Abstract
Investigations into the mechanisms of memory formation have abided by the central tenet of the consolidation theory—that memory formation occurs in stages which differ in their requirement for protein synthesis. The current most widely accepted hypothesis posits that new memories are encoded as neural activity-induced changes in synaptic efficacy, and stabilization of these changes requires de novo protein synthesis. However, the basic assumptions of this view have been challenged by concerns regarding the specificity of the methods used to support the claim. Studies on immediate-early genes (IEGs), in particular Arc, provide a distinct and independent perspective on the issue of the requirement of new protein synthesis in synaptic plasticity and memory consolidation. The IEG Arc and its protein are dynamically induced in response to neuronal activity, and are directly involved in synaptic plasticity and memory consolidation. Although we provide extensive data on Arc’s properties to address the requirement of genomic and proteomic responses in memory formation, Arc is merely one element in a network of genes that interact in a coordinated fashion to serve memory consolidation. From gene expression and other studies, we propose the view that the stabilization of a memory trace is a continuous and ongoing process, which does not have a discrete endpoint and cannot be reduced to a single deterministic “molecular cascade”. Rather, memory traces are maintained within metastable networks, which must integrate and update past traces with new ones. Such an updating process may well recruit and use many of the plasticity mechanisms necessary for the initial encoding of memory.
INTRODUCTION
The theory of memory consolidation, based on the idea that memory formation proceeds in stages and the stability and strength of newly formed memories increase with passage of time, has guided contemporary investigations into the neurobiological basis of learning and memory. The hypothesis originated from observations in human subjects in which interference introduced during a limited time after learning disrupted retention of learned information (Müller & Pilzecker, 1900). The term consolidation was adopted to describe the post-learning processes of memory stabilization. The idea was elaborated in later experiments when electroconvulsive shock administered to rodents at various time points post-training confirmed susceptibility of memory traces to interference at early, but not later, time points after learning (Duncan, 1949; Gerard, 1949). Thereafter, many investigations focused on identifying the molecular, cellular, and systems events and interactions, at successive time points post-learning, to address the mechanisms of memory consolidation (McGaugh, 2000).
Protein synthesis inhibitors (PSIs) became an important tool in the research on memory and consolidation since the seminal work of Agranoff, Davis, and Brink (1965) who administered the PSI puromycin into goldfish and demonstrated impairment of long-term memory. The obvious inference from this study was that long-term memory requires de novo protein synthesis whereas short-term memory does not. This study and others (Davis & Squire, 1984; Flexner, Flexner, & Stellar, 1963; Goelet, Castellucci, Schacher, & Kandel, 1986) solidified the current widely accepted model of memory consolidation, in which initially weak connections between newly recruited neurons become strengthened and stable in a de novo protein synthesis-dependent manner. The necessary cellular responses involved include activation of second-messenger systems, new RNA transcription, and protein synthesis. The mechanisms of stabilizing memory traces at the cellular level are referred to as “synaptic consolidation” (Dudai, 2004; Frankland & Bontempi, 2005). This is distinct from another form of consolidation, “systems” consolidation (Frankland & Bontempi, 2005), which denotes a reorganization of memory traces between brain regions. Although the terms “synaptic” and “systems” consolidation describe phenomena at different levels of analysis, the two processes may share similar mechanisms and occur in parallel. Reorganization of memory traces between brain regions (“systems”) may require modifications of connections (“synaptic”) within those networks. Here, we review the role of gene expression in memory with a focus at the “synaptic” consolidation level.
