Abstract
Microcystins are the most common cyanobacterial toxins found in freshwater lakes and reservoirs throughout the world. They are frequently produced by the unicellular, colonial cyanobacterium Microcystis aeruginosa; however, the role of the peptide for the producing organism is poorly understood. Differences in the cellular aggregation of M. aeruginosa PCC 7806 and a microcystin-deficient ΔmcyB mutant guided the discovery of a surface-exposed protein that shows increased abundance in PCC 7806 mutants deficient in microcystin production compared to the abundance of this protein in the wild type. Mass spectrometric and immunoblot analyses revealed that the protein, designated microcystin-related protein C (MrpC), is posttranslationally glycosylated, suggesting that it may be a potential target of a putative O-glycosyltransferase of the SPINDLY family encoded downstream of the mrpC gene. Immunofluorescence microscopy detected MrpC at the cell surface, suggesting an involvement of the protein in cellular interactions in strain PCC 7806. Further analyses of field samples of Microcystis demonstrated a strain-specific occurrence of MrpC possibly associated with distinct Microcystis colony types. Our results support the implication of microcystin in the colony specificity of and colony formation by Microcystis.
Microcystis aeruginosa is a unicellular colonial cyanobacterium frequently producing mass developments and surface scums in freshwater habitats. Microcystis cyanobacteria are widely known for their production of the potent hepatotoxin microcystin. Microcystins are a family of cyclic heptapeptides that potently inhibit protein phosphatases of the eukaryotic protein phosphatase P family. Several cases of human and animal poisonings have been attributed to the presence of these toxins in water supplies and recreational lakes (6, 15). Microcystins are synthesized by a large enzyme complex comprising nonribosomal peptide synthetases, polyketide synthases, and tailoring enzymes (32).
In the environment, Microcystis occurs as a mixture of morphotypes that differ in their cell and sheath characteristics (17). The formation of large colonies embedded in mucilage and the presence of gas vesicles enable Microcystis colonies to regulate their buoyancy (35). The ability to migrate vertically in lakes provides a significant advantage over many other phytoplankton species (1). Several studies have shown a correlation of Microcystis morphotypes with the presence of specific peptides. Microcystins are most frequently associated with M. aeruginosa and M. viridis, whereas other Microcystis morphotypes, such as M. wesenbergii and M. ichthyoblabe, usually lack these peptides (8, 34). A possible correlation of colony form and the nonribosomal peptide microcystin was further supported by the finding that microcystin influences the expression and oligomerization of a surface-exposed lectin that may facilitate the recognition and attachment of Microcystis cells (16). In addition, there is increasing evidence that microcystin released from dead cells may serve as an infochemical in the Microcystis community, thereby enhancing the fitness of surviving cells (26). The microcystin-dependent expression of the two microcystin-related proteins MrpA and MrpB that show similarity to the quorum sensing-controlled RhiA and RhiB proteins in Rhizobium leguminosarum (11) further supports the idea that microcystin may be perceived as an intercellular signal (4).
In the present study, the correlation of microcystin with a novel surface-exposed component, a glycoprotein, is reported. In the past few decades, an increasing number of bacterial proteins have been shown to be glycosylated, including a wide range of different cell envelope components such as membrane-associated glycoproteins, surface-associated glycoproteins, and crystalline surface layers (S-layers), as well as secreted glycoproteins and exoenzymes (21, 33). Examples of bacterial glycoproteins include, among others, the flagellins of Pseudomonas aeruginosa (30) and Campylobacter spp. (7), the type IV pili of P. aeruginosa (3) and Neisseria meningitides (23), the Fap1 fimbrial adhesin of Streptococcus parasanguinis (28), the high-molecular-weight protein (HmwA) of Haemophilus influenzae (10), and the autotransporter protein Ag43 of Escherichia coli (27). In general, carbohydrate modifications of bacterial proteins can be diverse in structure and are linked to either asparagines or serine and threonine residues (33). So far, not much information about protein glycosylation in cyanobacteria is available. Two cyanobacterial glycoproteins seem to play roles in different types of motility. The motile cyanobacterium Phormidium unicatum was shown previously to contain fibrillar arrays of a glycoprotein, oscillin, on top of its S-layer. The protein is conserved in motile filamentous cyanobacteria and seems to play a role in gliding motility (13). The S-layer glycoprotein SwmA was shown previously to be required in Synechococcus sp. strain WH8102 for nonflagellar swimming (2).
Here, we report a strong increase in the amount of a novel protein, MrpC (microcystin-related protein C), as a consequence of directed knockout mutagenesis in microcystin biosynthesis genes in M. aeruginosa PCC 7806 and a greater tendency of the microcystin-deficient cells than of the wild-type (WT) cells to aggregate. Further data indicate that the MrpC protein may be a potential target of an O-glycosyltransferase of the SPINDLY (SPY) family that is encoded downstream of the mrpC gene. The MrpC protein appears to be specific to distinct Microcystis colony types in field samples. Taken together, our data indicate that MrpC plays a role in cell-cell interaction in microcystin-producing strains of Microcystis.
MATERIALS AND METHODS
Cultivation of cyanobacteria and field sampling.
The axenic strain M. aeruginosa PCC 7806 came from the Pasteur Culture Collection of Cyanobacteria (Institut Pasteur, Paris, France). Mutants of this strain unable to produce microcystin were obtained by the insertion of a chloramphenicol resistance cartridge into the genes mcyB and mcyH (5, 22). WT and mutant cultures were grown at 23°C in batch cultures with Z8 medium (25) under continuous low light (30 microeinsteins m−2 s−1) until they reached optical densities at 750 nm (OD750) of 0.5 and 1.0, respectively. Light intensities were measured using a Li-Cor LI250 light meter (Walz, Effeltrich, Germany). Cell densities of cultures were determined by measuring the OD750 with a Uvikon 930 spectrometer (Contron Instruments, Schlieren, Germany). For the light experiment, cells were grown as described above and exposed for 2 h to either 70 μEm−2 s−1 or complete darkness. For microcystin addition experiments, a 10-μg/liter concentration of microcystin-LR (kindly provided by Keishi Ishida) was added to culture replicates. Field sampling was performed in summer 2007 at Lake Dollgow by net hauling using a plankton net (40-μm mesh size). For preservation, samples were fixed with 4% formaldehyde and stored at 4°C.
Extraction and fractionation of Microcystis proteins.
