Abstract
Bacteria can reduce toxic and carcinogenic Cr(VI) to insoluble and less toxic Cr(III). Thermus scotoductus SA-01, a South African gold mine isolate, has been shown to be able to reduce a variety of metals, including Cr(VI). Here we report the purification to homogeneity and characterization of a novel chromate reductase. The oxidoreductase is a homodimeric protein, with a monomer molecular mass of approximately 36 kDa, containing a noncovalently bound flavin mononucleotide cofactor. The chromate reductase is optimally active at a pH of 6.3 and at 65°C and requires Ca2+ or Mg2+ for activity. Enzyme activity was also dependent on NADH or NADPH, with a preference for NADPH, coupling the oxidation of approximately 2 and 1.5 mol NAD(P)H to the reduction of 1 mol Cr(VI) under aerobic and anaerobic conditions, respectively. The Km values for Cr(VI) reduction were 3.5 and 8.4 μM for utilizing NADH and NADPH as electron donors, respectively, with corresponding Vmax values of 6.2 and 16.0 μmol min−1 mg−1. The catalytic efficiency (kcat/Km) of chromate reduction was 1.14 × 106 M−1 s−1, which was >50-fold more efficient than that of the quinone reductases and >180-fold more efficient than that of the nitroreductases able to reduce Cr(VI). The chromate reductase was identified to be encoded by an open reading frame of 1,050 bp, encoding a single protein of 38 kDa under the regulation of an Escherichia coli σ70-like promoter. Sequence analysis shows the chromate reductase to be related to the old yellow enzyme family, in particular the xenobiotic reductases involved in the oxidative stress response.
Hexavalent chromium [Cr(VI)] is acutely toxic and is a known carcinogen (6). Cr(VI) has become a common environmental metal contaminant through the widespread use of Cr(VI) compounds by various industries as well as through their incorrect disposal (17). Cr(VI) usually occurs as the strongly oxidizing oxyanions chromate (CrO42−) and dichromate (Cr2O72−) (5), which are highly mobile within groundwater systems (17). In contrast, Cr(III) is relatively immobile in soils, as it is readily adsorbed by soil or forms insoluble oxides and hydroxides (5) considered to be relatively innocuous.
The structural similarity of chromate to SO42− allows for the uptake of Cr(VI) into bacteria via the sulfate transport system (5). Intracellular reduction through physiological reducing agents to transiently formed intermediates, such as Cr(V), and related free radical generation are considered to be major causes of Cr(VI) toxicity and carcinogenesis (6, 16). A variety of bacteria have been shown to mediate Cr(VI) reduction, and several enzymes with diverse physiological functions (25), including glutathione reductase, lipoyl reductase, and ferredoxin-NADP+ reductase, have been shown to be able to catalyze chromate reduction. Fortuitous chromate reduction via the nitroreductases (NfsA) of Pseudomonas ambigua, Escherichia coli, and Vibrio harveyi has also been demonstrated (1, 14, 27). The chromate reductases (ChrR) of Pseudomonas putida and E. coli reduce Cr(VI) to Cr(III) (2, 22), but catalytic and sequence similarities indicate these enzymes to be probable quinone reductases. Regardless, these reductases have been shown to be able to protect against chromate toxicity through decreased reactive oxygen species (ROS) formation (2).
Old yellow enzyme (OYE) from Saccharomyces carlsbergensis was the first flavin oxidoreductase identified (32). OYE homologues usually occur as homodimers with a noncovalently bound flavin mononucleotide (FMN) cofactor. The enzyme follows a bi-bi ping-pong mechanism, with NAD(P)H as the presumed physiological reductant. Despite over 70 years of research, the physiological oxidants for OYE as well as most members of the OYE family are still unknown (33). Although molecular oxygen can serve as an electron acceptor, the low reaction rate as well as the production of H2O2 makes it an unlikely physiological oxidant. Other substrates capable of reoxidizing OYE include methylene blue, Fe3+, quinones, cytochrome c, and ferricyanide. Bacterial homologues include, among others, the morphinone reductase (8), xenobiotic reductases (9), and, more recently, the Bacillus subtilis YqjM protein (7). The involvement of many of the OYE homologues in the oxidative stress response as well as the range of electrophilic substrates during reactions, such as the reduction of the olefinic bond of α,β-unsaturated carbonyl compounds, the reductive denitration of aliphatic nitro esters, and the reduction of nitroaromatic compounds, could suggest a detoxification enzyme in the antioxidant defense system (4, 7, 9, 12, 15).
The South African gold mine isolate Thermus scotoductus SA-01 can reduce a variety of heavy metals (11, 18), including Cr(VI) (21). This paper reports the purification, characterization, and sequence analysis of a novel cytoplasmic chromate reductase from T. scotoductus SA-01 related to OYE.
MATERIALS AND METHODS
Organism, growth conditions, and plasmids.
Thermus scotoductus SA-01 (ATCC 700910; American Type Culture Collection) was cultured in a complex organic medium, TYG (5 g tryptone [Biolab, Wadeville, South Africa], 3 g yeast extract [Saarchem, Wadeville, South Africa], and 1 g glucose in 1 liter double-distilled H2O), at 65°C, pH 7.0, with aeration (200 rpm). pTrueBlue (GenomicsOne, Laval, Canada) vector (25) and One Shot TOP10 Escherichia coli competent cells (Invitrogen, Carlsbad, CA) were used for Thermus scotoductus SA-01 genomic library construction and cultured on Luria-Bertani (LB) liquid medium with an appropriate antibiotic. For the expression of the recombinant protein, pET-22b(+) (Novagen, Darmstadt, Germany) in E. coli (BL21)DE3 (Lucigen, Middleton, MI) was used.