Some of the key support for the consolidation hypothesis came from studies examining the effects of PSIs on long-term memory. However, concerns about the technical issues and limits associated with the use of PSIs have been raised and brought on alternative explanations/hypotheses (Davis & Squire, 1984; Gold, 2006; Routtenberg & Rekart, 2005). For example, non-specific and toxic effects of PSIs (Gold, 2006; Routtenberg & Rekart, 2005; Rudy, Biedenkapp, Moineau, & Bolding, 2006) render uncertainties about whether the memory impairments observed in such studies are in fact due to direct inhibition of de novo protein synthesis. PSIs may not just inhibit new synthesis of proteins, but also induce kinase activation and apoptosis along with other unspecified effects (Rudy et al., 2006). As such, PSIs may selectively target active neurons made susceptible by their recent activity at the time of encoding and produce permanent alterations manifested as poor performance on retention testing (Rudy et al., 2006). Several studies reported a pharmacological “rescue” of PSI-induced amnesia and a lack of effects of PSIs on memory retention when training parameters were adjusted (reviewed in: Routtenberg and Reckart, 2005; Gold, 2006). These findings cast doubt on the requirement of de novo protein synthesis in memory consolidation. An alternative hypothesis proposes “post-translational protein modification (PTM)” of existing proteins as the only critical mechanisms for long-term memory (Routtenberg & Rekart, 2005). The PTM model suggests that modifications of proteins already present at activated synapses is necessary and sufficient for long-term memory, and that de novo transcription and translation merely serve a replenishment role. Another alternative suggests that de novo protein synthesis is critical in modulation, rather than consolidation, of memory, and it does not constitute the actual “substrate” of the memory trace (Gold, 2006). This suggestion explains the rescue of PSI-induced amnesia by pharmacological and training parameter manipulations. One must caution, however, that such pharmacological rescues of PSI induced amnesia with drugs such as amphetamine result in an altered brain state, and do not necessarily speak to how the brain normally processes information to form memories. The central issue of discussion in this article is whether newly synthesized proteins play an “instructive” role in the form of enabling plastic processes, as opposed to a “permissive” role, in the form of replenishment.
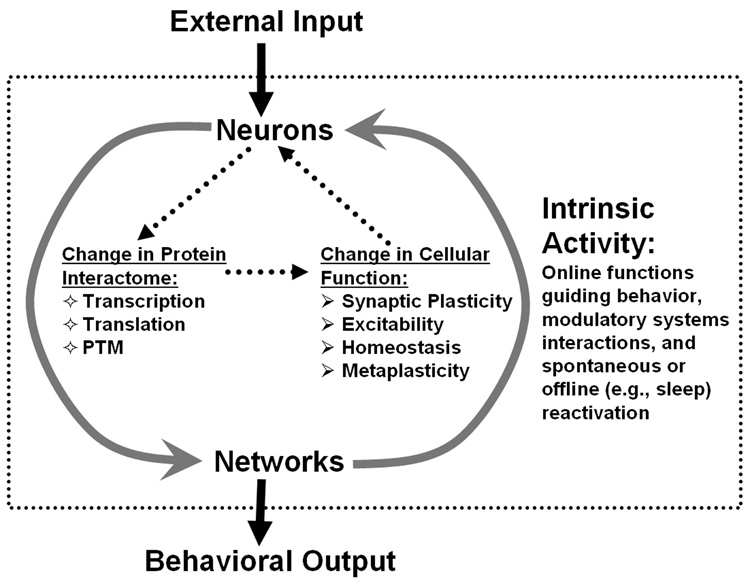
Nonspecific global and noxious effects of PSIs do confound interpretations of studies using these agents, but they do not necessarily rule out the requirement of de novo protein synthesis for formation of long-term memory. While the discussion over the methodological limitations associated with use of PSIs could ensue endlessly, contemporary studies employing sophisticated molecular biology techniques offer alternative approaches to test the question of whether memory consolidation requires “instructive” protein synthesis induced by neuronal activity. Specifically, studies examining the role of dynamically expressed immediate-early genes (IEGs) and proteins in memory processes address the issue of requirement for genomic and proteomic responses to activity in formation of long-term memory. Whereas concerns about non-specific targets of PSIs have been raised to dispute the contribution of de novo translation to synaptic plasticity, IEG studies counter these arguments by showing memory impairments after selectively blocking expression of specific IEG proteins, thus minimizing global toxic effects. Here, we demonstrate that IEG studies provide an independent perspective on the validity of the memory consolidation hypothesis and support for the requirement of activity/experience-dependent genomic responses for long-term memory. We start with an overview of IEGs and show how their induction profiles and cellular functions serve synaptic plasticity mechanisms thought to be necessary for long-term memory. Then, we review studies examining one particular IEG, Arc, and how the findings provide support for the requirement of a genomic response in memory consolidation. Furthermore, we describe how IEG/Arc studies can transcend levels of analysis, from the molecular to systems levels, to form an integrated view of memory function. Finally, we discuss how gene expression studies stimulate the idea that orchestrated expression of multiple activity-regulated genes is critical for gating synaptic plasticity and the ability of neurons to encode and store new information. Based on gene expression and other studies, we propose a dynamic model of memory, which integrates molecular, cellular, and systems level interactions underlying long-term memory (Fig. 1).
