Abstract
According to in silico analysis, the genome of Pseudomonas putida KT2440 encodes at least four Zn/Cd/Pb efflux transporters—two P-type ATPases (CadA1 and CadA2) and two czc chemiosmotic transporters (CzcCBA1 and CzcCBA2). In this study we showed that all these transporters are functional, but under laboratory conditions only two of them were involved in the mediation of heavy metal resistance in P. putida KT2440. CadA2 conferred Cd2+ and Pb2+ resistance, whereas CzcCBA1 was involved in export of Zn2+, Cd2+, and possibly Pb2+. CadA1, although nonfunctional in P. putida, improved Zn2+ resistance and slightly improved Cd2+ resistance when it was expressed in Escherichia coli. CzcCBA2 contributed to Zn resistance of a czcA1-defective P. putida strain or when the CzcA2 subunit was overexpressed in a transporter-deficient strain. It seemed that CzcA2 could complex with CzcC1 and CzcB1 subunits and therefore complement the loss of CzcA1. The CzcCBA2 transporter itself, however, did not function. Expression of cadA1, cadA2, and czcCBA1 was induced by heavy metals, and the expression levels were dependent on the growth medium and growth phase. Expression of cadA2 and czcCBA1 was nonspecific; both genes were induced by Zn2+, Cd2+, Pb2+, Ni2+, Co2+, and Hg2+. On the other hand, remarkably, expression of cadA1 was induced only by Zn2+. Possible roles of distinct but simultaneously functioning transporters are discussed.
Although trace concentrations of some heavy metals, such as zinc and copper, are essential for growth, all heavy metals are toxic to bacteria when high concentrations are present. Therefore, in order to survive in the presence of heavy metals, bacteria must have heavy metal detoxification mechanisms. Metal resistance systems in bacteria are abundant and widespread, and the frequencies range from a few percent of the isolates in clean environments to nearly all isolates in a heavily polluted environment (33). The most widespread heavy metal resistance mechanism is extrusion of metal ions from the cell by active transport, but metal binding factors and enzymatic transformations (oxidation, reduction, methylation, and demethylation) also play roles as second lines of defense against toxic metals. Currently, three main export mechanisms for heavy metals are known: (i) P-type ATPases, which use ATP energy to pump metal ions out of the cytoplasm (25, 29); (ii) CBA transporters, which are three-component transenvelope pumps of gram-negative bacteria and act as chemiosmotic antiporters (10, 13, 23); and (iii) cation diffusion facilitator (CDF) family transporters, which act as chemiosmotic ion-proton exchangers (1, 11, 37). Based on an analysis of 64 prokaryotic genomes, it was concluded that the presence of a CBA transporter (an RND protein) is exceptional and indicates high-level resistance to heavy metal ions. Whereas CDF transporters and P-type ATPases are commonly found among different bacterial species, these transporters thus form part of a cellular homeostasis system (22).
The genus Pseudomonas is composed of a large group of metabolically versatile bacteria adapted to diverse environments. The pseudomonads have received much attention because of their pathogenicity and drug resistance (Pseudomonas aeruginosa), their associations with plants (Pseudomonas syringae and Pseudomonas fluorescens), and their ability to use diverse carbon sources and therefore act as models for the biodegradation of organic compounds (Pseudomonas putida). Comparatively little attention has been paid, however, to the heavy metal resistance of pseudomonads. Yet the ability of pseudomonads to inhabit polluted environments (35) suggests that resistance mechanisms should be present.
The Zn-Cd-Pb resistance mechanisms characterized in the genus Pseudomonas include a CzrCBA (CzcCBA) transporter in P. aeruginosa which confers resistance to zinc, cadmium, and/or cobalt (13, 27). The expression of this transporter is controlled by the CzcRS two-component system, and expression is induced by zinc and copper (5). A similar transporter complex in P. putida CD2 confers zinc and cadmium resistance and is also induced by zinc and copper (15). Additionally, P-type ATPases responsible for divalent heavy metal efflux have been found in pseudomonads. The first such ATPase to be discovered was CadA in P. putida 06909; this system facilitates cadmium resistance and partial zinc resistance and is controlled by a transcriptional regulator of the MerR family (18). Similar but less characterized systems have been described for P. fluorescens strain ATCC 13525 (30) and P. putida CD2 (15). However, despite reports that genomes of pseudomonads contain several divalent heavy metal resistance mechanisms (6, 34), no studies on simultaneous functioning of different transporters in the same strain have been described.
In silico studies of the complete genome of the soil bacterium P. putida KT2440 revealed at least four systems for divalent cation detoxification—two P-type ATPases and at least two czc chemiosmotic antiporters (6). However, no data on the preferred substrates and functionality of these systems have been published. It would be interesting to know how these different systems, seemingly with the same substrates, are integrated into an effective heavy metal resistance network. In this paper, we report on the functions and substrate ranges of two P-type ATPases, CadA1 and CadA2, and two CBA transporters, CzcCBA1 and CzcCBA2, all in P. putida KT2440. We also show which metals induce these transporters and under what conditions they do so, and we try to explain the hierarchy of transporter functioning in the cell.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in the study are listed in Table 1. For construction and maintenance of plasmids, Escherichia coli strains XL-1 Blue and MC1061 were used. Bacteria were routinely grown in Luria-Bertani (LB) medium (31) at 37°C (E. coli) or 30°C (P. putida). For growth inhibition and induction assays heavy metal morpholinepropanesulfonic acid (MOPS) (HMM) medium (17) supplemented with 0.05% (wt/vol) casein hydrolysate was used.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Escherichia coli strains | ||
| XL-1 Blue | supE44 hsdR17 recA1 endA1 gyrA46 thi relA1 lac[F′ proAB+lacIqlacZΔM15 Tn10 (Tetr)] | Stratagene |
| MC1061 | araD139 Δ(ara leu)7697 ΔlacX74 galU galK hsdR2 strA mcrA mcrB1 | 7 |
| TH201 | K-12 ΔzntA ΔzitB | This study |
| Pseudomonas putida strains | ||
| KT2440 | Wild type | 2 |
| KT2440.1 | KT2440 ΔczcA1 | This study |
| KT2440.2 | KT2440 ΔczcA2 | This study |
| KT2440.3 | KT2440 ΔcadA1 | This study |