The proteins of different cellular compartments were isolated by successive fractionations. For the analysis of the secretome, culture supernatants were purified by a 0.2-μm-pore-size filter (Schleicher and Schüll, Dassel, Germany). Proteins from the purified supernatants were precipitated using trichloroacetic acid (10% [wt/vol] final concentration), washed four times with 80% (vol/vol) ice-cold acetone, and dissolved in urea-containing buffer (8 M urea, 100 mM NaH2PO4, 10 mM Tris-HCl, pH 8.0). The cell pellets were washed in a mixture of 10 mM Tris-HCl (pH 7.8), 2 mM NaCl, and 1 mM phenylmethylsulfonyl fluoride without any mechanic or heat treatment. Whole-cell extracts were prepared using a French press. For this purpose, cells were resuspended in 10 mM Tris-HCl buffer (pH 7.8) containing 1 mM EDTA and 1 mM phenylmethylsulfonyl fluoride. Cells were broken under a pressure of 900 lb/in2 at 4°C and subjected to an ultracentrifugation step at 20,000 × g for 45 min at 4°C. After centrifugation, the supernatant contained the cytosolic proteins. The extracellular proteins, including cell surface and periplasmic proteins, were obtained by incubating the cells with lysozyme and sucrose as described in reference 31. Protein concentrations were determined using the method of Lowry et al. (20). Extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting according to standard protocols.
Mass spectrometric analyses.
The protein spots or bands were excised from the gels, washed once with 30% (vol/vol) acetonitrile in 25 mM ammonium bicarbonate and once with 50% (vol/vol) acetonitrile in 10 mM ammonium bicarbonate, dehydrated in acetonitrile, and dried at 37°C. The gel pieces were reswollen with an appropriate volume of sequencing-grade trypsin solution (10 mg/liter in 3 mM Tris-HCl, pH 8.5; Promega, Madison, WI) or endoproteinase AspN solution (5 ng/μl in 3 mM Tris-HCl, pH 8.5; Roche, Mannheim, Germany) and incubated for 5 to 8 h at 37°C. Thereafter, extraction solution (50% [vol/vol] acetonitrile, 0.3% [vol/vol] trifluoroacetic acid) in a volume equal to that of the gel mixture was added, and the samples were agitated for 30 to 60 min. The resulting peptide-containing solution was applied to the matrix-assisted laser desorption ionization (MALDI) target (384-matrix-spot, 600-μm AnchorChip; Bruker Daltonik, Bremen, Germany) and analyzed by MALDI-time of flight (TOF) mass spectrometry and MALDI-TOF mass spectrometry-post-source decay (PSD) analysis using a Reflex III mass spectrometer (Bruker Daltonik) as described previously (9).
Gene identification.
The partial peptide sequences obtained from the quadrupole (Q)-TOF analysis were used as query sequences for a BlastP analysis of the genome database of M. aeruginosa PCC 7806. This database is part of a current genome sequencing project at Génopole-Ile de France (Institut Pasteur, Paris, France). The complete draft of the genome is available under accession numbers AM778843 to AM778958.
PCR.
PCR amplifications were performed using AmpliTaq according to the recommendations of the manufacturer (Qiagen, Hilden, Germany). Chromosomal DNA of Microcystis strains was isolated as described previously (12). mcyA was amplified using the primer pair McyA-Cd1F and McyA-Cd1R as described previously (12). mrpC was amplified with the primer pair MrpC169.fw (5′-CTGCAGCCACCAATAGCTT-3′) and MrpC387.rv (5′-CTGCGAAATAGCATCAGCAG-3′).
Purification of MrpC and antibody generation.
For the purification of MrpC, the supernatant of the ΔmcyB mutant was filtered and subjected to fractionation by ammonium sulfate precipitation. Ammonium sulfate powder was added to the protein solution under conditions of constant stirring at 4°C until saturation (10 to 30 min). The resulting solution was centrifuged at 10,000 × g for 10 min. The pellet was resolved in Tris-buffered saline. MrpC was found to precipitate at a concentration of 10% (wt/vol) ammonium sulfate at a high purity as confirmed by SDS-PAGE analysis. Purified MrpC (0.4 mg) was used to immunize two guinea pigs (Pineda Antikörper Service, Berlin, Germany).
Immunodetection of proteins and detection of carbohydrate modifications.
Protein fractions were separated using SDS-PAGE and blotted onto nitrocellulose filters (Hybond-C Extra; Amersham). The immunodetection of MrpC was performed with polyclonal antibodies (dilution, 1:1,000) raised in guinea pigs as mentioned above. Detection was performed using the SuperSignal West Pico chemiluminescent substrate kit (Pierce) according to the standard protocol. Glycoproteins were detected using the digoxigenin-glycan detection kit according to the instructions of the manufacturer (Roche, Mannheim, Germany). O-linked N-acetylglucosamine (O-GlcNAc) modifications of proteins were detected using a specific monoclonal antibody against O-GlcNAc (Alexis Biochemicals, San Diego, CA).
Gel filtration fast-performance liquid chromatography.
Aliquots (1.0 ml) of whole-cell extracts of the PCC 7806 WT and the ΔmcyB mutant were applied to a Superdex 75 prep-grade HiLoad 16/60 column (GE Healthcare). Isocratic elution was performed at 10°C with a flow rate of 1 ml·min−1, and 1.0-ml fractions were collected. Fractions (0.5 ml per sample) were analyzed on immunoblots (using the dot blot unit from Schleicher and Schüll) with the antibody to MrpC. Sizes of multimeric MrpC isoforms were determined using the retention times of standard proteins (thyroglobulin, bovine gamma globulin, chicken ovalbumin, equine myoglobin, and vitamin B12; Bio-Rad).
Immunofluorescence microscopy.
Cells of mid-logarithmic growth phase (OD750, 0.5) were harvested by centrifugation at 4,000 × g at room temperature for 5 min, washed once in phosphate-buffered saline (PBS; 140 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4), and resuspended in PBS. Cells were fixed in 3.7% (vol/vol) formaldehyde in PBS on ice for 1 h. After three PBS washing steps, cells were resuspended in GTE buffer (50 mM glucose, 20 mM Tris-HCl [pH 7.5], 10 mM EDTA [pH 8.0]) and spread onto poly-l-lysine-coated glass slides to dry. Glass slides with immobilized cells were dipped into methanol at −20°C for 5 min and subsequently into −20°C acetone for 30 s. Preparations were blocked with 2% (wt/vol) bovine serum albumin (BSA) in PBS and incubated with the primary antibody to MrpC at a dilution of 1:500 in 2% (wt/vol) BSA in PBS for 1 h at room temperature. After two PBS washes, preparations were incubated with a fluorescein isothiocyanate (FITC)-labeled goat-anti-guinea pig antibody (at a dilution of 1:100 in 2% [wt/vol] BSA in PBS; Sigma-Aldrich, St. Louis, MO) for 1 h. Cells were mounted in a drop of 4% (vol/vol) n-propylgallate dissolved in 87% (vol/vol) glycerol and stored for up to 4 weeks at −20°C.
The preparation of field samples for immunofluorescence microscopy was identical to the preparations of laboratory batch cultures, except for the fixation step. Field samples were already fixed with formaldehyde directly after the sampling, as described above.