Figure 1. A dynamic model of the molecular, cellular, and systems interactions involved in the establishment and maintenance of memory.
Neuronal activation driven by external inputs (e.g, sensory experience) and intrinsic brain network activity (information of the internal state of the organism) activate second messenger systems in brain neurons which modify the protein-interaction network (“interactome”). These changes occur via rapid post-translational modification (PTM) of pre-existing proteins and through alterations in gene expression (via regulated transcription and translation). Modifications of the protein interactome alter functional properties of neurons within an ensemble to entrain them into a representation encoding a discrete experience. While necessary for initially establishing memory, these initial changes are likely not sufficient for lasting memory. Rather, repeated intrinsic network activity may continue to reinforce a memory trace, using many of the same molecular mechanisms that were required initially. In this model, memories may never become fully “consolidated”, but remain dynamic for as long as they persist.
A GENERAL INTRODUCTION TO IMMEDIEATE-EARLY GENES
Neurons are distinct from other cells in that they actively propagate electrical impulses over distance (action potentials) and communicate across specialized connections (synapses) using chemical messengers (neurotransmitters). Complex neuronal signal transduction machinery converts the chemical signal back to electrical potentials and induces long-lasting modification of cellular properties including the machinery itself. These mechanisms contribute to unique features of neurons as networked signaling elements. The activation of a neuron and its recruitment into an ensemble representing a particular memory initiates a mechanism for long-term modification of synaptic responses aimed to maintain the coherence of that ensemble (Figure 1). IEG expression is one of the steps in the cascade of cellular events comprising this mechanism (Clayton, 2000; Guzowski, 2002; Tischmeyer & Grimm, 1999). Operationally, IEGs are defined as those genes of which transcription can be induced in the presence of PSIs, thereby do not require de novo protein synthesis or previous activation of any other responsive genes. The tight coupling of IEG expression to patterned synaptic activity and their putative cellular functions make IEGs attractive candidates for critical components of synaptic plasticity processes underlying long-term memory.
IEG induction
IEG transcription is initiated by patterned synaptic activity that induces long-term synaptic plasticity (Abraham, Mason, Demmer, Williams, Richardson, Tate, Lawlor, & Dragunow, 1993; Worley, Bhat, Baraban, Erickson, McNaughton, & Barnes, 1993). Neurotransmitter binding to postsynaptic receptors, or action potential firing following integration of postsynaptic potentials (Adams & Dudek, 2005), results in an influx of extracellular Ca2+ or release of Ca2+ from internal stores (Berridge, 1998) and sets off a signal transduction cascade involving postsynaptic second-messengers and activation of protein kinases. Some of these kinases regulate nuclear gene expression, including protein kinase A (Delghandi, Johannessen, & Moens, 2005; Shaywitz & Greenberg, 1999), mitogen-activated protein kinase (Cammarota, Bevilaqua, Ardenghi, Paratcha, Levi de Stein, Izquierdo, & Medina, 2000; Davis & Laroche, 2006; Davis, Vanhoutte, Pages, Caboche, & Laroche, 2000; Sweatt, 2001; Wu, Deisseroth, & Tsien, 2001), and calcium- and calmodulin-dependent kinase IV (Deisseroth, Heist, & Tsien, 1998; Ginty, 1997; Soderling, Chang, & Brickey, 2001). Activated kinases target specific constitutive regulatory transcription factors (RTFs) in the nucleus, such as the cAMP response element binding protein (CREB) and serum response factor (SRF). RTFs then bind to promoter regions of IEGs (CREB to CRE and SRF to SRE) and confer responsiveness to second messengers or growth factors (Finkbeiner & Greenberg, 1998; Shaywitz & Greenberg, 1999). Activated RTFs are then capable of recruiting the transcriptional machinery (Ginty, 1997; Finkbeiner and Greenberg, 1998), thereby initiating IEG transcription. Increased levels of IEG transcription have been observed within minutes of stimulation both in vitro (Greenberg, Ziff, & Greene, 1986) and in vivo (Guzowski, McNaughton, Barnes, & Worley, 1999; Vazdarjanova, McNaughton, Barnes, Worley, & Guzowski, 2002).