| KT2440.4 | KT2440 ΔcadA2 | This study |
| KT2440.21 | KT2440 ΔczcA2 ΔczcA1 | This study |
| KT2440.23 | KT2440 ΔczcA2 ΔcadA1 | This study |
| KT2440.24 | KT2440 ΔczcA2 ΔcadA2 | This study |
| KT2440.14 | KT2440 ΔczcA1 ΔcadA2 | This study |
| KT2440.13 | KT2440 ΔczcA1 cadA1::FRTkan | This study |
| KT2440.34 | KT2440 ΔcadA1 cadA2::FRTkan | This study |
| KT2440.241 | KT2440 ΔczcA2 ΔcadA2 ΔczcA1 | This study |
| KT2440.243 | KT2440 ΔczcA2 ΔcadA2 ΔcadA1 | This study |
| KT2440.143 | KT2440 ΔczcA1 ΔcadA2 cadA1::FRTkan | This study |
| KT2440.231 | KT2440 ΔczcA2 ΔcadA1 czcA1::FRTkan | This study |
| KT2440.2431 | KT2440 ΔczcA2 ΔcadA2 ΔcadA1 czcA1::FRTkan | This study |
| Plasmids | ||
| pKD4 | Ampr, oriR[R6Kγ], FRT-kan-FRT | 9 |
| pFLP2 | AmprsacB, encodes the Flp recombinase | 14 |
| pEX18Tc | Tcr, oriT sacB, gene replacement vector with multiple-cloning site from pUC18 | 14 |
| pSL1190 | Ampr, cloning vector | Amersham Pharmacia Biotech |
| pSLlux | Ampr, luxCDABE in pSL1190 | 19 |
| pDN18N | Tcr, T7 expression vector, RK2 replication origin | 26 |
| pEXcadA1FRTkan | Tcr Kmr, fragment of cadA1 gene interrupted by FRT-kan-FRT in pEX18Tc | This study |
| pEXcadA2FRTkan | Tcr Kmr, fragment of cadA2 gene interrupted by FRT-kan-FRT in pEX18Tc | This study |
| pEXczcA1FRTkan | Tcr Kmr, fragment of czcA1 gene interrupted by FRT-kan-FRT in pEX18Tc | This study |
| pEXczcA2FRTkan | Tcr Kmr, fragment of czcA2 gene interrupted by FRT-kan-FRT in pEX18Tc | This study |
| pDNPcadA1lux | Tcr, 682-bp fragment from cadA1 operator area in front of luxCDABE operon cloned into vector pDN18N | This study |
| pDNPcadA2lux | Tcr, 93-bp fragment from cadA2 operator area in front of luxCDABE operon cloned into vector pDN18N | This study |
| Tcr, 409-bp fragment from czcCBA1 operator area in front of luxCDABE operon cloned into vector pDN18N | This study | |
| pDNPczc2lux | Tcr, 991-bp fragment from czcCBA2 operator area in front of luxCDABE operon cloned into vector pDN18N | This study |
| pDNlux_2 | Tcr, luxCDABE in pDN18N opposite the T7 promoter | This study |
| pTIHis | Ampr, 5-kb PCR product of pFLP2 containing rightward λ promoter, cI857, and ori1600, self-ligated | This study |
| pTIcadA1His | Ampr, cadA1 in pTIHis | This study |
| pTIcadA2His | Ampr, cadA2 in pTIHis | This study |
| pTIczcA1His | Ampr, czcA1 in pTIHis | This study |
| pTIczcA2His | Ampr, czcA2 in pTIHis | This study |
| pTIczcCBA1His | Ampr, czcCBA1 in pTIHis | This study |
| pTIczcCBA2His | Ampr, czcCBA2 in pTIHis | This study |
| pTIluxHis | Ampr, luxCDABE in pTIHis | This study |
When required, antibiotics were used at the following final concentrations: for E. coli, 100 μg/ml ampicillin, 30 μg/ml kanamycin, and 12.5 μg/ml tetracycline; and for P. putida, 1,000 μg/ml carbenicillin in LB medium, 600 μg/ml carbenicillin in HMM, 30 μg/ml kanamycin, and 12.5 μg/ml tetracycline.
DNA manipulations.
Qiagen mini-prep, PCR purification, and gel purification kits (Qiagen, Germany) were used for DNA separation and purification. Bacterial genomic DNA was prepared using a GenElute bacterial genomic DNA kit (Sigma-Aldrich, Germany). Enzymes used for DNA manipulations were obtained from Fermentas (Lithuania) and New England Biolabs (United States). PCRs were carried out using Phusion DNA polymerase (Finnzymes, Finland). Primer sequences are shown in Table 2. Plasmids were constructed using standard recombinant DNA techniques and were introduced into E. coli and P. putida by electroporation (8, 31). The essential parts of the constructs were verified by DNA sequencing.
TABLE 2.
Sequences of the primers used for PCR amplification
| Primer pair | Sequence (5′-3′)a |
|---|---|
| A1 | ACTTAGGCATGCGTTGGAAGCGGTCCGTGAAT |
| ACTTAGAAGCTTGCTTGAACGCCCTGATGACC | |
| A2 | AGTGGTGAATTCGAGCCCTCATGGATCGCAAC |
| AGTGGTGAGCTCTCGCTGTGGAGTCGATGAGC | |
| A3 | AGTTGAGAGCTCCGCTCATGGTGATGACGACC |
| AGTTGAGAGCTCCTTCTTCGCCGCCTGGGTGA | |
| A4 | ACTTAGGAATTCATGACCAGCAGCAAGGCGAC |
| GCAAAGCTTCTGCCGAACAC | |
| A5 | AGTTGAGAGCTCGGCAGCTTTGTCCTTAAGTC |
| AGTTGAGAGCTCGGCAGAAATAGACAGCGGAA | |
| A6 | ACAACCTCCTTAGTACATGCAACC |
| CATCATCATCATCATCATTAACCCTGGCGTTACCCAACT | |
| A7 | ATGAAGAGTTTGCTTGAACG |
| GCTTCTGCGGTAGGCCAACA | |
| A8 | ATGAACCAGCCTGTCAGCCA |
| CAAAAGGCGCAAGCCGTTGA | |
| A9 | ATGTTTGAACGTATCATCAGATTCGCCA |
| AACCACTTCGGCCTCGTCGC | |
| A10 | CGGGGACACCTGCATGTTCG |
| CTGCTCATCCCGCCGATACG | |
| A11 | CAAAGTGCCCCGGTATAACC |
| AACCACTTCGGCCTCGTCGC | |
| A12 | ATGCTTCGCCCAGAGGACTGT |
| CTGCTCATCCCGCCGATACG | |
| A13 | CCGCTAAAGAGAAGCCCACA |
| AACTCGTCCCTTCTGGGGTC | |
| A14 | AGTACCTGCAGCACGAAATCTCCAGCAAGTG |
| CAAGTGGATCCGGGGTCATCCTTAAATTGAG | |
| A15 | CCTTACTGCAGATGCTCCTTTGATTCACGGC |
| ACTTCGGATCCTGATGGTCCTCGTGCACAAA | |
| A16 | CTTGCCGTCCTTGAGCACCA |
| GCCAGGGACTGTAGAAAGGG |
Underlining indicates restriction sites.
Construction of knockout strains.
For construction of chromosomal disruptions in P. putida KT2440, a sacB-based gene replacement system (14) was used. In short, gene replacement vectors were constructed as follows. A fragment of the gene was PCR amplified from the chromosomal DNA of P. putida KT2440 using primers pairs A1 and A2 for cadA1 (PP0041) and primer pairs A3, A4, and A5 for cadA2 (PP5139), czcA1 (PP0043), and czcA2 (PP2410), respectively. PCR products were ligated into the cloning vector pSL1190. Then the middle part of the gene fragment was removed by digestion with restriction enzymes and replaced with the FRT-kan-FRT fragment from plasmid pKD4. Cassettes containing the interrupted transporter genes were then transferred into plasmid pEX18Tc, which contains the sacB gene and cannot replicate in pseudomonads. In this manner we generated pEX-based gene replacement vectors which were electroporated into P. putida KT2440. Plasmid integrants were selected on LB medium plates containing kanamycin. Merodiploids were resolved by plating single colonies on LB medium plates containing kanamycin supplemented with 5% (wt/vol) sucrose; in the presence of sacB the sucrose was converted to toxic levan and bacteria were forced to undergo a second recombination event in order to lose sacB and to be able to grow on sucrose. Insertion mutants were verified by colony PCR; a colony from a plate was suspended in 50 μl of water, and 5 μl of the suspension was used as a template in a PCR.
The chromosomally integrated Kmr marker was deleted by Flp recombinase-catalyzed (acts on FRT sites) excision by electroporating the Flp-expressing pFLP2 plasmid into mutant strains. Excision of the Kmr marker was verified by colony PCR. Plasmid pFLP2 was cured by streaking cells on LB medium containing 5% sucrose.