Enzyme assays.
Chromate reductase activity was determined by measuring the decrease of hexavalent chromium by the s-diphenylcarbazide method (30). During purification, the enzyme was assayed in 1-ml reaction mixtures containing 20 mM MOPS-NaOH buffer, 10 mM CaCl2 (pH 6.5), 0.1 mM CrO3, 0.3 mM NADPH, and 0.05 ml of the enzyme preparation at 65°C. Samples (0.3 ml) were withdrawn and added to 2.5 ml of a 0.12 M H2SO4 stock solution. A 0.2-ml volume of the sym-diphenylcarbazide reagent (dissolved in acetone) was added to the reaction mixture to a final concentration of 0.4 mM. Absorbance was measured using a Spectronic Genesys 5 spectrophotometer (Milton Roy Company, NY) at 540 nm. One unit is defined as the amount of enzyme required to reduce 1 μmol of Cr(VI) per minute.
The pH optimum of the enzyme was determined in an equimolar morpholineethanesulfonic acid (MES)-morpholinepropanesulfonic acid (MOPS)-Bicine (20 mM) buffer at 65°C. The temperature optimum of the enzyme was analyzed by adjusting the pH of a 20 mM MOPS buffer to pH 6.5 at various temperatures. Kinetic parameters were determined at the optimum pH and temperature over a 5-min incubation.
Purification of Cr(VI) reductase.
All purification steps were carried out at room temperature with enzyme preparations stored at 4°C. Purification to homogeneity entailed five chromatographic steps, including anion-exchange, hydrophobic-interaction, dye-affinity, and size-exclusion chromatography, using an ACTA Prime purification system (GE Healthcare AB, Uppsala, Sweden).
Thermus scotoductus SA-01 was fractionated into periplasmic, cytoplasmic, and membrane fractions as previously described (23). Crude cytoplasmic proteins were applied to a DEAE-Toyopearl 650 M column (6 × 2.5 cm; Tosoh Corporation, Tokyo, Japan) equilibrated with 20 mM MOPS-NaOH buffer (pH 7; buffer A). Unbound proteins were eluted with 200 ml of the same buffer. Bound proteins were eluted with an isocratic NaCl concentration of 60 mM (100 ml) in the same buffer (flow rate, 300 ml h−1). Strongly bound proteins were eluted with buffer A containing 1 M NaCl (100 ml). Fractions with Cr(VI) reduction activity were pooled, dialyzed against 20 mM MOPS (pH 7.0), reloaded onto the DEAE column, and again eluted with a linear NaCl gradient of 0 to 0.1 M NaCl in buffer A (400 ml). Active fractions were pooled, and (NH4)2SO4 was added to a final concentration of 0.4 M.
For hydrophobic-interaction chromatography, a phenyl-Toyopearl 650 M (6 × 2.5 cm; Tosoh Corporation, Tokyo, Japan) column was equilibrated with 0.4 M (NH4)2SO4 in buffer A. Unbound proteins were eluted with the equilibration buffer (200 ml; flow rate, 300 ml h−1), after which bound proteins were eluted with a 200-ml linear gradient of 0.4 to 0 M (NH4)2SO4.
The dye-affinity chromatography step employed a Blue Sepharose CL-6B (Sigma-Aldrich, Steinheim, Germany) column (17 × 1.3 cm) equilibrated with buffer A (flow rate, 120 ml h−1). After binding of the dialyzed sample, the column was washed sequentially with 100 ml buffer A containing 0.1 M NaCl and buffer A containing 10 mM NAD+. The chromate reductase was then eluted with 100 ml of buffer A containing both 0.1 M NaCl and 10 mM NAD+. Active fractions were pooled and concentrated to 3 ml using ultrafiltration on a 10-kDa-cutoff cellulose membrane Vivaspin column (VivaScience, Hanover, Germany).
The final purification step was size-exclusion chromatography, whereby the native molecular mass of the chromate reductase was also determined. The concentrate was loaded onto a Sephacryl S-100 HR column (2.6 × 62 cm; Sigma-Aldrich, St. Louis, MO) equilibrated with 20 mM MOPS-NaOH (pH 7.0) containing 50 mM NaCl. Proteins were eluted with the same buffer at a flow rate of 0.5 ml min−1. Cytochrome c (12.4 kDa), chymotrypsin (25 kDa), and bovine serum albumin (66 kDa) were used as molecular mass standards.
Gel electrophoresis.
Electrophoresis under denaturing conditions or sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to the protocol described by Laemmli (15), using a 10% resolving gel and a 4% stacking gel. Precision Plus protein standards (Bio-Rad, Hercules, CA) were used as molecular mass markers, and proteins were visualized by staining of the polyacrylamide gels with Coomassie brilliant blue R-250.
Determination of flavin content.
UV-visible spectra of the purified enzyme were recorded on a Beckman Coulter DU-800 spectrophotometer (Fullerton, CA). Flavin cofactor content was determined through thin-layer chromatography. The cofactor was released from the enzyme by autoclaving the sample, followed by centrifugation (14,000 × g for 10 min). The supernatant was applied to Silicagel 60 thin-layer chromatography aluminum plates (Merck, Darmstadt, Germany), using butanol-acetic acid-water (12:3:5) as the solvent. FMN and flavin adenine dinucleotide (FAD) were used as standards, and fluorescence spots were visualized using UV light.