Sample observation, image acquisition, and processing were carried out using the DeltaVision Spectris system (Applied Precision, Issaquah, WA) with the preinstalled default softWorx software package. Images were acquired as stacks of z-sections with one image taken every 0.2 μm over the whole cell volume. Two sets of excitation and emission filters were used for visualization: the rhodamine-Texas Red-phycoerythrin filter set (excitation wavelength, 555 nm; emission wavelength, 617 nm) to visualize the red/orange autofluorescence of cyanobacteria and the FITC filter set (excitation wavelength, 490 nm; emission wavelength, 528 nm) to visualize FITC-coupled green immunostaining of extracellular MrpC. Acquired raw images were deconvolved by iterative constrained deconvolution to enhance image quality and contrast by using the algorithms implemented in the softWorx software package.
Nucleotide sequence accession number.
The sequence of the mrpC gene cluster is available in the EMBL database under the accession number AM778945 (IPF_2679-2677).
RESULTS
Identification of MrpC.
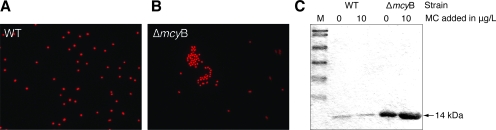
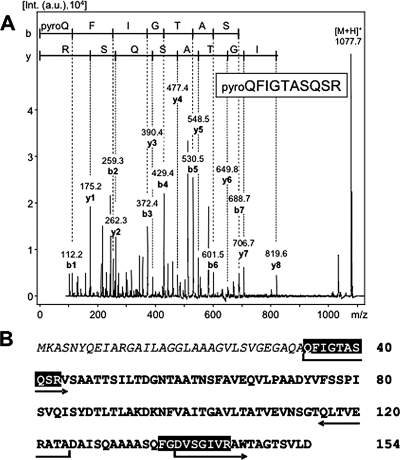
Compared to the WT, the microcystin-deficient ΔmcyB mutant shows an increased tendency to aggregate into little clusters of cells in laboratory batch cultures (5). The WT strain M. aeruginosa PCC 7806 never shows this feature under standard laboratory conditions (Fig. 1A). The aggregating phenotype can be observed both macroscopically and microscopically and was also noticed for the microcystin-deficient ΔmcyA and ΔmcyH mutants (Fig. 1B and data not shown). This phenomenon prompted us to compare the secreted protein fractions from M. aeruginosa PCC 7806 (WT) and the ΔmcyB mutant. These analyses revealed the differential accumulation of a predominant protein of approximately 14 kDa (Fig. 1C) in the media of the ΔmcyB mutant cells and the WT. The abundance of the protein in the mutant was strongly increased compared to that in the WT. The addition of trace amounts of microcystin to the cultures did not alter the amounts of the secreted protein (Fig. 1C). Similar results were obtained in several independent experiments at different cell densities and were also seen with two further mutants deficient in the production of microcystin (ΔmcyA and ΔmcyH mutants; data not shown). The protein was excised from the gels, digested with trypsin, and subjected to Q-TOF analysis. The comparison of the partial peptide sequences with sequences in the genome database for M. aeruginosa PCC 7806 led to the identification of a 15-kDa protein without significant similarities to known proteins in databases. According to its microcystin-dependent expression pattern in M. aeruginosa strain PCC 7806 and its possible relation to the observed phenotype of microcystin-deficient mutants, the protein was classified as microcystin-related protein (4) and designated MrpC. MALDI mass spectra of tryptic digests of MrpC showed only two very intense peaks at m/z 949.5 and m/z 1,077.5 (data not shown). Both ion signals did not match theoretical masses of tryptic peptides derived from the MrpC amino acid sequence. Therefore, these ions were subjected to further sequence analysis by the MALDI-TOF mass spectrometry-PSD technique. The PSD spectrum of the ion signal at m/z 1,077.5 showed nearly complete b- and y-ion series from which the sequence pyro-QFIGTASQSR (residues 34 to 43 of MrpC, where pyro-Q represents pyroglutamate) could be deduced (Fig. 2A). The modification of the N-terminal glutamine residue to pyroglutamate has been described previously for other extracellular proteins of both eukaryotes and prokaryotes (18). The identification of a modified glutamine at position 34 therefore clearly indicated N-terminal processing of MrpC during secretion. The experimental findings were further supported by an in silico analysis of the MrpC protein sequence for the presence of putative leader sequences for secretion (http://www.cbs.dtu.dk/services/SignalP/). In agreement with the mass spectrometric data, a signal sequence cleavage site between amino acids 33 and 34 of the protein was predicted with a high probability value. The second peptide ion signal at m/z 949.5 was assigned to the sequence stretch FGDVSGIVR (residues 136 to 144) (Fig. 2B) by PSD analysis and indicated a non-trypsin-specific cleavage site. Both peptides of the trypsin digest correspond to the partial sequences already determined by Q-TOF analysis that were used to initially identify MrpC (see Table S1 in the supplemental material).
FIG. 1.
Phenotype of microcystin-deficient ΔmcyB mutant cells and analysis of the secreted protein fractions from M. aeruginosa PCC 7806 and the ΔmcyB mutant. (A) Fluorescence micrograph of the M. aeruginosa PCC 7806 WT showing red autofluorescence of cells in a dispersed cell suspension. (B) Fluorescence micrograph of ΔmcyB mutant cells showing red autofluorescence of characteristic cell aggregates. (C) Comparative analysis of the secreted protein fractions by SDS-PAGE. Cultures were grown to late log phase (OD750, 1.0) and treated with 0 or 10 μg of microcystin (MC)/liter for 2 h as indicated.
FIG. 2.
(A) MALDI-TOF-PSD mass spectrum of the N-terminal peptide fragment obtained after trypsin digestion (m/z 1,077.5). The amino acid sequence could be assigned to residues 34 to 43 of MrpC (pyro-QFIGTASQSR), with a modification of the N-terminal glutamine residue to pyroglutamate. Int., intensity; a.u., arbitrary units. (B) Sequence coverage of MrpC after trypsin and AspN digestion. Black boxes indicate peptides obtained after trypsin digestion and sequenced by PSD analysis. Arrows indicate sequences of peptides obtained after AspN digestion that potentially carry posttranslational modifications. The signal sequence of MrpC is shown in italics.