Cellular Functions of IEGs
IEGs encode a diverse range of proteins including activity-induced RTFs, structural proteins, signal transduction proteins, growth factors, and enzymes. Initial studies of IEGs in the brain focused on activity-induced RTFs, such as c-fos, c-jun, and zif268 (also known as Krox-24, Egr1, NGFI-A, and zenk, in avian species)(Tischmeyer & Grimm, 1999). Subsequent cloning of brain/neuron-specific IEGs revealed those that encode proteins with a diverse range of cellular functions (Lanahan & Worley, 1998; Nedivi, Hevroni, Naot, Israeli, & Citri, 1993); these non-RTF IEGs are called “effector IEGs” and include Arc, Homer 1a, tissue-plasminogen activator, Narp, and BDNF. By definition, RTF IEGs regulate transcription of other genes. RTF IEGs are speculated to drive expression of delayed effector genes (Clayton, 2000; Morgan & Curran, 1991). Delayed effector genes are expressed after IEGs and may play specific roles in neurotransmission, cellular maintenance, and plasticity. It is also speculated that RTF IEGs play a role in enabling metaplasticity or function as coincidence detectors (Clayton, 2000; Guzowski, 2002; Kaczmarek, 2000). Effector IEGs have a wide range of cellular functions, including those related to cellular growth (BDNF, Narp), intracellular signaling (RheB, RGS-2, Homer 1a), synaptic modification or other structural changes (Arc, Homer 1a, Narp, tissue plasminogen activator, BDNF), metabolism (COX-2), or synaptic homeostasis (Arc, Homer 1a)(Guzowski, 2002; Lanahan & Worley, 1998; Shepherd, Rumbaugh, Wu, Chowdhury, Plath, Kuhl, Huganir, & Worley, 2006). These functions are compatible with the types of synaptic modifications that are thought to underlie synaptic plasticity. Notably, IEG expression is low in quiescent animals; even “basal” expression is dependent on synaptic input (Lyford, Yamagata, Kaufmann, Barnes, Sanders, Copeland, Gilbert, Jenkins, Lanahan, & Worley, 1995).
A PARTICULARLY INSTRUCTIVE IEG: ARC
Of the IEGs investigated in the field of learning and memory, the effector IEG Arc [activity-regulated cytoskeleton-associated protein (Lyford et al., 1995), which is also known as Arg3.1 (Link, Konietzko, Kauselmann, Krug, Schwanke, Frey, & Kuhl, 1995)] has received particular attention because of its tight experience-dependent regulation in behaviorally defined neural networks (Guzowski et al., 1999; Vazdarjanova et al., 2002), its mRNA transport to and expression at activated synapses (Moga, Calhoun, Chowdhury, Worley, Morrison, & Shapiro, 2004; Steward, Wallace, Lyford, & Worley, 1998; Steward & Worley, 2001a; Steward & Worley, 2001b), its requisite for both late-LTP and long-term memory (Guzowski, Lyford, Stevenson, Houston, McGaugh, Worley, & Barnes, 2000; Plath, Ohana, Dammermann, Errington, Schmitz, Gross, Mao, Engelsberg, Mahlke, Welzl, Kobalz, Stawrakakis, Fernandez, Waltereit, Bick-Sander, Therstappen, Cooke, Blanquet, Wurst, Salmen, Bosl, Lipp, Grant, Bliss, Wolfer, & Kuhl, 2006), and its capacity for modification of synaptic function (Plath et al., 2006; Rial Verde, Lee-Osbourne, Worley, Malinow, & Cline, 2006). The following review of Arc’s transcription properties, putative functions, and regulation at cellular and network levels provides further evidence for a requirement of an instructive genomic (and proteomic) response for long-term memory formation.