Determination of N-terminal amino acid sequences.
N-terminal amino acid sequencing was performed using an Applied Biosystems 4774A gas-phase sequencer (Foster City, CA) at the Protein Chemistry Facility of the Centro de Investigaciones Biologicas (CSIC, Madrid, Spain).
DNA library construction.
Total genomic DNA (gDNA) of SA-01 was isolated using a modified SDS-proteinase K treatment method (29) and restriction digested using the endonuclease Sau3AI (New England BioLabs, Beverly, MA), using serial dilutions of enzyme ranging from 250 U ml−1 to 4.6 U ml−1 (15 min at 37°C). Digested DNA fragments ranging from 3 to 6 kbp were excised from the agarose gel and purified using a GFX PCR DNA and gel band purification kit (GE Healthcare, Buckinghamshire, United Kingdom).
pTrueBlue was linearized using BamHI (New England Biolabs) for 3 h at 37°C (1 U μl−1), followed by dephosphorylation using Antarctic phosphatase (New England BioLabs) (0.93 U μl−1; 4 h at 37°C). Size-fractionated DNA fragments were ligated into pTrueBlue at 1:2 and 1:3 vector/insert ratios, using an average 4.5-kb insert size for conversion of molar ratios to mass ratios, with T4 DNA ligase (New England BioLabs) (6 Weiss units) at 16°C for 12 h. Again, the plasmids were purified using a GFX PCR DNA and gel band purification kit.
Ligation mixtures were transformed into One Shot TOP10 E. coli competent cells (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations. Transformed cells were plated onto LB medium plates (100 μl plate−1) containing ampicillin (0.06 mg ml−1), isopropyl-β-d-thiogalactopyranoside (IPTG; 0.01 mg ml−1), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (0.04 mg ml−1) and incubated overnight at 37°C to verify DNA ligation using blue-white selection.
Screening of genome library.
An oligonucleotide probe was designed from the N-terminal amino acid sequence of the chromate reductase. A codon usage table was constructed from four complete protein coding sequences available for Thermus scotoductus from the GenBank DNA sequence database. Codon preference was analyzed using Kazusa CountCodon (20). The degenerate oligonucleotide probe (5′ GCC CTS CTS TTY ACS CCC CTS GAD GGS 3′, where S = G/C, Y = C/T, and D = A/G/T) was labeled with digoxigenin (DIG) at both the 5′ and 3′ ends and purified by high-performance liquid chromatography (TIB Molbiol, Berlin, Germany).
Colony hybridization using the DIG-labeled oligonucleotide probe was done as described in the DIG application manual for filter hybridization (Roche, Mannheim, Germany). Membranes were hybridized for 4 h with 10 ml DIG Easy Hyb solution containing 25 ng ml−1 DIG-labeled DNA probe at 33°C. Anti-DIG antibodies conjugated with alkaline phosphatase were used for the detection of DIG-labeled oligonucleotide probes. The membranes were incubated overnight with 4-nitroblue tetrazolium chloride (0.34 mg ml−1) and 5-bromo-4-chloro-3-indolylphosphate (0.175 mg ml−1) for colorimetric detection, and corresponding positive clones were identified on the master plates. Positive clones were inoculated into 5 ml LB medium supplemented with 0.1 mg ml−1 ampicillin and grown overnight at 37°C. Plasmid extractions were performed using a GeneJET plasmid miniprep kit (Fermentas, Glen Burnie, MD) per the manufacturer's instructions.
DNA sequencing and analysis.
Plasmid inserts were sequenced by Inqaba Biotechnical Industries (South Africa), using a Spectrumedix SCE2410 genetic analysis system (SpectruMedix, State College, PA). Sequencing reactions were performed using a BigDye (v3.1) Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA), using the universal T7 promoter primer and the pTrueBlueRev primer (5′-GGGATGCGCAGCTAATC-3′), which were located on the plasmid vector. Further sequencing was performed using a primer walking strategy. Sequences were analyzed using Vector NTI 9.0.0 (InforMax, Frederick, MD).
Homology searches against the GenBank database were performed using the basic local alignment search tool (BLAST) (3). Multiple alignments were done using CLUSTAL W multiple sequence alignment software (28). Secondary structure prediction was done using 3D-PSSM (10).
Cloning and heterologous expression of the chromate reductase.
The complete open reading frame (ORF) for the chromate reductase was amplified by PCR from genomic DNA, using the Expand high-fidelity PCR system (Roche, Mannheim, Germany). PCRs were performed using the primers CrS_F_Nde (5′ CAT ATG GCC TTG CTC TTC ACC CCC CTG GAA CTC 3′) and CrS_R_Eco (5′ GAA TTC CTA AAA CCC CCT TTG GTA CTG GGG GGG TAC 3′), which contained an NdeI and an EcoRI restriction site, respectively (underlined). Reaction mixtures (50 μl) consisted of 10× Expand high-fidelity buffer with 15 mM MgCl2 (5 μl), deoxynucleoside triphosphates (0.8 mM), Expand high-fidelity enzyme mix (1 μl), 50 ng of gDNA, and 0.2 μM of both the forward and reverse primers. Reaction conditions consisted of an initial denaturing step at 95°C for 5 min, followed by 25 cycles of denaturing at 95°C (30 s), annealing at 65°C (40 s), and elongation at 72°C (1.5 min), with a final extension at 72°C for 10 min.
