic genes in Rhizobium leguminosaurm bv. trifolii: evidence for plant-inducible l-rhamnose locus involved in competition for nodulation. Mol. Plant Microbe Interact. 111175-1185. [Google Scholar]
- 25.Pape, T., and T. R. Schneider. 2004. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 37843-844. [Google Scholar]
- 26.Perrakis, A., R. Morris, and V. Lamzin. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6458-463. [DOI] [PubMed] [Google Scholar]
- 27.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 12715-21. [DOI] [PubMed] [Google Scholar]
- 28.Richardson, J. S., M. F. Hynes, and I. J. Oresnik. 2004. A genetic locus necessary for rhamnose uptake and catabolism in Rhizobium leguminosarum bv. trifolii. J. Bacteriol. 1868433-8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richardson, J. S., and I. J. Oresnik. 2007. l-Rhamnose transport is sugar-kinase (RhaK) dependent in Rhizobium leguminosarum bv. trifolii. J. Bacteriol. 1898437-8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryu, K. S., C. Kim, I. Kim, S. Yoo, B. S. Choi, and C. Park. 2004. NMR application probes a novel and ubiquitous family of enzymes that alter monosaccharide configuration. J. Biol. Chem. 27925544-25548. [DOI] [PubMed] [Google Scholar]
- 31.Ryu, K. S., J. I. Kim, S. J. Cho, D. Park, C. Park, H. K. Cheong, J. O. Lee, and B. S. Choi. 2005. Structural insights into the monosaccharide specificity of Escherichia coli rhamnose mutarotase. J. Mol. Biol. 349153-162. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. W. Russell. 2001. Moleclular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Sambrook, J., E. F. Fritsch, and T. A. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Schoepfer, R. 1993. The pRSET family of T7 promoter expression vectors for Escherichia coli. Gene 12483-85. [DOI] [PubMed] [Google Scholar]
- 35.Sheldrick, G. M. 2002. Macromolecular phasing with SHELXE. Z. Kristallogr. 217644-650. [Google Scholar]
- 36.Thoden, J. B., and H. M. Holden. 2002. High resolution X-ray structure of galactose mutarotase from Lactococcus lactis. J. Biol. Chem. 27720854-20861. [DOI] [PubMed] [Google Scholar]
- 37.Thoden, J. B., J. Kim, F. M. Raushel, and H. M. Holden. 2003. The catalytic mechanism of galactose mutarotase. Protein Sci. 121051-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thoden, J. B., J. Kim, F. M. Raushel, and H. M. Holden. 2002. Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis. J. Biol. Chem. 27745458-45465. [DOI] [PubMed] [Google Scholar]
- 39.Timson, D. J., and R. J. Reece. 2003. Identification and characterisation of human aldose 1-epimerase. FEBS Lett. 54321-24. [DOI] [PubMed] [Google Scholar]
- 40.Triplett, E. W., and M. J. Sadowsky. 1992. Genetics of competition for nodulation of legumes. Annu. Rev. Microbiol. 46399-428. [DOI] [PubMed] [Google Scholar]
- 41.Uson, I., and G. Sheldrick. 1999. Advances in direct methods for protein crystallography. Curr. Opin. Struct. Biol. 9643-648. [DOI] [PubMed] [Google Scholar]
- 42.Vincent, J. M. 1970. A manual for the practical study of root-nodule bacteria. Blackwell Scientific Publications, Oxford, England.