Activity-regulated Arc transcription
Using differential cloning techniques following maximal electroconvulsive seizures (MECS), Arc/Arg3.1 cDNA of approximately 3,000 base pairs was identified by both the Kuhl and Worley groups (Link et al., 1995; Lyford et al., 1995). The comparison of the sequence to the GenBank suggested that c-terminus of Arc (aa155-316) shares low to modest homology with the actin-binding α-spectrin, a cytoskeletal component. Other than that, Arc lacks homology with any other genes and does not belong to any gene family, whereas many other IEGs do (i.e. c-fos, Homer 1a, c-jun, jun-B; Egr 1–4 [Egr 1 is also known as zif268]). It is speculated that Arc may have appeared relatively late in evolution, as current evidence fails to show an obvious homolog in invertebrate species. This singularity in the genome suggests a highly specific function(s) for Arc. Northern blot analysis of Arc mRNA expression revealed a low basal level and a dramatically elevated level in rat hippocampus and neocortex following MECS. This increase was blocked by a systemic injection of a NMDA antagonist MK-801, demonstrating that Arc transcription is induced by excitatory synaptic activity via NMDA receptor activation (Link et al., 1995; Lyford et al., 1995). Moreover, unilateral intraocular injection of tetrodotoxin showed that the high basal levels of Arc in the visual cortex reflect sensory processing and not constitutive gene expression (Lyford et al., 1995).
The time course of Arc transcription has been analyzed using MECS and fluorescence in situ hybridization (FISH) (Guzowski et al., 1999). At rest, Arc RNA transcription is only detected in a few neurons in the hippocampus and neocortex. Within 1–2 minutes of MECS, however, intense intranuclear foci (INF) labeling of active transcription is present in most neurons, and it disappears within ~15 minutes. Subsequently, prominent labeling arises in cytoplasmic and dendritic regions, from 15 to 45 min after the stimulus. After that, Arc mRNA could be observed primarily in dendritic regions.
Arc is also expressed following stimuli that induce long-term potentiation (LTP) (Link et al., 1995; Lyford et al., 1995; Waltereit, Dammermann, Wulff, Scafidi, Staubli, Kauselmann, Bundman, & Kuhl, 2001) as well as after exploration of an environment and learning tasks in vivo (Guzowski et al., 1999; Guzowski, Miyashita, Chawla, Sanderson, Maes, Houston, Lipa, McNaughton, Worley, & Barnes, 2006; Vazdarjanova & Guzowski, 2004; Vazdarjanova et al., 2002). The time-course of behavioral induction of Arc in the hippocampus follows the same pattern as after MECS (Guzowski et al, 1999). Figure 2 illustrates this dynamic induction of Arc transcription in hippocampal and neocortical neurons in rats 25 min after exploration of an open-field environment. Whereas very low levels of Arc mRNA are found in rats taken directly from their home cages (“caged control”), Arc expression is robustly, yet transiently, induced by behavioral experience. A recent study showed that Arc is induced exclusively in α calcium-calmodulin kinase II (αCaMKII) expressing principal neurons in the striatum, hippocampus, and neocortex (Vazdarjanova, Ramirez-Amaya, Insel, Plummer, Rosi, Chowdhury, Mikhael, Worley, Guzowski, & Barnes, 2006). The tight association of Arc expression with αCaMKII supports the hypothesis that Arc and αCaMKII act as “plasticity partners”, which promote synaptic modifications that accompany learning in principal neurons (Vazdarjanova et al., 2006).