Multiple mutants were constructed by repeating the procedure described above with a previously mutated strain. In the case of strains in which both the cadA1 and czcA1 genes were mutated, the final excision of the Kmr marker by Flp recombinase was omitted, as these genes are located very close together on the chromosome (6). Performing Flp-mediated recombination twice for the same region of the chromosome could have resulted in the excision of a putative open reading frame located between these genes, in addition to removal of the Kmr marker. This unwanted mutation would have compromised our investigations.
The E. coli TH201 strain was constructed by using the method developed by Datsenko and Wanner (9). First, the zntA gene was knocked out in E. coli K-12, and then the zitB gene was removed.
Growth inhibition assays.
Growth inhibition assays were performed in 24-well microplates (Nunc, Denmark). P. putida strains were grown overnight in HMM medium, diluted 100-fold into fresh HMM medium containing various concentrations of metal salts [ZnCl2, CdCl2, Pb(CH3COO)2, CoCl2, NiCl2], and incubated for 24 h at 28°C with shaking. Optical density at 600 nm (OD600) was determined using a Victor Wallac 1420 multilabel counter (PerkinElmer, United States). The MIC was defined as the concentration of metal at which no growth was detected. The relative growth was determined by dividing the OD600 measured in metal-containing medium by the OD600 measured in metal-free medium. A relative growth of 1 corresponded to no growth inhibition, whereas a relative growth of 0 corresponded to no growth. At least three independent measurements were obtained for each strain.
Complementation of P. putida mutants and expression of transporter genes in E. coli.
A fragment of the pFLP2 plasmid containing the rightward λ promoter pR, cI857, and ori1600 was PCR amplified using primer pair A6. One of the primers contained six His codons and a stop codon. The cadA1, cadA2, czcA1, czcCBA1, czcA2, and czcCBA2 transporter genes and operons were PCR amplified with primer pairs A7, A8, A9, A10, A11, and A12, respectively, using P. putida chromosomal DNA as the template. The amplicons included the start codons of the original genes and ended just before the stop codons. PCR products were phosphorylated and ligated with the fragment of the pFLP2 plasmid that permitted gene control by the λ promoter, in order to create temperature-inducible expression plasmids. As a control, we used an empty vector, a self-ligated pFLP2 fragment designated pTIHis. The plasmids were electroporated into transporter-deficient P. putida KT2440.2431 and E. coli TH201, and growth inhibition assays were carried out at 28 and 37°C as described above.
Plasmid pTIluxHis was constructed in a similar manner; the luxCDABE operon was ligated with the PCR-amplified fragment from the pFLP2 plasmid.
In vivo measurement of inducer specificity.
Putative promoter areas of the transporters studied were PCR amplified with primer pair A13 for PcadA1, primer pair A14 for PcadA2, primer pair A15 for Pczc1, and primer pair A16 for Pczc2, using chromosomal DNA of P. putida KT2440 as the template. PCR products were ligated into the pSLlux plasmid to control the expression of the luxCDABE operon. Subsequently, fragments containing the putative promoter areas and luxCDABE operons were transferred into a low-copy-number vector, pDN18N, to generate plasmids pDNPcadA1lux, pDNPcadA2lux, pDNPczc1lux, and pDNPczc2lux. These plasmids were electroporated into P. putida KT2440 and E. coli MC1061. To measure inducer specificity, the cells were grown overnight in LB or HMM medium, diluted 100-fold into fresh medium, and then grown at 30°C with shaking. Aliquots of bacteria were removed at different time points and used in induction assays. A growth curve was constructed by determining OD600 values at 1-h intervals, and growth phases were assigned based on different stages of the growth curve. For induction assays, 50 μl of a bacterial suspension at a specific growth phase in a specific medium was plated into a 96-well microplate (Thermo Labsystems, Finland) and mixed with an equal volume of an aqueous metal solution [ZnCl2, CdCl2, Pb(CH3COO)2, CoCl2, NiCl2, or HgCl2] using various metal concentrations. Plates were then incubated for 2 h (LB medium) or 4 h (HMM medium) at 30°C, and luminescence was measured with a Victor Wallac 1420 multilabel counter. All measurements were obtained using two replicates in at least three independent experiments. Induction of a promoter by metals was expressed as an induction coefficient (IC), calculated as follows: IC = LM/LW, where LM is the luminescence in the metal solution and LW is the background luminescence in water. A metal was considered to induce expression from a promoter if the level of luminescence increased at least twofold (IC ≥ 2) in the presence of the metal compared to the background luminescence. A control plasmid, pDNlux_2, consisted of the luxCDABE operon without any promoter sequence in pDN18N. The control plasmid was used to distinguish nonspecific changes in luminescence caused by the physiological state of the cell from the changes caused by the effects of a metal on the promoter area being studied.
RESULTS AND DISCUSSION
Selection of genes.
We were interested in determining the basis of divalent heavy metal resistance in P. putida KT2440. Our targets were therefore two individual genes encoding the putative P-type ATPases CadA1 and CadA2 and two operons encoding the putative CBA transporters CzcCBA1 and CzcCBA2, as revealed in silico (6). To study the functionalities and substrate ranges of these transporters, we constructed mutant strains of P. putida KT2440 in which one or several of these genes were knocked out. It has been shown previously that disruption of the RND component (gene A) of the CBA transporter is sufficient to abolish its function (24). We therefore concentrated on the cadA1, cadA2, czcA1, and czcA2 genes. To avoid situations where one transporter might compensate for the loss of another transporter, we constructed all possible knockout combinations of these genes. In other words, we constructed strains in which one gene, two genes (altogether six combinations), three genes (four combinations), or all four genes were knocked out (Table 1).
Growth inhibition by heavy metals.
The functionalities and substrate ranges of the transporters were studied by determining the resistance of transporter-deficient strains to different divalent heavy metals. Mutations in the cadA2, czcA1, and czcA2 genes affected the sensitivity of P. putida KT2440 to Zn2+, Cd2+, and Pb2+ but had no effect on Co2+ or Ni2+ resistance (Table 3).
TABLE 3.
MICs of selected divalent heavy metals for P. putida KT2440 transporter-deficient strains
| P. putida strain | Genotype | MIC (μM) ofa:
|
||||
|---|---|---|---|---|---|---|
| Zn2+ | Cd2+ | Pb2+ | Co2+ | Ni2+ | ||
| KT2440 | Wild type | 630 | 380 | 230 | 75 | 190 |
| KT2440.1 | czcA1 | 160 | 190 | 230 | 75 | 190 |
| KT2440.2 | czcA2 | 630 | 380 | 230 | 75 | 190 |
| KT2440.3 | cadA1 | 630 | 380 | 230 | 75 | 190 |
| KT2440.4 | cadA2 | 630 | 23 | 120 | 75 | 190 |
| KT2440.21 | czcA2 czcA1 | 78 | 190 | 230 | 75 | 190 |
| KT2440.23 | czcA2 cadA1 | 630 | 380 | 230 | 75 | 190 |
| KT2440.24 | czcA2 cadA2 | 630 | 23 | 120 | 75 | 190 |
| KT2440.14 | czcA1 cadA2 | 160 | 12 | 120 | 75 | 190 |
| KT2440.13 | czcA1 cadA1 | 160 | 190 | 230 | 75 | 190 |
| KT2440.34 | cadA1 cadA2 | 630 | 23 | 120 | 75 | 190 |
| KT2440.241 | czcA2 cadA2 czcA1 | 78 | 12 | 120 | 75 | 190 |
| KT2440.243 | czcA2 cadA2 cadA1 | 630 | 23 | 120 | 75 | 190 |
| KT2440.143 | czcA1 cadA2 cadA1 | 160 | 12 | 120 | 75 | 190 |
| KT2440.231 | czcA2 cadA1 czcA1 | 78 | 190 | 230 | 75 | 190 |
| KT2440.2431 | czcA2 cadA2 cadA1 czcA1 | 78 | 12 | 120 | 75 | 190 |
MIC in HMM medium after 24 h of growth at 28°C.