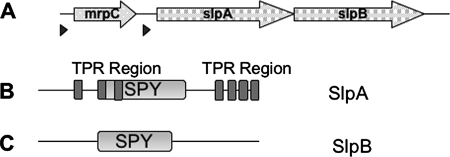
Two further open reading frames are located downstream of the mrpC gene (Fig. 3A and Table 1). The putative protein encoded next to mrpC exhibits 50% similarity to the SPY protein of Arabidopsis thaliana that is implicated in a gibberellin signaling cascade (14). Similar to the plant protein, the Microcystis protein is predicted to comprise seven tetratricopeptide repeat (TPR) domains and an O-GlcNAc transferase-SPY domain (Fig. 3B). Further downstream, a second protein containing a highly similar O-GlcNAc transferase-SPY signature but without TPR motifs is encoded (Fig. 3C). The two proteins were designated SPY-like proteins SlpA and SlpB. An alignment of the SPY domains of the Microcystis proteins with similar sequences from plants and bacteria is provided in the supplemental material (see Fig. S1). MrpC is rich in serine and threonine residues (19%). An in silico analysis of the proteins by using the PSORT subcellular prediction software (http://www.psort.org/psortb/) strongly suggested a cell surface localization pattern for MrpC. No clear localization patterns for SlpA and SlpB could be deduced. Taken together, mrpC and the flanking genes may constitute a gene cluster encoding a cell surface-associated protein and two proteins potentially involved in the transfer of GlcNAc to hydroxyl groups of serine and threonine residues (Table 1).
FIG. 3.
(A) Schematic representation of the mrpC gene cluster. Triangles indicate putative ribosome binding sites. Detailed information on the gene products is presented in Table 1. (B and C) Schematic representation of the SlpA (B) and SlpB (C) domain architectures. SPY and TPR domains are shown in light and dark gray, respectively.
TABLE 1.
Sequence similarities and deduced functions of all proteins encoded by the mrpC gene cluster
| Protein | No. of amino acids | Deduced function | Protein with sequence similarity | % Identity/% similarity (no. of relevant amino acids) | Predicted subcellular localization |
|---|---|---|---|---|---|
| MrpC | 154 | No deduced function | No significant similarity | Cell surface | |
| SlpA | 1,254 | O-GlcNAc transferase | SPY (gibberellin signal transduction protein of A. thaliana) | 32/49 (709) | |
| SlpB | 1,246 | O-GlcNAc transferase | TPR domain protein of Geobacter sulfurreducens | 43/60 (363) |
The MrpC protein occurs in two isoforms.
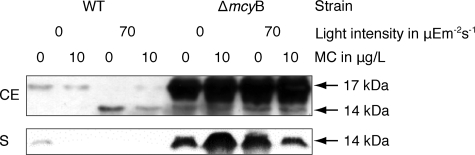
To further investigate the subcellular localization of MrpC and to study the expression under different environmental conditions, we generated an antibody against the protein purified from the supernatant of the ΔmcyB mutant. The antibody specifically recognized the 14-kDa MrpC protein in the secreted protein fraction; however, it also recognized a second protein of approximately 17 kDa in cellular extracts of M. aeruginosa PCC 7806 (WT) and the ΔmcyB mutant (Fig. 4). Similar to the 14-kDa MrpC protein, the 17-kDa protein showed increased accumulation in the ΔmcyB mutant compared to that in the WT. The large amounts of the 14-kDa MrpC protein in the secreted fraction and of the 17-kDa protein in cellular extracts from the ΔmcyB mutant were independent from the light conditions and from the addition of extracellular microcystin (Fig. 4 and further unpublished data); however, the accumulation patterns of the 14-kDa protein and the 17-kDa variant in whole-cell extracts of the WT differed under different light conditions (Fig. 4). The analysis of the tryptic digest of the 17-kDa protein using MALDI-TOF mass spectrometry and MALDI-TOF mass spectrometry-PSD analysis showed the presence of two dominant ions at m/z 949.5 and m/z 1,077.5 exhibiting a PSD pattern identical to that of the 14-kDa MrpC protein. It was therefore concluded that the 17-kDa protein represents a second isoform of MrpC that is contained in or bound to the cell. As the protein did not contain the N-terminal signal sequence but had the characteristic pyroglutamate moiety at its N terminus, it was concluded that the second isoform of MrpC is extracellular but more tightly bound to the cell surface than the 14-kDa variant. This assumption was supported by the fact that the 17-kDa protein could be washed off the cells by incubating the cells with lysozyme and sucrose as described in reference 31 (data not shown).
FIG. 4.
Immunoblot analyses of cell extracts and of secreted protein fractions of M. aeruginosa PCC 7806 (WT) and the ΔmcyB mutant using antiserum against MrpC. Cells were grown until late log phase (OD750, 1.0) and subsequently exposed to strong light (70 μEm−2 s−1) or darkness for 2 h. Parallel aliquots of cultures were spiked with 10 μg of microcystin (MC)/liter as indicated. CE, whole-cell extract; S, secreted fraction.
Glycosylated extracellular proteins in other bacteria typically form high-molecular-weight complexes and frequently form distinct surface structures such as pili or fimbriae (3, 10, 23). In order to see if MrpC forms higher oligomeric structures under native conditions, we performed size exclusion chromatography experiments with whole-cell protein extracts from M. aeruginosa PCC 7806 and the ΔmcyB mutant. A subsequent analysis of the protein fractions using the specific antibody to MrpC revealed MrpC oligomers in a size range between approximately 120 and 170 kDa for both the WT and the mutant (see Table S2 in the supplemental material). No MrpC signals in the size range below 50 kDa were obtained. We can thus conclude that the whole amount of MrpC is part of higher-molecular-mass complexes under native conditions. Furthermore, these data indicate that the differential accumulation patterns of the protein in the WT and the ΔmcyB mutant are not due primarily to differences in the oligomerization state.
Posttranslational modification of MrpC.
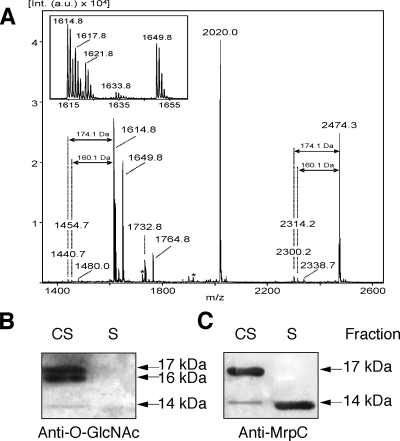
The molecular masses of both MrpC monomers detected in this study clearly exceed the theoretical mass of the processed MrpC protein of 12 kDa. This size difference may be attributed to posttranslational modifications of the two protein variants. Indeed, detailed analyses of AspN digests of MrpC using MALDI-TOF mass spectrometry and MALDI-TOF mass spectrometry-PSD analysis indicated a modification for several of the peptides and increased the overall sequence coverage (Fig. 5A and 2B). None of the observed ion signals in the MALDI mass spectrometry spectrum matched theoretical masses of AspN-derived peptides. The peptide ion at m/z 2,020.0 was unambiguously assigned by PSD analysis to the processed N terminus of MrpC, starting with the pyroglutamate residue (residues 34 to 44) (Fig. 2B), but must contain a modification in the C-terminal direction from residue V44. PSD analyses of the peptide ions at m/z 1,649.8 and m/z 1,764.8 yielded b-ion series that clearly matched the partial sequence DVSGIVRAW2ox (residues 138 to 146) (Fig. 2B; PSD data not shown). Although the C-terminal sequences could not be assigned, these analyses clearly indicate a modification in the C-terminal direction from residue 146 (Fig. 2B). The PSD spectrum of the peptide ion at m/z 2,474.3 showed a series of y-ions corresponding to the sequence stretch TQLTVERATA (residues 115 to 124) (Fig. 2B). However, the N-terminal sequence of the peptide could not be determined. The PSD spectrum exhibited extensive fragmentations, including the loss of 160 Da from the precursor ion and from many of the smaller fragments (data not shown). The loss of 160 Da was also observed in the MALDI mass spectrometry spectrum as a result of in-source decay. Here, it was accompanied by a loss of 174 Da (Fig. 5A). The same fragmentations were observed for the ion signal at m/z 1,614.8. Similar patterns for glycosylated proteins in other bacteria, e.g., the O-linked glycan of Flavobacterium meningosepticum (24), have been reported previously. The peptide fragmentation pattern therefore indicates a putative glycosylation site upstream of residue 115 of MrpC (Fig. 2B). Taken together, the peptide fragmentation patterns of both MrpC isoforms strongly indicate posttranslational modifications, presumably including protein glycosylation. However, none of the putative modifications could be related to the size difference of the two MrpC isoforms. Despite detailed mass spectrometric analyses, the observed sequence coverage (Fig. 2B) remained remarkably low. We therefore speculate that extensive modifications, including differences between the cell surface and secreted MrpC isoforms, are located in the part of the MrpC sequence that was not covered by mass spectrometry.