The PCR product was ligated into pGEM-T Easy (Promega, Madison, WI) vector at a 1:1 molar ratio overnight at 4°C according to the manufacturer's instructions and proliferated in One Shot TOP10 E. coli competent cells (Invitrogen, Carlsbad, CA). Plasmids were isolated using a GeneJet miniprep kit (Fermentas, Glen Burnie, MD). Plasmids containing inserts were double digested with the restriction enzymes NdeI (0.5 U μl−1; Fermentas) and EcoRI (0.5 U μl−1; Fermentas) at 37°C (3 h) for ligation into pET22b(+), which was similarly digested. Inserts and digested pET22b(+) vectors were cleaned from a low-melting-point agarose gel by using a GFX PCR DNA and gel band purification kit. Cohesive end ligations were performed at a 1:1 molar ratio, using 40 ng of vector. Ligations were performed in 20-μl reaction volumes overnight at 4°C with 1.5 Weiss U μl−1 T4 DNA ligase (New England Biolabs, Beverly, MA). Ligation mixtures were again transformed into TOP10 E. coli cells, and positive clones were identified through plasmid isolation and restriction digestion as described above.
The pET22-chromate reductase construct was transformed into E. coli BL21(DE3) competent cells (Lucigen, Middleton, WI) for expression. Positive clones were identified through selection on LB plates containing 100 μg ml−1 ampicillin and inoculated into LB medium also containing the same appropriate antibiotic. Cells were incubated with shaking (200 rpm) until an optical density at 600 nm of approximately 0.8 to 1 was reached. IPTG was added as an inducer to a final concentration of 1 mM, and cells were grown for an additional 4 h. Cells were harvested through centrifugation (8,000 × g, 10 min) and washed using 20 mM MOPS-NaOH (pH 7).
Harvested cells were resuspended in 20 mM MOPS-NaOH (pH 7) and were broken by ultrasonic treatment for 5 min (100 W), after which unbroken cells and debris were removed through centrifugation (8,000 × g for 10 min). The soluble fraction (cytoplasm) was separated from the insoluble fraction (membranes) through ultracentrifugation (100,000 × g, 90 min). The recombinant chromate reductase was purified from the E. coli proteins through a single heat denaturation step of 1.5 h at 70°C.
Analytical techniques.
Protein concentrations were determined using the bicinchoninic acid (BCA) method (26). A BCA protein assay kit and a Micro BCA protein assay kit from Pierce (Rockford, IL) were used according to the manufacturer's instructions, with bovine serum albumin as the standard.
DNA concentrations were determined using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE).
NAD(P)H consumption was related to Cr(VI) reduction by measuring the oxidation of NADH through the decrease in absorbance at 340 nm, using a Beckman Coulter DU-800 spectrophotometer or a Beckman Coulter DU-650 spectrophotometer (Fullerton, CA) within an anoxic Coy anaerobic chamber (Coy Laboratory Products, Grass Lake, MI). The NAD(P)H concentration was determined from the A340 by using the extinction coefficient 6.3 mM cm−1 (34).
Nucleotide sequence accession number.
The DNA sequence reported here has been submitted to the EMBL nucleotide sequence database under accession number AM902709.
RESULTS
Purification of chromate reductase.
Previous studies have shown that Thermus scotoductus SA-01 has more than one protein capable of chromate reduction (21). In addition to the dihydrolipoamide dehydrogenase fortuitously catalyzing Cr(VI) reduction (22), SA-01 also has a constitutively produced cytoplasmic protein capable of chromate reduction.
The cytoplasmic chromate reductase was purified from the crude soluble fraction of T. scotoductus to homogeneity through DEAE-Toyopearl, phenyl-Toyopearl, Blue Sepharose, and Sephacryl S100HR chromatography (Table 1), with an overall purification of 450-fold relative to the crude cytoplasmic fraction and with a final yield of 9.1%.
TABLE 1.
Purification of cytoplasmic chromate reductase from T. scotoductus
| Purification step | Volume (ml) | Total protein (mg) | Total activity (U)a | Sp act (U mg−1) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| Cytoplasm | 150 | 480.0 | 9.42 | 0.02 | 100.0 | 1.0 |
| DEAE-Toyopearl chromatography | 49 | 79.4 | 4.81 | 0.06 | 51.0 | 3.1 |
| 84 | 15.4 | 2.80 | 0.18 | 29.7 | 9.3 | |
| Phenyl-Toyopearl chromatography | 88 | 2.0 | 2.55 | 1.25 | 27.1 | 63.7 |
| Blue Sepharose chromatography | 39.5 | NDb | 0.63 | NDb | 6.7 | NDb |
| Sephacryl S100HR chromatography | 17.5 | 0.1 | 0.85 | 8.84 | 9.1 | 450 |
One unit (U) of activity is defined as the amount of enzyme required to reduce 1 μmol of Cr(VI) per minute, using NADH as the electron donor.
NAD+ interference.
Physical characterization.
The molecular mass of the protein was determined through SDS-PAGE analysis to be approximately 36 kDa. The native molecular mass of the chromate reductase was determined to be 72.4 kDa through size-exclusion chromatography, suggesting a homodimeric quaternary structure for the native protein.