The most remarkable changes in resistance were observed with cadmium. Removal of czcA1 had minor effects on Cd2+ resistance, decreasing the MIC twofold, whereas cadA2 mutants were 16 times more sensitive to Cd than the P. putida KT2440 wild-type strain (Fig. 1A and Table 3). Both CadA2 and CzcCBA1 were required for full resistance to cadmium, because strains in which both cadA2 and czcA1 were interrupted could tolerate 32-fold less Cd than the wild-type strain. The cumulative loss of resistance to cadmium in cadA2 czcA1 double mutants is caused by independent transporter functions in the cell. Deletion of only one of these transporters causes a decrease in cadmium resistance, as one of the transporters cannot take over the functions of the other transporter. CBA transporters probably bind their substrates in the periplasm (20), whereas the substrates for the P-type ATPases are thiol-bound cations from the cytoplasm (32). The results indicate that the CzcCBA1 transporter serves as a first line of defense against cadmium, exporting metal ions even before they have entered the cytoplasm. Only a small amount of Cd2+ can be detoxified in this manner, as the CzcCBA1 effect on cadmium resistance was relatively small. Ions which escape the CzcCBA1 transport system and enter the cell are exported by CadA2, which is a more efficient Cd2+ transporter. Also, CzcCBA1 might be responsible for subsequent export of periplasmic Cd2+ ions that have been transported there by CadA2. Deletion of either cadA1 or czcA2 had no effect on cadmium resistance.
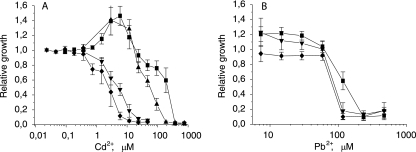
FIG. 1.
Effects of cadmium (A) and lead (B) on the growth of P. putida KT2440 transporter-deficient strains. Dose-response curves for P. putida wild-type strain KT2440 (▪), czcA1 mutant strains KT2440.1, KT2440.21, KT2440.13, and KT2440.231 (▴), cadA2 mutant strains KT2440.4, KT2440.24, KT2440.34, and KT2440.243 (▾), and czcA1 cadA2 double-mutant strains KT2440.14, KT2440.241, KT2440.143, and KT2440.2431 (⧫) are shown. The relative growth indicates the growth efficiency in the presence of a metal compared to the growth in medium without the metal. The data are the means ± standard deviations of at least three independent experiments.
An unexpected phenomenon, enhanced bacterial growth of strains with functional CadA2, was observed at Cd2+ concentrations ranging from 1 to 20 μM (Fig. 1A). This was the concentration range that inhibited the growth of cadA2 mutant strains. In other words, this was the concentration range at which CadA2 normally protects the cell from the toxic effects of cadmium by exporting the cation. As CadA2 is an ATPase, it requires energy to remove Cd2+ ions from the cell. It may be that the continuous usage of ATP forces the cells into some altered growth rhythm, which, in the long term, results in enhanced growth. Although this hypothesis may seem unusual, it was supported by the fact that when an energy-consuming bacterial luminescence operon was transformed into P. putida KT2440, bacterial growth improved by up to twofold in the presence of cadmium compared to the growth in cadmium-free medium (data not shown). This improved growth was seen exclusively in the strains with functional CadA2 and never in cadA2 knockout strains.
Alternatively, Cd2+ may have been a nonspecific substrate for some other efflux pump. When intracellular Cd2+ concentrations were sufficiently high, this hypothetical pump exported Cd2+ in preference to its normal substrate, which therefore remained in the cell and may have promoted growth by acting as a micronutrient. The shortcoming of this hypothesis is that cadA2 knockout strains should also show enhanced growth (because similar intracellular cadmium concentrations are reached in all strains before cadmium becomes toxic), although at different concentrations than the wild-type strain. Finally, contamination of the Cd salt with a micronutrient might be hypothesized.
Both the CzcCBA1 and CzcCBA2 transporters were found to influence zinc resistance. The czcA1 mutants could tolerate fourfold-lower Zn2+ concentrations and the czcA1 czcA2 double mutants could tolerate eightfold-lower Zn2+ concentrations than wild-type strain P. putida KT2440 (Table 3). Removal of only czcA2, however, had no effect on zinc resistance, probably because CzcCBA1 alone could remove excess zinc from the cell. Neither deletion of cadA1 nor deletion of cadA2 had any effect on Zn2+ resistance. This indicates that CzcCBA1 is the main Zn2+ efflux system in P. putida KT2440.
When growing the mutant strains in the presence of lead, we observed small but reproducible differences in the growth of different strains. Strains with a deletion in the cadA2 gene were twice as sensitive to lead as wild-type strain P. putida KT2440 (Table 3). Although an additional deletion in the czcA1 gene had no effect on the MIC of lead, this deletion inhibited the growth of bacteria at concentrations at which the wild-type and cadA2 single-mutant strains attained their full growth potential (relative growth, >1) (Fig. 1B). Deletion of cadA1, czcA2, or only czcA1 (without deletion of cadA2) had no effect on the lead resistance of P. putida (data not shown). Therefore, lead resistance in P. putida KT2440 occurs at least partially through the action of CadA2, and CzcCBA1 might also be involved in lead detoxification.
Complementation tests.
To confirm the functionality data for the CadA1, CadA2, CzcCBA1, and CzcCBA2 transporters described above, we tried to complement deleted transporters. The cadA1, cadA2, czcA1, and czcA2 transporter genes were cloned into plasmids under the temperature-inducible pR promoter and transformed into the knockout strain P. putida KT2440.2431. As a control, the same vector without an inserted transporter gene was used. The pR promoter activity depends on the CI857 repressor. This repressor tightly represses transcription at temperatures up to 30°C but is inactivated at higher temperatures, allowing strong expression from the promoter (16). However, in Pseudomonas this promoter is leaky, and some expression also occurs at temperatures lower than 30°C (14). For testing we used 28°C (low-level expression) and 37°C (high-level expression). Although P. putida KT2440 is usually grown at 30°C, it also grows well at 37°C. In order to minimize adverse effects that a temperature shift might have on bacteria, the growth inhibition assay results were always normalized to a positive growth control which contained no metal.
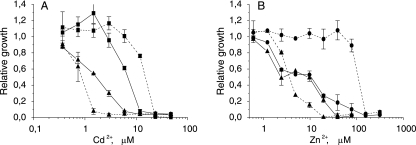
A complementation assay confirmed the role of CadA2 as a Cd transporter. Even at 28°C, a temperature at which expression from the pR promoter should be minimal, a twofold increase in Cd resistance was observed. An increase in the temperature to 37°C improved Cd resistance 16-fold (Fig. 2A and Table 4). Overexpression of other transporters in strain KT2440.2431 had no effect on Cd resistance (Table 4).
FIG. 2.
Effects of cadmium (A) and zinc (B) on the growth of the P. putida KT2440.2431 transporter-deficient strain complemented with transporter genes from a temperature-inducible plasmid. Dose-response curves for control strain P. putida KT2440.2431(pTIHis) (▴), a strain complemented with the cadA2 gene [P. putida KT2440.2431(pTIcadA2His)] (▪), and a strain complemented with the czcA2 gene [P. putida KT2440.2431(pTIczcA2His)] (•) grown at 28°C (solid lines) and 37°C (dashed lines) are shown. The relative growth indicates the growth efficiency in the presence of a metal compared to the growth in medium without the metal. The data are the means ± standard deviations of at least three independent experiments.