FIG. 5.
Posttranslational modification of MrpC. (A) MALDI-TOF mass spectrum of the AspN digest of MrpC from the cell surface fraction. The peptide ions at m/z 2,474.3 and m/z 1,614.8 show losses of 160 and 174 Da, characteristic of glycan modifications of proteins. Peptides derived from the autoproteolysis of AspN are indicated by asterisks. (B and C) Immunoblot analyses of subcellular fractions of the ΔmcyB mutant using antiserum against O-GlcNAc (B) and MrpC (C). CS, cell surface fraction; S, secreted fraction.
Detection of O-GlcNAc modifications of MrpC.
Two candidate proteins potentially facilitating protein O glycosylation, SlpA and SlpB, are encoded downstream of mrpC. To further examine the possibility of MrpC glycosylation, we analyzed the different cellular fractions of the M. aeruginosa PCC 7806 WT and the mutant on Western blots using digoxigenin-glycan reagents that detect carbohydrate moieties on proteins. However, although signals potentially corresponding to both MrpC isoforms could be obtained, the general intensities of the signals were only slightly above the detection limit.
To extend these findings, we applied an antibody against O-linked GlcNAc residues to Western blots of cell surface and supernatant extracts from the PCC 7806 WT and the mutant. The commercial monoclonal antibody applied was previously shown to be highly specific for O-GlcNAc modifications in eukaryotic protein extracts (37). Employed for the analysis of Microcystis extracts, the antibody specifically reacted with two proteins of approximately 16 and 17 kDa in the cell surface fraction of the ΔmcyB mutant. A faint band at approximately 14 kDa was also observed (Fig. 5B). The same blot was subsequently hybridized with the antibody to the MrpC protein. The sizes and intensities of the 14- and 17-kDa bands detected in the extracts from the ΔmcyB mutant were in agreement with the O-GlcNAc signals (Fig. 5C). The signal detected at a position of about 16 kDa by using the antibody against O-linked GlcNAc residues did not correspond to an MrpC signal and may therefore represent a different protein. Alternatively, the glycosylation of this protein variant may prevent recognition by the MrpC antibody.
The secreted variant of MrpC that was detected in the ΔmcyB mutant with the antibody to the MrpC protein (Fig. 5C) did not show any O-GlcNAc signal (Fig. 5B).
In contrast to that of the ΔmcyB mutant, the cell surface fraction of the WT showed no clear signals with the antibody against O-linked GlcNAc residues, the amount of MrpC being presumably too low for detection (data not shown).
These data suggest O-GlcNAc modifications of MrpC in the cell surface fraction, whereas the secreted variant, at least in the mutant, seems not to carry these carbohydrate modifications. The precise chemical structure of the glycan is presently unknown. Apart from the putative MrpC glycosylation described above, no further putative O-GlcNAc signals were observed in Microcystis protein extracts. The specificity of the antibody and the presence of putative O-GlcNAc transferase genes in the mrpC gene cluster suggest that these modifications are most likely related to serine and threonine residues. The verification of these findings and the elucidation of the full structure of the carbohydrate modifications attached to MrpC would require a complete structural elucidation of the proteins and were not further objectives of this study.
In situ detection of MrpC.
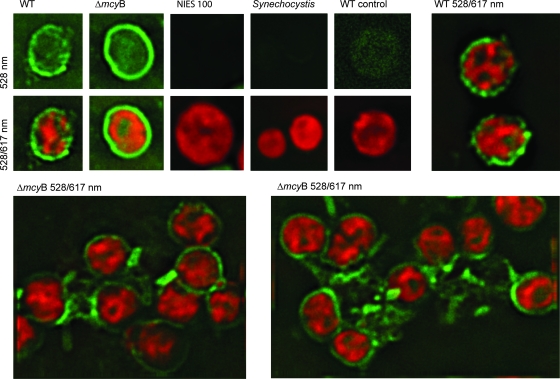
Glycosylated proteins in bacteria are frequently implicated in cell adhesion and cellular interactions (33). To test this possibility for MrpC in Microcystis, we constructed plasmid vectors for insertional mutagenesis in the corresponding gene and the neighboring slpA gene. However, several attempts to mutagenize the genes failed. In general, only a few mutagenesis attempts with M. aeruginosa PCC 7806 have been successful so far, most likely due to the extensive restriction barrier in Microcystis (29). In order to get more insights into the functionality of the MrpC protein, we therefore performed immunofluorescence microscopy studies to detect the MrpC protein in situ. WT and ΔmcyB mutant cells were fixed and permeabilized and subsequently incubated with the MrpC antibody and a secondary FITC-labeled antibody. Cells of the nontoxic Microcystis strain Nies 100 and of Synechocystis sp. strain PCC 6803 were treated in the same way and served as negative controls. We further included a control sample of the PCC 7806 WT without the primary antibody incubation step. All cells showed bright red autofluorescence under a 617-nm emission filter (Fig. 6, middle and bottom panels and top right panel). In the green fluorescence channel, only PCC 7806 WT and ΔmcyB mutant cells, but none of the samples used as negative controls, showed strong and specific signals above the autofluorescence background (Fig. 6, left five top panels). A strong but punctually distributed fluorescence at the outer peripheries of the PCC 7806 WT cells was observed (Fig. 6, top and middle far-left panels and top right panel). A similar distribution, but clearly increased intensity, was observed for the ΔmcyB mutant cells. The signals at the outer peripheries of the cells frequently formed closed rings in the two-dimensional micrographs of all sections examined, thus indicating that the entire cell surface is covered by MrpC (Fig. 6, top and middle ΔmcyB panels). Moreover, in the ΔmcyB mutant samples, assemblages of cells that were connected by mucus-like structures displaying a bright MrpC fluorescence signal were prevalent (Fig. 6, bottom panels). No such signals were observed in samples of the WT strain (Fig. 6, top right panel). The immunofluorescence data therefore provide evidence for an involvement of MrpC in cellular interactions and may account for the increased aggregation tendency observed for the mcyB mutant (Fig. 1A and B).