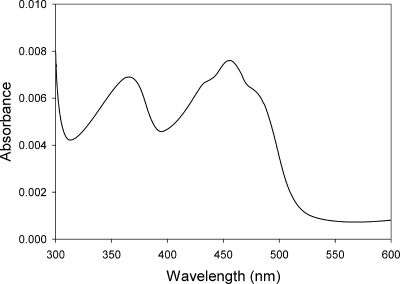
Activity corresponded with a light yellow color resistant to ultrafiltration, with UV-visible absorbance maxima at 375 nm and 460 nm, indicative of flavoproteins (Fig. 1). The noncovalently bound flavin was identified as FMN through thin-layer chromatography (data not shown).
FIG. 1.
UV-visible absorbance spectrum of the purified cytoplasmic chromate reductase from Thermus scotoductus SA-01 (0.02 mg ml−1).
Enzymatic properties.
Chromate reductase activity was optimal at a pH of 6.3. The enzyme was optimally active at 65°C but was also active over a wide temperature range, retaining >70% of its activity at 80°C. Activity was dependent on the presence of the divalent metal Ca2+ or Mg2+, which increased the activity >4-fold, whereas Zn2+, Mn2+, and EDTA inhibited the reaction (Table 2). The chromate reductase could accept electrons from both NADH and NADPH, with a preference toward NADPH.
TABLE 2.
Effects of metals and EDTA on specific activity of chromate reductase
| Metal or EDTA (10 mM) | Sp act (mean ± SD [μmol min−1 mg−1]) |
|---|---|
| None | 3.1 ± 0.5 |
| Ca | 14.5 ± 2.2 |
| Mg | 13.6 ± 0.7 |
| Zn | NAa |
| Mn | 1.5 ± 0.1 |
| EDTA | 1.0 ± 0.6 |
NA, no activity.
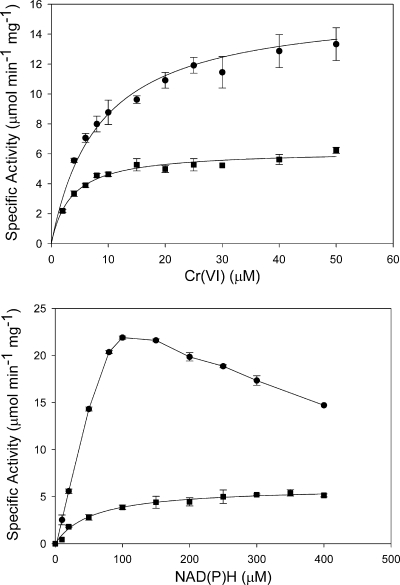
The apparent Km values obtained for Cr(VI) were 3.5 ± 0.3 μM and 8.4 ± 1.1 μM with NADH and NADPH as electron donors, respectively, with corresponding Vmax values of 6.2 ± 0.2 μmol min−1 mg−1 and 16.0 ± 0.6 μmol min−1 mg−1 (Fig. 2, top panel). At concentrations above 0.1 mM, NADPH showed substrate inhibition, but it was still more efficient than NADH at these concentrations (Fig. 2, bottom panel).
FIG. 2.
Steady-state kinetics of purified chromate reductase. Reaction mixtures contained 20 mM MOPS-NaOH (pH 6.5), 10 mM CaCl2, and 0.3 mM NADPH (•) or 0.3 mM NADH (▪) (top) or 0.1 mM Cr(VI) with various NADPH (•) or NADH (▪) concentrations (bottom) and were incubated under optimal conditions. Error bars indicate standard errors.
NAD(P)H consumption and stoichiometry.
Stoichiometric analysis relating NAD(P)H oxidation to Cr(VI) reduction showed that 2.1 ± 0.1 mol NADH or 1.9 ± 0.2 mol NADPH is consumed per mol Cr(VI) reduced under aerobic conditions, whereas only 1.3 ± 0.2 mol NADH or 1.5 ± 0.2 mol NADPH is consumed under anaerobic conditions. Since the reduction of Cr(VI) to Cr(III) requires only three electrons, the fourth electron is most probably involved in ROS formation through the reduction of molecular oxygen. NAD(P)H oxidation with FMN and FAD as electron acceptors was determined due to sequence similarities of the chromate reductase (see below) with predicted NADH-dependent flavin oxidoreductases. Although an increase in NAD(P)H oxidation was observed with FAD and FMN as substrates, the reaction rates were significantly lower than that observed for Cr(VI) (Table 3) and were more comparable to the NAD(P)H oxidase rates, where electrons are donated to oxygen, suggesting that the reactions are fortuitous.
TABLE 3.
Rates of NAD(P)H oxidation by different substrates
| Substrate | Sp act (mean ± SD [μmol min−1 mg−1])
|
|
|---|---|---|
| NADH | NADPH | |
| Cr(VI) | 12.0 ± 1.1 | 122.5 ± 13.0 |
| FAD | 1.0 ± 0.4 | 2.0 ± 0.02 |
| FMN | 1.8 ± 1.0 | 4.2 ± 1.4 |
| Molecular oxygen | 1.1 ± 0.3 | 1.0 |
Sequencing of the gene.