TABLE 4.
MICs of divalent heavy metals for P. putida KT2440 and E. coli TH201 transporter-deficient strains expressing transporter genes in trans at 28 and 37°C
| Strain | MIC (μM) ofa:
|
|||||
|---|---|---|---|---|---|---|
| Zn2+
|
Cd2+
|
Co2+
|
||||
| 28°C | 37°C | 28°C | 37°C | 28°C | 37°C | |
| P. putida strains | ||||||
| KT2440(pTIHis) | 310 | 310 | 190 | 190 | NTb | NT |
| KT2440.2431(pTIHis) | 39 | 20 | 6.0 | 1.5 | NT | NT |
| KT2440.2431(pTIcadA1His) | 39 | 20 | 6.0 | 1.5 | NT | NT |
| KT2440.2431(pTIcadA2His) | 39 | 20 | 12 | 23 | NT | NT |
| KT2440.2431(pTIczcA1His) | 39 | 20 | 6.0 | 1.5 | NT | NT |
| KT2440.2431(pTIczcA2His) | 160 | 160 | 6.0 | 1.5 | NT | NT |
| E. coli strains | ||||||
| TH201(pTIHis) | 10 | 2.5 | 0.094 | 0.047 | 38 | 38 |
| TH201(pTIcadA1His) | 10 | 10 | 0.094 | 0.094 | 38 | 38 |
| TH201(pTIcadA2His) | 40 | 20 | 0.19 | 0.75 | 38 | 38 |
| TH201(pTIczcCBA1His) | 10 | 2.5 | 0.094 | 0.047 | 38 | 38 |
| TH201(pTIczcCBA2His) | 10 | 2.5 | 0.094 | 0.047 | 38 | 38 |
MIC in HMM medium after 24 h of growth at 28 or 37°C.
NT, not tested.
A remarkable increase in the Zn resistance of P. putida KT2440.2431 was observed when czcA2 was overexpressed (Fig. 2B). Although there was a fourfold increase in the MIC at 28°C, the overall growth did not improve considerably. At 37°C, however, the MIC was eightfold greater, and normal (uninhibited) growth of P. putida KT2440.2431 overexpressing czcA2 was possible even with 100 μM Zn2+, whereas the strain carrying the control plasmid did not exhibit full growth at Zn2+ concentrations more than 1 μM. Overexpression of other transporter genes, even czcA1, had no effect on the zinc resistance of P. putida KT2440.2431 (Table 4).
Expression of cadA2 and czcA2 in trans partially complemented the loss of chromosomal transporter genes even at 28°C. However, despite the improvements in resistance described above, neither the zinc resistance level nor the cadmium resistance level of the wild-type strain P. putida KT2440 were observed in the complementation studies (Table 4). We assumed that this might have been because of the lower level of expression of transporter proteins from the expression plasmids than from the normal chromosome of the wild-type strain. To test this idea, we constructed a plasmid in which a luxCDABE operon was controlled by the same temperature-inducible pR promoter and compared the luminescence of strains P. putida KT2440(pTIluxHis) and KT2440(pDNPcadA2lux) grown in the presence of cadmium. In the first case the level of luminescence indicated the level of expression from the temperature-inducible promoter that we used for complementation studies. In the second case the level of luminescence reflected the level of expression of cadA2 from the bacterial chromosome. Indeed, the level of luminescence was about three- to fivefold lower in the strain carrying the pTIluxHis plasmid, indicating that we were unable to express the protein at the wild-type level (data not shown). This probably also explains why we did not observe any complementation by czcA1. Normally, czcA1 expression is increased more than 1,000-fold in the presence of zinc (Table 5, Pczc1 induction with Zn2+), indicating that a high concentration of protein is needed to obtain resistance. Obviously, expression from plasmid pTIczcA1His was not enough to complement the loss of chromosomal czcA1. As overexpression of czcA2 was able to confer some zinc resistance on the knockout strain, it is possible that the CzcA2 subunit in a complex with the CzcC1 and CzcB1 proteins (that were expressed at the normal Zn-induced level, and therefore the limiting factor was subunit A of the transporter complex) was a more effective zinc transporter than CzcCBA1. Equal amounts of CzcCBA1 and CzcCB1A2 could be formed; however, only overexpression of CzcA2 improved the resistance of a quadruple-knockout mutant. Lower levels of the CzcCB1A2 protein complex were therefore adequate to increase zinc resistance, showing that CzcCB1A2 is a more effective zinc transporter than CzcCBA1. These results indicate that subunits of the CzcCBA1 and CzcCBA2 transporters might be interchangeable and that czcA2 can, to some extent, compensate for the loss of czcA1 without the CzcCBA2 transporter complex itself functioning.
TABLE 5.
Inducibility of promoters regulating the transcription of transporter genes of P. putida KT2440 in HMM and LB media at different stages of growth
| Medium, promoter, and phase | Background luminescence (RLU)a | Maximal induction coefficient (fold) withb:
|
|||||
|---|---|---|---|---|---|---|---|
| Zn2+ | Cd2+ | Pb2+ | Co2+ | Ni2+ | Hg2+ | ||
| HMM medium | |||||||
| PcadA1 | |||||||
| Lag phase | 30 | 4.0 ± 1.3 | |||||
| Early exponential phase | 45 | 22 ± 13 | |||||
| Exponential phase | 30 | 26 ± 5 | 2.2 ± 0.2 | ||||
| Late exponential phase | 30 | 27 ± 6 | |||||
| Stationary phase | 25 | 11 ± 2 | |||||
| PcadA2 | |||||||
| Lag phase | 9,000 | 3.7 ± 0.1 | 6.6 ± 2.5 | ||||
| Early exponential phase | 20,000 | 13 ± 7 | 5.1 ± 1.7 | 3.1 ± 1.0 | |||
| Exponential phase | 35,000 | 9.4 ± 0.1 | 6.1 ± 1.0 | 5.1 ± 0.1 | 2.7 ± 0.6 | ||
| Late exponential phase | 40,000 | 2.2 ± 0.1 | 6.8 ± 1.4 | 3.5 ± 0.8 | 2.2 ± 0.7 | ||
| Stationary phase | 50,000 | 2.1 ± 0.1 | 5.1 ± 1.2 | 2.1 ± 0.6 | 3.0 ± 0.3 | 2.5 ± 0.5 | |
| Pczc1 | |||||||
| Lag phase | 200 | 69 ± 24 | 16 ± 7 | 8.0 ± 2.3 | 5.4 ± 1.4 | 13 ± 8 | |
| Early exponential phase | 400 | 280 ± 60 | 52 ± 8 | 15 ± 5 | 13 ± 2 | 40 ± 15 | 3.4 ± 0.2 |
| Exponential phase | 250 | 590 ± 70 | 120 ± 20 | 7.8 ± 5.6 | 220 ± 80 | ||
| Late exponential phase | 200 | 1100 ± 200 | 23 ± 10 | 4.7 ± 0.1 | |||
| Stationary phase | 200 | 950 ± 220 | 2.4 ± 0.5 | 2.6 ± 1.4 | |||
| LB medium | |||||||
| PcadA1 | |||||||
| Lag phase | 50 | 35 ± 16 | |||||
| Early exponential phase | 250 | 78 ± 20 | 5.5 ± 2.2 | ||||
| Exponential phase | 400 | 78 ± 23 | |||||
| Stationary phase | 400 | 77 ± 30 | |||||
| PcadA2 | |||||||
| Lag phase | 8,000 | 2.3 ± 0.1 | 3.3 ± 0.5 | 3.7 ± 0.3 | 2.0 ± 0.3 | 2.9 ± 1.3 | 2.3 ± 0.1 |
| Early exponential phase | 85,000 | 3.9 ± 0.5 | 3.3 ± 0.3 | 2.6 ± 0.7 | 5.6 ± 1.4 | ||
| Exponential phase | 400,000 | 2.0 ± 0.4 | 3.1 ± 0.2 | 3.2 ± 0.3 | 2.5 ± 0.1 | ||
| Stationary phase | 350,000 | 2.6 ± 0.1 | 3.9 ± 0.2 | 3.8 ± 0.9 | 2.1 ± 0.2 | ||
| Pczc1 | |||||||
| Lag phase | 300 | 120 ± 10 | 30 ± 8 | 7.5 ± 0.7 | 3.2 ± 0.1 | ||
| Early exponential phase | 3,000 | 130 ± 30 | 43 ± 1 | 14 ± 3 | 6.4 ± 0.4 | 30 ± 6 | 17 ± 3 |
| Exponential phase | 4,000 | 450 ± 110 | 18 ± 12 | 3.1 ± 0.4 | 4.6 ± 1.7 | 3.4 ± 1.5 | 2.7 ± 0.1 |
| Stationary phase | 1,500 | 510 ± 180 | |||||
Bacterial luminescence in a metal-free environment (water), representing the noninduced level of expression from the promoter. RLU, relative light units.