FIG. 6.
Immunofluorescence micrographs of the ΔmcyB mutant and WT strains of M. aeruginosa PCC 7806 obtained by using the antibody against MrpC and a FITC-coupled secondary antibody. Images of cells were acquired either with a green fluorescence emission filter (first five top panels) or with both green and red fluorescence emission filters (remaining panels). Nies 100, Microcystis sp. strain Nies 100; Synechocystis, Synechocystis sp. strain PCC 6803; WT control, PCC 7806 WT without primary antibody incubation.
Distribution of MrpC in laboratory strains and field samples of Microcystis.
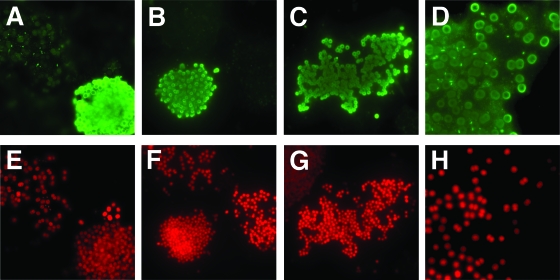
In order to evaluate the distribution of the mrpC region in different Microcystis strains, a PCR survey with DNA from 22 Microcystis strains from different culture collections was performed. For eight of the strains, a PCR product was obtained with mrpC-specific primers (see Fig. S2 in the supplemental material). These data indicate that the presence of mrpC may be highly strain specific and is not a common feature of Microcystis cells. Seven of the strains testing positive in the mrpC screening are known to produce microcystin and yielded a PCR product with primers specific for the mcyA gene (12), whereas one of the strains, namely, M. aeruginosa PCC 7005, did not contain the toxin biosynthesis genes (see Fig. S2 in the supplemental material). To further verify the strain specificity on the protein level and to get further insights into a possible association of MrpC with specific Microcystis morphotypes, we performed immunofluorescence studies of field samples isolated from Lake Dollgow, which is situated north of Berlin, Germany. At the time of the study, Microcystis was a predominant phytoplankton species in the lake. Formaldehyde-fixed net samples collected from the lake were permeabilized and hybridized using the MrpC antibody and a secondary FITC-labeled antibody as described above. About 30% of the Microcystis colonies yielded very bright green fluorescence, whereas the remaining colonies revealed only their autofluorescence, similar to controls without secondary antibody treatment. Representative colonies are shown in Fig. 7. Colonies reacting with the MrpC antibody showed strong fluorescence signals on the surfaces of cells but also in the surrounding sheath and showed ring-like structures in two-dimensional micrographs, as observed for the laboratory strain M. aeruginosa PCC 7806 (Fig. 7A to D). Neighboring colonies not reacting with the antibody showed signals neither at the cell surfaces nor in the surrounding sheath and were thus visible only under the 617-nm emission filter of the microscope due to their red autofluorescence (Fig. 7E to H). These data indicate that MrpC is completely lacking in more than half of the Microcystis colonies in Lake Dollgow and confirm the strain specificity of MrpC. The Microcystis colonies collected from Lake Dollgow did not show very pronounced morphotypes, probably due to the sampling period in early summer. Nevertheless, a closer inspection of the colonies reacting with the MrpC antibody revealed that they could be assigned to two distinct colony subtypes. One subtype was a group of globular colonies with densely packed cells that was designated Microcystis sp. and that is exemplified in the micrographs in Fig. 7A and B. The second subtype reacting with the MrpC antibody that is represented in Fig. 7C was assigned to the morphotype M. aeruginosa. These preliminary results indicate that the MrpC protein may be specific to certain Microcystis colony types. In order to support this hypothesis, more field samples from different lakes exhibiting pronounced morphotypes will have to be tested in the future.
FIG. 7.
Immunofluorescence micrographs of Microcystis field colonies from samples obtained from Lake Dollgow (Germany) in 2007 produced by using the antibody against MrpC and a FITC-coupled secondary antibody. Images of cells were acquired either with a green fluorescence emission filter (A to D) in the blue channel of the microscope or with a red fluorescence emission filter (E to H) in the green channel of the microscope. Panels D and H show enlarged sections of an individual colony.
DISCUSSION
In this study, we provide further evidence for the role of microcystin in the cellular aggregation of Microcystis PCC 7806 (16). We unambiguously show that a small extracellular protein, MrpC, is present in increased abundance in different subcellular fractions from a microcystin-deficient mutant compared to that in fractions from the WT M. aeruginosa PCC 7806. We have discovered that the cell surface-associated protein occurs in high oligomeric forms under native conditions and provide evidence for the posttranslational glycosylation of the protein. Immunofluorescence microscopy data indicate an implication of MrpC in the cell-cell attachment of M. aeruginosa PCC 7806, which is in accordance with the observation of an increased aggregation tendency of the microcystin-deficient mutants compared to that of the WT laboratory strain. Results from in situ studies of field samples containing a mixture of Microcystis morphotypes, as well as the screening of laboratory strains by PCR, suggest that the cell surface protein is highly strain specific and may be connected to distinct colony types of Microcystis.
Relationship between MrpC and microcystin and putative role of glycosylation.
So far, the nature of the relationship between microcystin and MrpC in M. aeruginosa PCC 7806 remains elusive. The strong increase in the abundance of the protein is not correlated with a corresponding increase of the transcript accumulation level of mrpC in microcystin-deficient mutants, suggesting similar transcription levels as well as similar levels of transcript stability (Y. Zilliges and E. Dittmann, unpublished data). As shown in this study, the lack of microcystin did not significantly alter the subcellular localization of MrpC or the oligomerization state of the protein. We could furthermore rule out the possibility that microcystin influences the expression of the protein through an interaction with a yet-unknown signaling cascade, as could be shown for the microcystin biosynthesis protein McyB that responds to the addition of microcystin (26). Microcystin may therefore rather affect the general stability of the MrpC protein. The stability may be related either to the glycosylation status of MrpC or to the expression level of a putative binding partner of the protein. At least one other cell surface protein, the cell surface-exposed lectin microvirin, shows differential accumulation and oligomerization patterns at the surfaces of ΔmcyB mutant cells compared to those at the surfaces of the microcystin-producing WT cells (16). However, so far there is no evidence for direct interaction of the two proteins. The increased accumulation of MrpC in microcystin-deficient mutants may thus be a secondary effect of the loss of microcystin and is possibly specific to the M. aeruginosa strain PCC 7806.