The N-terminal sequence of the purified chromate reductase, determined through Edman degradation, was ALLFTPLELGG. A degenerate oligonucleotide probe labeled with DIG was used to screen a genomic DNA library of Thermus scotoductus for the reductase gene. Colony hybridization of the gDNA library gave two positive clones, both containing inserts of approximately 5 kbp. Sequence analysis of the clones confirmed the presence of the oligonucleotide probe sequence and revealed an ORF of 1,050 bp. A GenBank similarity search (BLASTn) showed the ORF to have the highest sequence identity to a probable NADH-dependent flavin oxidoreductase from Thermus thermophilus HB8 (86%), from whole-genome sequencing. Computational analysis of the chromate reductase gene showed it to encode a single protein of 349 amino acids with a predicted molecular mass of 37.98 kDa, consistent with the experimental molecular mass, and a theoretical pI of 7.83.
A putative ribosome binding site (GGAGG) was identified directly upstream of the ORF but uncharacteristically close to the start codon, with only a 1-nucleotide (nt) separation. The 5′-flanking region of the ORF also included putative E. coli σ70-like −10 and −35 hexamer promoter elements approximately 100 nt upstream of the translation initiation site. A 9-bp inverted repeat was detected immediately downstream of the ORF, indicating a putative transcription termination sequence.
Amino acid sequence comparison.
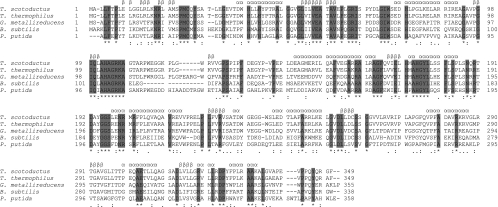
BLAST analysis of the predicted amino acid sequence against the RCSB Protein Data Bank showed the highest sequence identity (50%) to be with an OYE homologue (YqjM) from Bacillus subtilis involved in the oxidative stress response through the reductive degradation of various xenobiotics (7, 12). High identity (45%) with the xenobiotic reductase (XenA) from Pseudomonas putida 86, catalyzing the NAD(P)H-dependent reduction of electrophilic xenobiotics (9), was also found. Multiple alignments of the gene product with the predicted flavin oxidoreductase from T. thermophilus HB8 (NCBI:YP 143423), XenA from P. putida 86 (PDB:2H8X), YqjM from B. subtilis (PDB:1Z41A), and a predicted NADH:flavin oxidoreductase from Geobacter metallireducens (NCBI:YP 385521) are shown in Fig. 3.
FIG. 3.
Multiple alignment (Clustal W) of the chromate reductase with predicted NADH:flavin oxidoreductases from Thermus thermophilus (NCBI:YP 143423) and Geobacter metallireducens (NCBI:YP 385521), the xenobiotic reductase from Pseudomonas putida 86 (PDB:2H8X), and YqjM of Bacillus subtilis (PDB:1Z41A). Predicted secondary structure elements are indicated by α (helix) or β (sheet). Symbols indicate identical residues (*), conserved substitutions (:), and semiconserved substitutions (.).
Heterologous expression of the chromate reductase.
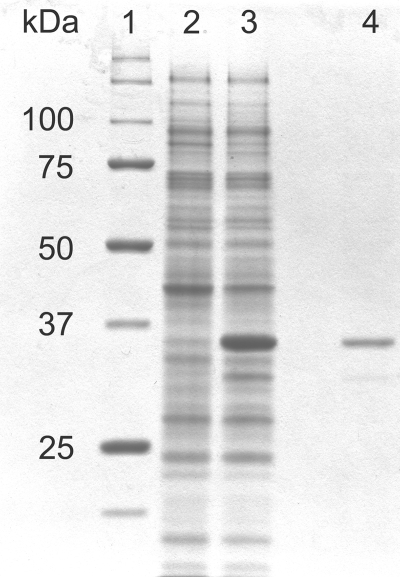
The cytoplasmic chromate reductase was cloned and expressed in E. coli BL21(DE3) cells at 37°C through induction of exponentially growing cells with 1 mM IPTG. SDS-PAGE analysis showed the accumulation of a major protein band of the expected size of approximately 36 kDa in the soluble (cytoplasmic) fraction of E. coli (Fig. 4), which also showed chromate reduction activity at 65°C that was not present in the control cells harboring only the pET22b(+) vector. Heat denaturation of the host proteins at 70°C resulted in the rapid purification of the inherently thermostable recombinant chromate reductase to near homogeneity (Fig. 4).
FIG. 4.
SDS-PAGE analysis of the recombinant chromate reductase from Thermus scotoductus SA-01 expressed in E. coli. Lane 1, standard molecular mass marker; lane 2, soluble fraction (10 μg) of E. coli transformed with pET22b(+); lane 3, soluble fraction (10 μg) of E. coli transformed with pET22b(+) containing the chromate reductase gene; lane 4, soluble fraction (1 μg) after heat denaturation of host proteins. Proteins were visualized by Coomassie blue staining.
DISCUSSION
We report here the purification, characterization, and heterologous expression of a novel cytoplasmic chromate reductase from Thermus scotoductus SA-01. The cytoplasmic chromate reductase from T. scotoductus is a homodimeric enzyme with a monomer molecular mass (apoprotein) of approximately 36 kDa containing a noncovalently bound FMN cofactor as well as the requirement of Ca2+ for activity. The chromate reductase is optimally active at 65°C and a pH of 6.3, coupling the oxidation of NAD(P)H, with a preference for NADPH, to the reduction of Cr(VI). The catalytic efficiency of the chromate reductase with NADPH as an electron donor (kcat/Km) was 1.14 × 106 M−1 s−1, which is at least 180-fold more efficient than the NfsA proteins (1, 14) and >50-fold more efficient than the ChrR proteins (2). Low levels of NAD(P)H oxidase as well as flavin reductase activity were observed.