The data indicate by how much the bacterial luminescence in metal solutions exceeded the background luminescence. The maximal induction coefficient is shown, and in different cases it was measured with different metal concentrations; only induction coefficients greater than 2 are shown. The data are the means ± standard deviations of three independent experiments.
We did not test complementation of the loss of lead resistance. Although there is no inorganic phosphate in HMM medium, precipitation of lead still occurs. As our growth inhibition assay was based on measurements of the OD600, lead precipitation interfered with the results and made it practically impossible to detect small changes in bacterial growth.
Regulation of the expression of the cadA1, cadA2, czcCBA1, and czcCBA2 genes in P. putida KT2440.
In order to determine the expression profiles of the transporters studied, we fused their promoter regions to reporter genes and measured the responses in wild-type strain P. putida KT2440 growing in the presence of different heavy metals. Reporter plasmids contained the upstream regions of the transporter genes or operons, cloned in front of the bacterial luciferase (lux) operon. The transcription factors acting on the promoter region were not encoded in the plasmid but were encoded in the chromosome. The expression patterns and inducer specificities were determined by measuring changes in luminescence. All promoters were tested for inducibility with different concentrations of Zn2+ (tested at concentrations ranging from 10 nM to 3 mM with a dilution factor of 3.3), Cd2+ (tested at concentrations ranging from 10 nM to 0.3 mM), Pb2+ (tested at concentrations ranging from 10 nM to 3 mM), Co2+ (tested at concentrations ranging from 100 nM to 0.3 mM), Ni2+ (tested at concentrations ranging from 300 nM to 1 mM), and Hg2+ (tested at concentrations ranging from 10 nM to 10 μM). As bioluminescence is an energy-demanding process, the amount of nutrients in the medium may have an influence on the level of luminescence and consequently also on the measurable induction; therefore, we tested inducibility in both rich and minimal media. The results were confirmed to be due to the effects of metals on specific promoter regions by using a control plasmid with no promoter region in front of the lux operon. No nonspecific increase in luminescence was observed in any of the conditions tested (data not shown).
Rather surprisingly, the inducer specificities and the efficiencies of the promoters studied were found to be dependent on the bacterial growth phase and also somewhat on the growth medium (Table 5). To our knowledge, this is the first time that a clear dependence of expression of metal resistance mechanisms on the growth phase has been shown.
The promoter of the cadA1 gene, PcadA1, was inducible with Zn2+ in all growth phases. The highest levels of induction (27-fold in minimal HMM medium and 78-fold in LB medium) were observed in the exponential growth phase. Cd2+ also acted as an inducer of the PcadA1 promoter, but only in the exponential phase and at a very low level; other metals had no effect on the expression from PcadA1. This promoter exhibited very low background luminescence, indicating that it was very strongly controlled with practically no leakage. In contrast, the promoter for the other ATPase, PcadA2, was inducible by all metals tested. The highest level of expression occurred with Cd2+, which acted as an inducer throughout growth in both minimal and rich media. Pb2+ notably induced expression in the lag phase and during exponential growth in minimal medium, whereas in rich medium there was stable low-level induction by Pb2+ in all growth phases. Co2+ and Hg2+ acted as inducers in earlier growth phases in rich medium and later during growth in minimal medium. Surprisingly, Hg2+ was the most effective inducer of PcadA2 in rich medium. Zn2+ was a poor inducer of PcadA2, causing only around twofold induction. Ni2+ induced the expression of PcadA2 only in rich medium and only in the lag phase. In general, the maximal level of induction of PcadA2 was rather low, 13-fold in minimal medium and only 5-fold in rich medium. This could be explained by the relatively high level of background luminescence or expression (1,000-fold higher than that for PcadA1), which set limits on the additional observable expression. In other words, the PcadA2 promoter was not very strongly controlled and was expressed quite highly even without metal induction.
As observed with PcadA2, expression from the promoter of the czcCBA1 operon, Pczc1, was nonspecific. This promoter was inducible with every metal tested. However, the expression profile with Zn2+ induction clearly differed from the expression profiles obtained using other metals. In minimal medium Zn2+ caused more than 1,000-fold induction and was the only metal which acted as an effective inducer in stationary phase (Table 5). All other metals exhibited maximal induction in the exponential growth phase, and thereafter the induction efficiency gradually decreased. The results were quite similar in rich medium, with some exceptions. In minimal medium, the Pczc1 promoter inducer efficiency decreased in the order Zn ≫ Ni > Cd ≫ Pb > Co > Hg, whereas in rich medium the efficiencies were somewhat different and inducer efficiency decreased in the order Zn ≫ Cd > Ni > Hg > Pb > Co. A lack of specificity of heavy metal-regulated promoters has also been reported previously (4, 18, 28, 38).
The fourth promoter studied, Pczc2, was not inducible with any metal tested. Although metals did not cause any increase in the level of expression from Pczc2, this promoter showed relatively high background expression that was 10-fold higher than that observed with the Pczc1 promoter (data not shown). This implies that some amounts of the CzcC2, CzcB2, and CzcA2 proteins might be continuously synthesized by the cell.
The expression profiles showed that the expression of cadA1, cadA2, and czcCBA1 was regulated in a metal-responsive manner. In silico studies have shown that cadA2 and czcCBA1 are associated with transcription factor genes (cadR2 and czcRS1, respectively) located close to transporter genes in the chromosome (6). No transcription factor has been associated with CadA1, and no open reading frame coding for a putative transcription factor has been found close to the cadA1 gene in the chromosome. Our results showed clearly, however, that expression from the cadA1 promoter is regulated in a Zn-responsive manner. Expression of the fourth transporter studied, czcCBA2, was not affected by the presence of metals in the growth environment. This might have been due to a nonfunctional promoter in terms of heavy metal regulation or to the lack of the transcription factor. As also found for cadA1, no transcription factor gene is associated with the czcCBA2 operon. CBA transporters are regulated by two-component mechanisms consisting of a membrane-localized sensor kinase (the S component) and the cognate response regulator (the R component) (12, 21, 36). Although three czcR genes and two czcS genes have been found in the chromosome of P. putida KT2440 (6), they appear to have no influence on expression from Pczc2.