Much of the precise information on the effects of glycosylation on protein functions is derived from studies with eukaryotic glycoproteins. The effects of the addition and removal of O-GlcNAc modifications at serine and threonine residues of proteins in animal systems, where O glycosylation is emerging as a key regulator of nuclear and cytoplasmic protein activities, are particularly well-studied. O-GlcNAc modifications in animals are connected to histone remodeling, transcription, proliferation, apoptosis, and proteasomal degradation (19). There is not much information available on the functional importance of this type of glycosylation in bacteria. Most studies addressing this question were performed with pathogenic bacteria. In H. influenzae, a glycosyltranferase comprising a SPY domain, HMW1C, was shown previously to be associated with the glycosylation of the high-molecular-weight surface adhesin HMW1A that mediates attachment to human epithelial cells (10). In the absence of glycosylation, HMW1A is partially degraded and is efficiently released from the surface, resulting in reduced adherence. As suggested above, the glycosylation of MrpC may have a similar effect on protein stability. In many cases, bacterial glycoproteins form discrete structures on the cell surface, such as flagella, pili, and fimbriae (33). It is generally not well understood why glycoproteins frequently mediate attachment between cells or to the substrate. It may be speculated, however, that MrpC interacts with an unknown sugar binding protein or with lipopolysaccharides and, thus, may be part of a network mediating Microcystis colony formation.
Implications of MrpC for colony formation by and ecostrategy of Microcystis.
There are many indications that colony formation by Microcystis is a tightly balanced process (reference 16 and unpublished data). Batch cultures are artificial systems that enrich factors that would be diluted very fast in the field. Molecular studies of proteins that are potentially involved in cellular interactions are therefore only the first step toward an understanding of colony formation and colony type. Despite these limitations, the results of our studies clearly show that MrpC is a dominant cell surface-exposed protein in about 30% of Microcystis strains and is influenced by the presence or absence of microcystin in the M. aeruginosa strain PCC 7806. It remains to be shown if the protein is involved in cellular interactions in specific Microcystis colony types. As pointed out above, more and more data from the literature support a correlation of Microcystis chemotypes with certain morphotypes (8, 16, 34). There is increasing evidence that oligosaccharides attached to surfaces of cells provide specific information within biological systems and do not play only a structural role for the producing cells (33). MrpC as a surface-exposed glycoprotein may be one of the major factors discriminating among Microcystis ecotypes and, as such, may be a useful marker protein for future field studies.
Colony formation is an important precondition for the vertical migration behavior of Microcystis. Field studies have indicated that different Microcystis morphotypes may show different ecostrategies in the field (36). MrpC, as a potential colony type-specific factor, may be highly important for the competitive advantage of specific Microcystis ecotypes in the field. Future studies, including microarray analyses, will help to elucidate the complete subset of genes specific for the different Microcystis ecotypes. Applied to field situations, such analyses may provide a deeper understanding of the success of Microcystis in freshwater lakes worldwide.
Supplementary Material
Acknowledgments
We thank Stefan Börno, Jana Müller, Katrin Hinrichs, and Ramona Günther for technical assistance. We are further grateful to Arthur Guljamow and Harald Saumweber for their help with immunofluorescence microscopy analysis. We thank K. Ishida for providing microcystin-LR. We further thank S. Ferris, A. M. Castets, C. Pichon, L. Frangeul, A. Marcel, P. Glaser, and S. Cole (Plate-forme Génomique—Pasteur Génopole), who were involved in the genome sequencing project financed by the Institut Pasteur, the Ministère de l'Education Nationale, de la Recherche et de la Technologie (MENRT), and the Centre National de la Recherche Scientifique (URA 2172). We are further grateful to Julia Kehr for performing the Q-TOF analysis of the protein.
This work was supported by a grant of the German-Israeli-Foundation (GIF) to E.D. and T.B.
Footnotes
Published ahead of print on 15 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bonnet, M. P., and M. Poulin. 2002. Numerical modelling of the planktonic succession in a nutrient-rich reservoir: environmental and physiological factors leading to Microcystis aeruginosa dominance. Ecol. Modell. 15693-112. [Google Scholar]
- 2.Brahamsha, B. 1996. An abundant cell-surface polypeptide is required for swimming by the nonflagellated marine cyanobacterium Synechococcus. Proc. Natl. Acad. Sci. USA 936504-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castric, P. 1995. pilO, a gene required for glycosylation of Pseudomonas aeruginosa 1244 pilin. Microbiology 1411247-1254. [DOI] [PubMed] [Google Scholar]
- 4.Dittmann, E., M. Erhard, M. Kaebernick, C. Scheler, B. A. Neilan, H. von Döhren, and T. Börner. 2001. Altered expression of two light-dependent genes in a microcystin-lacking mutant of Microcystis aeruginosa PCC 7806. Microbiology 1473113-3119. [DOI] [PubMed] [Google Scholar]
- 5.Dittmann, E., B. A. Neilan, M. Erhard, H. von Döhren, and T. Börner. 1997. Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Mol. Microbiol. 26779-787. [DOI] [PubMed] [Google Scholar]
- 6.Dittmann, E., and C. Wiegand. 2006. Cyanobacterial toxins: occurrence, biosynthesis and impact on human affairs. Mol. Nutr. Food Res. 507-17. [DOI] [PubMed] [Google Scholar]
- 7.Doig, P., N. Kinsella, P. Guerry, and T. J. Trust. 1996. Characterization of a post-translational modification of Campylobacter flagellin: identification of a sero-specific glycosyl moiety. Mol. Microbiol. 19379-387. [DOI] [PubMed] [Google Scholar]
- 8.Fastner, J., M. Erhard, and H. von Döhren. 2001. Determination of oligopeptide diversity within a natural population of Microcystis spp. (cyanobacteria) by typing single colonies by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 675069-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fulda, S., S. Mikkat, F. Huang, J. Huckauf, K. Marin, B. Norling, and M. Hagemann. 2006. Proteome analysis of salt stress response in the cyanobacterium Synechocystis sp. strain PCC 6803. Proteomics 62733-2745. [DOI] [PubMed] [Google Scholar]