N-terminal sequencing of the chromate reductase revealed the absence of an N-terminal methionine residue. Whether this is due to the uncharacteristically closely spaced ribosome binding site or to posttranslational modification is unknown. The chromate reductase is encoded by an ORF of 1,050 bp with putative promoter elements and a ribosome binding site located in the 5′ regulatory flanking region. The putative −10 and −35 hexamers showed high similarity to E. coli σ70 promoter consensuses (TATAAT and TTGACA, respectively). Comparison of the primary structure of the chromate reductase with the PDB showed the highest homology to the OYE homologues YqjM from Bacillus subtilis (7, 12) and XenA from Pseudomonas putida (9). Both of these reductases have been shown to reduce a variety of xenobiotic substrates. Due to the broad substrate specificity of YqjM and other members of the family, as well as their involvement in the oxidative stress response, Fitzpatrick and coworkers (12) speculated that it is unlikely that these enzymes have a single specific physiological substrate in vivo and that these enzymes could function in the maintenance of the cellular redox state. Reduction of Cr(VI) by cellular components to Cr(V) as well as a transiently formed Cr(V) intermediate before complete reduction to Cr(III) by some flavoenzymes is thought to be a major cause of chromate oxidative stress and carcinogenesis through the formation of ROS (16, 24). The NfsA (nitroreductase) from E. coli has been shown to be able to preempt ROS generation and minimize oxidative stress during chromate reduction, protecting against chromate toxicity as well as increasing Cr(VI) tolerance (1).
The high degree of similarity between the chromate reductase and the OYE homologues suggests the characteristic (βα)8-barrel (TIM barrel) fold. Secondary structure prediction also indicated eight alternating β-sheets and α-helixes (10). (βα)8-Barrel-folded proteins catalyze a variety of biochemical reactions and are among the most frequent folds observed (19, 31).
Multiple alignments of the chromate reductase with these enzymes showed that the sequence identities corresponded to the functionally important amino acids identified in YqjM and XenA involved in lining the active site, especially those involved in the interaction between the apoprotein and FMN. In YqjM, a sulfate ion from the crystallization solution occupied the catalytic site on the isoalloxazine ring of the FMN, and in XenA a sulfate ion could be modeled into the same position. The residues capable of interaction with the sulfate ion are also conserved in the chromate reductase (specifically His173, His176, and Tyr178) (Fig. 4). The structural similarity between sulfate and chromate could be an indication of how chromate binds as a substrate in the catalytic site. In addition, Kitzing and coworkers (12) showed that the active site of YqjM is hydrophobic, wide open, and easily accessible and that the huge substrate binding pocket allows the binding of a variety of different substrates (including the possibility of other proteins) to control the cellular redox state.
Acknowledgments
This research was supported by the National Research Foundation, South Africa.
We thank Victor Parro (Centro de Astrobiologica, Spain) for help with the N-terminal sequencing and for helpful advice as well as José Berenguer (Centro de Biología Molecular, Spain) for help with heterologous expression studies.
Footnotes
Published ahead of print on 8 February 2008.
REFERENCES
- 1.Ackerley, D. F., C. F. Gonzalez, M. Keyhan, R. Blake II, and A. Matin. 2004. Mechanisms of chromate reduction by the Escherichia coli protein, NfsA, and the role of different chromate reductases in minimizing oxidative stress during chromate reduction. Environ. Microbiol. 6851-860. [DOI] [PubMed] [Google Scholar]
- 2.Ackerley, D. F., C. F. Gonzalez, C. H. Park, R. Blake II, M. Keyhan, and A. Matin. 2004. Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli. Appl. Environ. Microbiol. 70873-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 4.Blehert, D. S., B. G. Fox, and G. H. Chambliss. 1999. Cloning and sequence analysis of two Pseudomonas flavoprotein xenobiotic reductases. J. Bacteriol. 1816254-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cervantes, C., J. Campos-García, S. Devars, F. Gutiérrez-Corona, H. Loza-Tavera, J. C. Torres-Guzmán, and R. Moreno-Sánchez. 2001. Interactions of chromium with microorganisms and plants. FEMS Microbiol. Rev. 25335-347. [DOI] [PubMed] [Google Scholar]
- 6.Codd, R., C. T. Dillon, T. Levina, and P. A. Lay. 2001. Studies on the genotoxicity of chromium: from the test tube to the cell. Coord. Chem. Rev. 216537-582. [Google Scholar]
- 7.Fitzpatrick, T. B., N. Amrhein, and P. Macheroux. 2003. Characterization of YqjM, and old yellow enzyme homolog from Bacillus subtilis involved in the oxidative stress response. J. Mol. Biol. 27819891-19897. [DOI] [PubMed] [Google Scholar]