Although expression from PcadA2 and Pczc1 was nonspecific, these two promoters each had a preferred metal inducer in the minimal medium (Cd and Zn, respectively). This was expected based on the substrate preferences of the transporters; CadA2 protects cells mainly from cadmium toxicity, and CzcCBA1 protects cells mainly from zinc toxicity. The very high level of background expression (not induced) from PcadA2 suggests that CadA2 is quite abundant in the cell even when extracellular metals are absent. It is probable that CadA2 is continuously expressed and quickly exports lower concentrations of Cd2+ and Pb2+, thus giving the host the ability to colonize areas polluted with low levels of metal without any change in the CadA2 transcription level. When metal concentrations increase, additional transporter synthesis is initiated. Unlike other promoters, PcadA1 was specific, responding at a significant level only to Zn2+. The level of expression remained very low, however, compared to the levels of expression of the cadA2 and czcCBA1 promoters. It may be that CadA1 is functional but that the CadA1 level in the cell is so low, even after induction by zinc, that CadA1 does not contribute much to metal resistance. The expression from PcadA1 differed in many ways from that from other promoters. First, PcadA1 was very tightly regulated, and practically no expression was detected in metal-free environments. Second, PcadA1 was very specific, responding only to Zn2+. Finally, PcadA1 was the only promoter which showed better induction in rich medium. These differences indicate that some type of transcription factor other than a factor belonging to the MerR family, to which, for example, CadR2 belongs, might be involved in the regulation of PcadA1.
Functionality of P. putida transporters in E. coli.
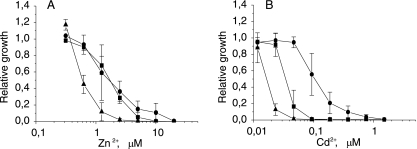
We also tested P. putida KT2440 transporter functions and promoter expression in E. coli to see if the genes or operons and promoters were functional in a recombinant host. To express the czcCBA transporters in E. coli, we constructed new overexpression plasmids, in which the C and B components of the transporter were included in addition to the A subunit. For testing, we used E. coli TH201, in which both known zinc resistance mechanisms (zntA and zitB) were disrupted. In addition to resistance to Zn and Cd, which had been shown to be substrates in P. putida, we also tested resistance to Co, as this metal is normally a substrate of czc transporters (23, 27).
Expression of cadA2 improved resistance to Zn and Cd, while expression of cadA1 conferred resistance to Zn and slight resistance to Cd in E. coli (Fig. 3), indicating that CadA1 is a fully functional transporter, which could not be proven with P. putida KT2440. CadA1 and CadA2 were equally efficient in detoxifying zinc in E. coli as the levels of resistance to Zn observed were similar (Fig. 3A). This contradicts the results obtained with P. putida KT2440, in which these transporters were not involved in zinc resistance at all, showing that the transporters have different functions in different hosts. The CBA transporters CzcCBA1 and CzcCBA2 had no effect on E. coli heavy metal resistance, indicating that these transporters are not functional in E. coli (Table 4). The promoters of czcCBA1, czcCBA2, and cadA1 were not inducible in E. coli. In addition, their levels of expression were very low (data not shown), indicating that they were not recognized as promoters in E. coli. On the other hand, PcadA2 was found to be inducible with Zn2+, Cd2+, and Pb2+ in E. coli. The levels of expression were very low, however, compared to the levels in P. putida KT2440 (data not shown). The functionality of CadA2 in E. coli could be explained by the fact that it is very similar to ZntA, a Zn/Cd/Pb-translocating P-type ATPase from E. coli (29). Also, the CadA2 promoter region and the controlling transcription factor resemble those of ZntA (3, 4), explaining the inducibility of PcadA2 in E. coli.
FIG. 3.
Effects of zinc (A) and cadmium (B) on the growth of the E. coli TH201 transporter-deficient strain overexpressing P. putida KT2440 transporter genes from a temperature-inducible plasmid. Dose-response curves for control strain E. coli TH201(pTIHis) (▴), a strain overexpressing the cadA2 gene [E. coli TH201(pTIcadA2His)] (•), and a strain overexpressing the cadA1 gene [E. coli TH201(pTIcadA1His)] (▪) grown at 37°C are shown. The relative growth indicates the growth efficiency in the presence of a metal compared to the growth in medium without the metal. The data are the means ± standard deviations of at least three independent experiments.
Conclusions.
Our results indicate that in one way or another all four transporters studied, CadA1, CadA2, CzcCBA1, and CzcCBA2, are functional. CadA2 is constantly expressed at a high level and acts as a housekeeping resistance mechanism against Cd and Pb in P. putida KT2440. When the substrate metals are present in the growth environment, the expression of CadA2 increases. Although we were unable to show that CadA2 participates in zinc resistance in P. putida KT2440, CadA2 clearly conferred resistance to zinc in E. coli. In P. putida KT2440, zinc resistance was facilitated by CzcCBA1. Although several metals induced CzcCBA1 expression, the best inducer was undoubtedly zinc, which was also the main substrate of the transporter. Also, cadmium and possibly lead were transported by CzcCBA1. It is probable that CzcCBA1 is rather inefficient when it acts as a cadmium or lead transporter and that it is able to transport only small amounts of periplasmic Cd2+ or Pb2+. All the ions which escape CzcCBA1 are removed from the cytoplasm by CadA2. In addition, CzcCBA2 also conferred zinc resistance. The CzcCBA2 functionality was observed only in a czcA1-defective background or with overexpression of CzcA2 in a transporter-deficient strain. This suggests that although CzcCBA2 is a functional Zn transporter, the levels of it are too low to markedly affect Zn resistance properties of the cell. The level of expression of CzcCBA2 could not be altered by any metal tested, indicating that there was not a metal-responsive transcription operator for the czcCBA2 promoter. It is unclear whether the subunits of the two CBA transporters are interchangeable (that is, whether CzcA2 can complex with CzcCB1). If they are, then it seems that the CzcCB1A2 complex is much more efficient at Zn transportation than CzcCBA1, as lower levels of the CzcCB1A2 transporter are required to confer efficient zinc resistance. The fourth transporter, CadA1, did not confer tolerance to any metal in P. putida KT2440. CadA1 expression was, however, inducible by zinc, and when CadA1 was overexpressed in E. coli, Zn resistance was apparent. We therefore assumed that CadA1 is a Zn-responsive Zn transporter, but the level of CadA1, even after induction, is so low in P. putida KT2440 that CadA1 cannot significantly reduce the intracellular concentration of zinc. However, the possibility that CadA1 is highly expressed under certain growth conditions not examined in this study cannot be ruled out. Further studies of the transcription factor active on the cadA1 promoter would clarify this point.
Finally, although there are four functional transporters with overlapping functions, all encoded in the chromosome, which is quite a high number for a bacterium which should keep its chromosome as small as possible, the possibility that there are additional divalent heavy metal resistance mechanisms that remain to be discovered in P. putida KT2440 cannot be ruled out. Compared to E. coli, the quadruple-knockout strain P. putida KT2440.2431 can tolerate 100-fold-higher concentrations of cadmium and 4-fold-higher concentrations of zinc (Table 4). Some possible candidates for additional resistance mechanisms are CzcD, a CDF transporter, metallothionein, and two additional czc chemiosmotic antiporters, as found in silico by Canovas et al. (6). The functionalities and substrates of these candidate mechanisms remain to be determined.
Acknowledgments
We thank Herbert P. Schweizer for providing plasmids pEX18Tc and pFLP2. Triinu Simkin is thanked for technical assistance.