- 10.Grass, S., A. Z. Buscher, W. E. Swords, M. A. Apicella, S. J. Barenkamp, N. Ozchlewski, and J. W. St. Geme III. 2003. The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol. Microbiol. 48737-751. [DOI] [PubMed] [Google Scholar]
- 11.Gray, K. M., J. P. Pearson, J. A. Downie, B. E. Boboye, and E. P. Greenberg. 1996. Cell-to-cell signaling in the symbiotic nitrogen-fixing bacterium Rhizobium leguminosarum: autoinduction of a stationary phase and rhizosphere-expressed genes. J. Bacteriol. 178372-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hisbergues, M., G. Christiansen, L. Rouhiainen, K. Sivonen, and T. Börner. 2003. PCR-based identification of microcystin-producing genotypes of different cyanobacterial genera. Arch. Microbiol. 180402-410. [DOI] [PubMed] [Google Scholar]
- 13.Hoiczyk, E., and W. Baumeister. 1997. Oscillin, an extracellular, Ca2+-binding glycoprotein essential for the gliding motility of cyanobacteria. Mol. Microbiol. 26699-708. [DOI] [PubMed] [Google Scholar]
- 14.Jacobsen, S. E., K. A. Binkowski, and N. E. Olszewski. 1996. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. USA 939292-9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jochimsen, E. M., W. W. Carmichael, J. S. An, D. M. Cardo, S. T. Cookson, C. E. Holmes, M. B. Antunes, D. A. de Melo Filho, T. M. Lyra, V. S. Barreto, S. M. Azevedo, and W. R. Jarvis. 1998. Liver failure and death after exposure to microcystins at a hemodialysis center in Brazil. N. Engl. J. Med. 338873-878. [DOI] [PubMed] [Google Scholar]
- 16.Kehr, J. C., Y. Zilliges, A. Springer, M. D. Disney, D. D. Ratner, C. Bouchier, P. H. Seeberger, N. T. de Marsac, and E. Dittmann. 2006. A mannan binding lectin is involved in cell-cell attachment in a toxic strain of Microcystis aeruginosa. Mol. Microbiol. 59893-906. [DOI] [PubMed] [Google Scholar]
- 17.Komarek, J., and K. Anagnostidis. 1999. Cyanoprokaryota, p. 1-548. In H. Ettl, G. Gartner, H. Heynig, and D. Mollenhauer (ed.), Süβwasserflora von Mitteleuropa, vol. 1. Gustav Fischer, Jena, Germany. [Google Scholar]
- 18.Lambert-Buisine, C., E. Willery, C. Locht, and F. Jacob-Dubuisson. 1998. N-terminal characterization of the Bordetella pertussis filamentous haemagglutinin. Mol. Microbiol. 281283-1293. [DOI] [PubMed] [Google Scholar]
- 19.Love, D. C., and J. A. Hanover. 2005. The hexosamine signaling pathway: deciphering the “O-GlcNAc code.” Sci. STKE 2005re13. [DOI] [PubMed] [Google Scholar]
- 20.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193265-275. [PubMed] [Google Scholar]
- 21.Messner, P. 1997. Bacterial glycoproteins. Glycoconj. J. 143-11. [DOI] [PubMed] [Google Scholar]
- 22.Pearson, L. A., M. Hisbergues, T. Börner, E. Dittmann, and B. A. Neilan. 2004. Inactivation of an ABC transporter gene, mcyH, results in loss of microcystin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Appl. Environ. Microbiol. 706370-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Power, P. M., L. F. Roddam, K. Rutter, S. Z. Fitzpatrick, Y. N. Srikhanta, and M. P. Jennings. 2003. Genetic characterization of pilin glycosylation and phase variation in Neisseria meningitidis. Mol. Microbiol. 49833-847. [DOI] [PubMed] [Google Scholar]
- 24.Reinhold, B. B., C. R. Hauer, T. H. Plummer, and V. N. Reinhold. 1995. Detailed structural analysis of a novel, specific O-linked glycan from the prokaryote Flavobacterium meningosepticum. J. Biol. Chem. 27013197-13203. [DOI] [PubMed] [Google Scholar]
- 25.Rippka, R. 1988. Isolation and purification of cyanobacteria. Methods Enzymol. 1673-27. [DOI] [PubMed] [Google Scholar]
- 26.Schatz, D., Y. Keren, A. Vardi, A. Sukenik, S. Carmeli, T. Börner, E. Dittmann, and A. Kaplan. 2007. Towards clarification of the biological role of microcystins, a family of cyanobacterial toxins. Environ. Microbiol. 9965-970. [DOI] [PubMed] [Google Scholar]
- 27.Sherlock, O., U. Dobrindt, J. B. Jensen, R. Munk Vejborg, and P. Klemm. 2006. Glycosylation of the self-recognizing Escherichia coli Ag43 autotransporter protein. J. Bacteriol. 1881798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephenson, A. E., H. Wu, J. Novak, M. Tomana, K. Mintz, and P. Fives-Taylor. 2002. The Fap1 fimbrial adhesin is a glycoprotein: antibodies specific for the glycan moiety block the adhesion of Streptococcus parasanguis in an in vitro tooth model. Mol. Microbiol. 43147-157. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi, I., D. Hayano, M. Asayama, F. Masahiro, M. Watahiki, and M. Shirai. 1996. Restriction barrier composed of an extracellular nuclease and restriction endonuclease in the unicellular cyanobacterium Microcystis sp. FEMS Microbiol. Lett. 145107-111. [DOI] [PubMed] [Google Scholar]
- 30.Takeuchi, K., F. Taguchi, Y. Inagaki, K. Toyoda, T. Shiraishi, and Y. Ichinose. 2003. Flagellin glycosylation island in Pseudomonas syringae pv. glycinea and its role in host specificity. J. Bacteriol. 1856658-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tielker, D., S. Hacker, R. Loris, M. Strathmann, J. Wingender, S. Wilhelm, F. Rosenau, and K. E. Jaeger. 2005. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology 1511313-1323. [DOI] [PubMed] [Google Scholar]
- 32.Tillett, D., E. Dittmann, M. Erhard, H. von Döhren, T. Börner, and B. A. Neilan. 2000. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chem. Biol. 7753-764. [DOI] [PubMed] [Google Scholar]
- 33.Upreti, R. K., M. Kumar, and V. Shankar. 2003. Bacterial glycoproteins: functions, biosynthesis and applications. Proteomics 3363-379. [DOI] [PubMed] [Google Scholar]
- 34.Via-Ordorika, L., J. Fastner, R. Kurmayer, M. Hisbergues, E. Dittmann, J. Komarek, M. Erhard, and I. Chorus. 2004. Distribution of microcystin-producing and non-microcystin-producing Microcystis sp. in European freshwater bodies: detection of microcystins and microcystin genes in individual colonies. Syst. Appl. Microbiol. 27592-602. [DOI] [PubMed] [Google Scholar]
- 35.Walsby, A. E. 1994. Gas vesicles. Microbiol. Rev. 5894-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welker, M., L. Sejnohova, D. Nemethova, H. von Döhren, J. Jarkovsky, and B. Marsalek. 2007. Seasonal shifts in chemotype composition of Microcystis sp. communities in the pelagial and the sediment of a shallow reservoir. Limnol. Oceanogr. 52609-619. [Google Scholar]
- 37.Zhang, X., and V. Bennett. 1996. Identification of O-linked N-acetylglucosamine modification of ankyrinG isoforms targeted to nodes of Ranvier. J. Biol. Chem. 27131391-31398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.