- 8.French, C. E., and N. C. Bruce. 1995. Bacterial morphinone reductase is related to old yellow enzyme. Biochem. J. 312671-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griese, J. J., R. P. Jakob, S. Schwarzinger, and H. Dobbek. 2006. Xenobiotic reductase A in the degradation of quinoline by Pseudomonas putida 86: physiological function, structure and mechanism of 8-hydroxycoumarin reduction. J. Mol. Biol. 361140-152. [DOI] [PubMed] [Google Scholar]
- 10.Kelley, L. A., R. M. MacCallum, and M. J. E. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299499-520. [DOI] [PubMed] [Google Scholar]
- 11.Kieft, T. L., J. K. Fredrickson, T. C. Onstott, Y. A. Gorby, H. M. Kostandarithes, T. J. Bailey, D. W. Kennedy, S. W. Li, A. E. Plymale, C. M. Spadoni, and M. S. Gray. 1999. Dissimilatory reduction of Fe(III) and other electron acceptors by a Thermus isolate. Appl. Environ. Microbiol. 651214-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitzing, K., T. B. Fitzpatrick, C. Wilken, J. Sawa, G. P. Bourenkov, P. Macheroux, and T. Clausen. 2005. The 1.3 Å crystal structure of the flavoprotein YqjM reveals a novel class of old yellow enzymes. J. Biol. Chem. 28027904-27913. [DOI] [PubMed] [Google Scholar]
- 13.Kohli, R. M., and V. Massey. 1998. The oxidation half-reaction of old yellow enzyme. J. Biol. Chem. 27332763-32770. [DOI] [PubMed] [Google Scholar]
- 14.Kwak, Y. H., D. S. Lee, and H. B. Kim. 2003. Vibrio harveyi nitroreductase is also a chromate reductase. Appl. Environ. Microbiol. 694390-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 16.Liu, K. J., and X. Shi. 2001. In vivo reduction of chromium (VI) and its related free radical generation. Mol. Cell. Biochem. 22241-47. [PubMed] [Google Scholar]
- 17.Losi, M. E., C. Amrhein, and W. T. S. Frankenberger. 1994. Environmental biochemistry of chromium. Rev. Environ. Contam. Toxicol. 13691-121. [DOI] [PubMed] [Google Scholar]
- 18.Möller, C., and E. van Heerden. 2006. Isolation of a soluble and membrane-associated Fe(III) reductase from the thermophile, Thermus scotoductus (SA-01). FEMS Microbiol. Lett. 265237-243. [DOI] [PubMed] [Google Scholar]
- 19.Nagano, N., C. A. Orengo, and J. M. Thornton. 2002. One fold with many functions: the evolutionary relationship between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 321741-765. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence database: status for the year 2000. Nucleic Acids Res. 28292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Opperman, D. J., and E. van Heerden. 2007. Aerobic Cr(VI) reduction by Thermus scotoductus strain SA-01. J. Appl. Microbiol. 1031907-1913. [DOI] [PubMed] [Google Scholar]
- 22.Opperman, D. J., and E. van Heerden. 22 January 2008, posting date. A membrane-associated protein with Cr(VI)-reducing activity from Thermus scotoductus SA-01. FEMS Microbiol. Lett. doi: 10.1111/j.1574-6968.2007.01063.x. (Subsequently published, FEMS Microbiol. Lett. 208210-218, 2008.) [DOI] [PubMed] [Google Scholar]
- 23.Park, C. H., M. Keyhan, B. Wielinga, S. Fendorf, and A. Matin. 2000. Purification to homogeneity and characterization of a novel Pseudomonas putida chromate reductase. Appl. Environ. Microbiol. 661788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi, X., and N. S. Dalal. 1990. NADPH-dependent flavoenzymes catalyze one electron reduction of metal ions and molecular oxygen and generate hydroxyl radicals. FEBS Lett. 276189-191. [DOI] [PubMed] [Google Scholar]
- 25.Slilaty, S. N., and S. Lebel. 1998. Accurate insertional inactivation of lacZα: construction of pTrueBlue and M13TrueBlue cloning vectors. Gene 21383-91. [DOI] [PubMed] [Google Scholar]
- 26.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 15076-85. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki, T., N. Miyata, H. Horitsu, K. Kawai, K. Takamizawa, Y. Tai, and M. Okazaki. 1992. NAD(P)H-dependent chromium(VI) reductase of Pseudomonas ambigua G-1: a Cr(V) intermediate is formed during the reduction of Cr(VI) to Cr(III). J. Bacteriol. 1745340-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Towner, P. 1991. Isolation of DNA by SDS-proteinase K treatment, p. 52-53. In T. A. Brown (ed.) Essential molecular biology, vol. 1. A practical approach. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 30.Urone, P. F. 1955. Stability of colorimetric reagent for chromium, s-diphenylcarbazide, in various solvents. Anal. Chem. 271354-1355. [Google Scholar]
- 31.Vega, M. C., E. Lorentzen, A. Linden, and M. Wilmanns. 2003. Evolutionary markers in the (β/α)8-barrel fold. Curr. Opin. Chem. Biol. 7694-701. [DOI] [PubMed] [Google Scholar]
- 32.Warburg, O., and W. Christian. 1933. Über das gelbe Ferment und seine Wirkungen. Biochem. Z. 266377-411. [Google Scholar]
- 33.Williams, R. E., and N. C. Bruce. 2002. “New uses for an old enzyme”—the old yellow enzyme family of flavoenzymes. Microbiology 1481607-1614. [DOI] [PubMed] [Google Scholar]
- 34.Ziegenhorn, J., M. Senn, and T. Bücher. 1976. Molar absorptivities of β-NADH and β-NADPH. Clin. Chem. 22151-160. [PubMed] [Google Scholar]