The study was financially supported by the EnSTe Graduate School (Finland), the Archimedes Foundation (Estonia), and the Academy of Finland (grants 201677 and 207258). Additional support was provided by the Estonian Science Foundation, by Estonian Ministry of Education targeted funding grant 0222601Bs03, and by the French-Estonian collaborative PARROT program.
Footnotes
Published ahead of print on 7 December 2007.
REFERENCES
- 1.Anton, A., C. Grosse, J. Reissmann, T. Pribyl, and D. H. Nies. 1999. CzcD is a heavy metal ion transporter involved in regulation of heavy metal resistance in Ralstonia sp. strain CH34. J. Bacteriol. 1816876-6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagdasarian, M., R. Lurz, B. Ruckert, F. C. Franklin, M. M. Bagdasarian, J. Frey, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16237-247. [DOI] [PubMed] [Google Scholar]
- 3.Binet, M. R., and R. K. Poole. 2000. Cd(II), Pb(II) and Zn(II) ions regulate expression of the metal-transporting P-type ATPase ZntA in Escherichia coli. FEBS Lett. 47367-70. [DOI] [PubMed] [Google Scholar]
- 4.Brocklehurst, K. R., J. L. Hobman, B. Lawley, L. Blank, S. J. Marshall, N. L. Brown, and A. P. Morby. 1999. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol. Microbiol. 31893-902. [DOI] [PubMed] [Google Scholar]
- 5.Caille, O., C. Rossier, and K. Perron. 2007. A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa. J. Bacteriol. 1894561-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canovas, D., I. Cases., and V. de Lorenzo. 2003. Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ. Microbiol. 51242-1256. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138179-207. [DOI] [PubMed] [Google Scholar]
- 8.Choi, K. H., A. Kumar, and H. P. Schweizer. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64391-397. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franke, S., G. Grass, C. Rensing, and D. H. Nies. 2003. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 1853804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grass, G., B. Fan, B. P. Rosen, S. Franke, D. H. Nies, and C. Rensing. 2001. ZitB (YbgR), a member of the cation diffusion facilitator family, is an additional zinc transporter in Escherichia coli. J. Bacteriol. 1834664-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grosse, C., G. Grass, A. Anton, S. Franke, A. N. Santos, B. Lawley, N. L. Brown, and D. H. Nies. 1999. Transcriptional organization of the czc heavy-metal homeostasis determinant from Alcaligenes eutrophus. J. Bacteriol. 1812385-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassan, M. T., D. van der Lelie, D. Springael, U. Romling, N. Ahmed, and M. Mergeay. 1999. Identification of a gene cluster, czr, involved in cadmium and zinc resistance in Pseudomonas aeruginosa. Gene 238417-425. [DOI] [PubMed] [Google Scholar]
- 14.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 21277-86. [DOI] [PubMed] [Google Scholar]
- 15.Hu, N., and B. Zhao. 2007. Key genes involved in heavy-metal resistance in Pseudomonas putida CD2. FEMS Microbiol. Lett. 26717-22. [DOI] [PubMed] [Google Scholar]
- 16.Jechlinger, W., J. Glocker, W. Haidinger, A. Matis, M. P. Szostak, and W. Lubitz. 2005. Modulation of gene expression by promoter mutants of the lambda cI857/pRM/pR system. J. Biotechnol. 11611-20. [DOI] [PubMed] [Google Scholar]
- 17.LaRossa, R. A., D. R. Smulski, and T. K. Van Dyk. 1995. Interaction of lead nitrate and cadmium chloride with Escherichia coli K-12 and Salmonella typhimurium global regulatory mutants. J. Ind. Microbiol. 14252-258. [DOI] [PubMed] [Google Scholar]
- 18.Lee, S. W., E. Glickmann, and D. A. Cooksey. 2001. Chromosomal locus for cadmium resistance in Pseudomonas putida consisting of a cadmium-transporting ATPase and a MerR family response regulator. Appl. Environ. Microbiol. 671437-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leedjärv, A., A. Ivask, M. Virta, and A. Kahru. 2006. Analysis of bioavailable phenols from natural samples by recombinant luminescent bacterial sensors. Chemosphere 641910-1919. [DOI] [PubMed] [Google Scholar]
- 20.Legatzki, A., G. Grass, A. Anton, C. Rensing, and D. H. Nies. 2003. Interplay of the Czc system and two P-type ATPases in conferring metal resistance to Ralstonia metallidurans. J. Bacteriol. 1854354-4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munson, G. P., D. L. Lam, F. W. Outten, and T. V. O'Halloran. 2000. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 1825864-5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nies, D. H. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27313-339. [DOI] [PubMed] [Google Scholar]
- 23.Nies, D. H., and S. Silver. 1989. Plasmid-determined inducible efflux is responsible for resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus. J. Bacteriol. 171896-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nies, D. H., A. Nies, L. Chu, and S. Silver. 1989. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc. Natl. Acad. Sci. USA 867351-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nucifora, G., L. Chu, T. K. Misra, and S. Silver. 1989. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc. Natl. Acad. Sci. USA 863544-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nunn, D., S. Bergman, and S. Lory. 1990. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J. Bacteriol. 1722911-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perron, K., O. Caille, C. Rossier, C. Van Delden, J. L. Dumas, and T. Kohler. 2004. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J. Biol. Chem. 2798761-8768. [DOI] [PubMed] [Google Scholar]
- 28.Ralston, D. M., and T. V. O'Halloran. 1990. Ultrasensitivity and heavy-metal selectivity of the allosterically modulated MerR transcription complex. Proc. Natl. Acad. Sci. USA 873846-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rensing, C., B. Mitra, and B. P. Rosen. 1997. The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc. Natl. Acad. Sci. USA 9414326-14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossbach, S., T. L. Wilson, M. L. Kukuk, and H. A. Carty. 2000. Elevated zinc induces siderophore biosynthesis genes and a zntA-like gene in Pseudomonas fluorescens. FEMS Microbiol. Lett. 19161-70. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Sharma, R., C. Rensing, B. P. Rosen, and B. Mitra. 2000. The ATP hydrolytic activity of purified ZntA, a Pb(II)/Cd(II)/Zn(II)-translocating ATPase from Escherichia coli. J. Biol. Chem. 2753873-3878. [DOI] [PubMed] [Google Scholar]
- 33.Silver, S., and L. T. Phung. 2005. A bacterial view of the periodic table: genes and proteins for toxic inorganic ions. J. Ind. Microbiol. Biotechnol. 32587-605. [DOI] [PubMed] [Google Scholar]
- 34.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406959-964. [DOI] [PubMed] [Google Scholar]
- 35.Timmis, K. N. 2002. Pseudomonas putida: a cosmopolitan opportunist par excellence. Environ. Microbiol. 4779-781. [DOI] [PubMed] [Google Scholar]
- 36.van der Lelie, D., T. Schwuchow, U. Schwidetzky, S. Wuertz, W. Baeyens, M. Mergeay, and D. H. Nies. 1997. Two-component regulatory system involved in transcriptional control of heavy-metal homoeostasis in Alcaligenes eutrophus. Mol. Microbiol. 23493-503. [DOI] [PubMed] [Google Scholar]
- 37.Xiong, A., and R. K. Jayaswal. 1998. Molecular characterization of a chromosomal determinant conferring resistance to zinc and cobalt ions in Staphylococcus aureus. J. Bacteriol. 1804024-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon, K. P., T. K. Misra, and S. Silver. 1991. Regulation of the cadA cadmium resistance determinant of Staphylococcus aureus plasmid pI258. J. Bacteriol. 1737643-7649. [DOI] [PMC free article] [PubMed] [Google Scholar]