Figure 2. Robust induction of Arc mRNA expression in hippocampus and neocortex by behavioral experience.
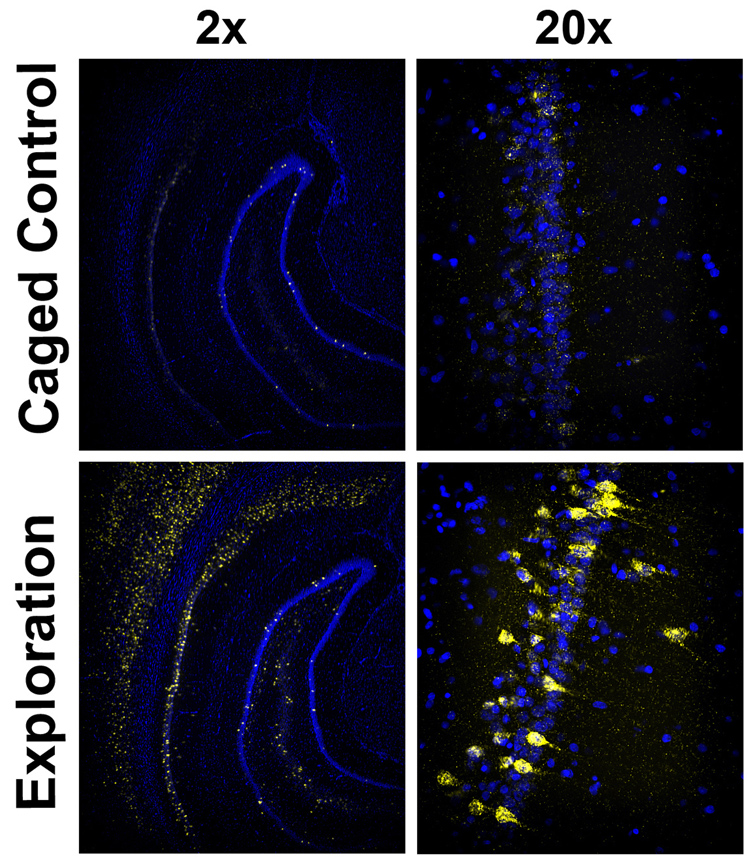
Fluorescence in situ hybridization to Arc mRNA in 20 micron brain sections of rats sacrificed directly from the home cage (upper panel; “caged control”) or 25 min after a 5 min exposure to a novel environment (lower panel; “exploration”). Cell nuclei are shown in blue and Arc mRNA is shown in yellow. The left panels show low magnification images (2x objective) of ventral hippocampus and surrounding neocortex, and the right panels show higher magnification images (20x objective) of the CA1 region of hippocampus from the same field. Note the extremely low levels of Arc mRNA in the caged control and the dramatic increase throughout the neocortex and each subregion of the hippocampus.
The transient activation of Arc transcription described above is seen in many regions of the brain including the pyramidal cell layers of the hippocampus, the neocortex, the striatum, and the amygdala. Interestingly, in the granule cells of the dentate gyrus, Arc RNA and protein exhibit a greatly prolonged period of expression extending to 8 hours following stimulation (Ramirez-Amaya, Vazdarjanova, Mikhael, Rosi, Worley, & Barnes, 2005). The significance of this sustained expression is not yet known.
Arc protein is expressed at very low levels in the brains of caged control animals, and its expression peaks around 1 hr following behavioral induction (Ramirez-Amaya et al., 2005). Like RNA, Arc protein is expressed in a subset of hippocampal neurons in an all-or-none fashion following behavioral activation (Fig. 3; left panels). Consistent with past reports of a short half-life of Arc protein, treatment of rats with the PSI anisomycin before training results in essentially no Arc protein expression within 90 minutes (Figure 3; right panels).
Figure 3. “Digital” expression of Arc protein in hippocampal neurons.
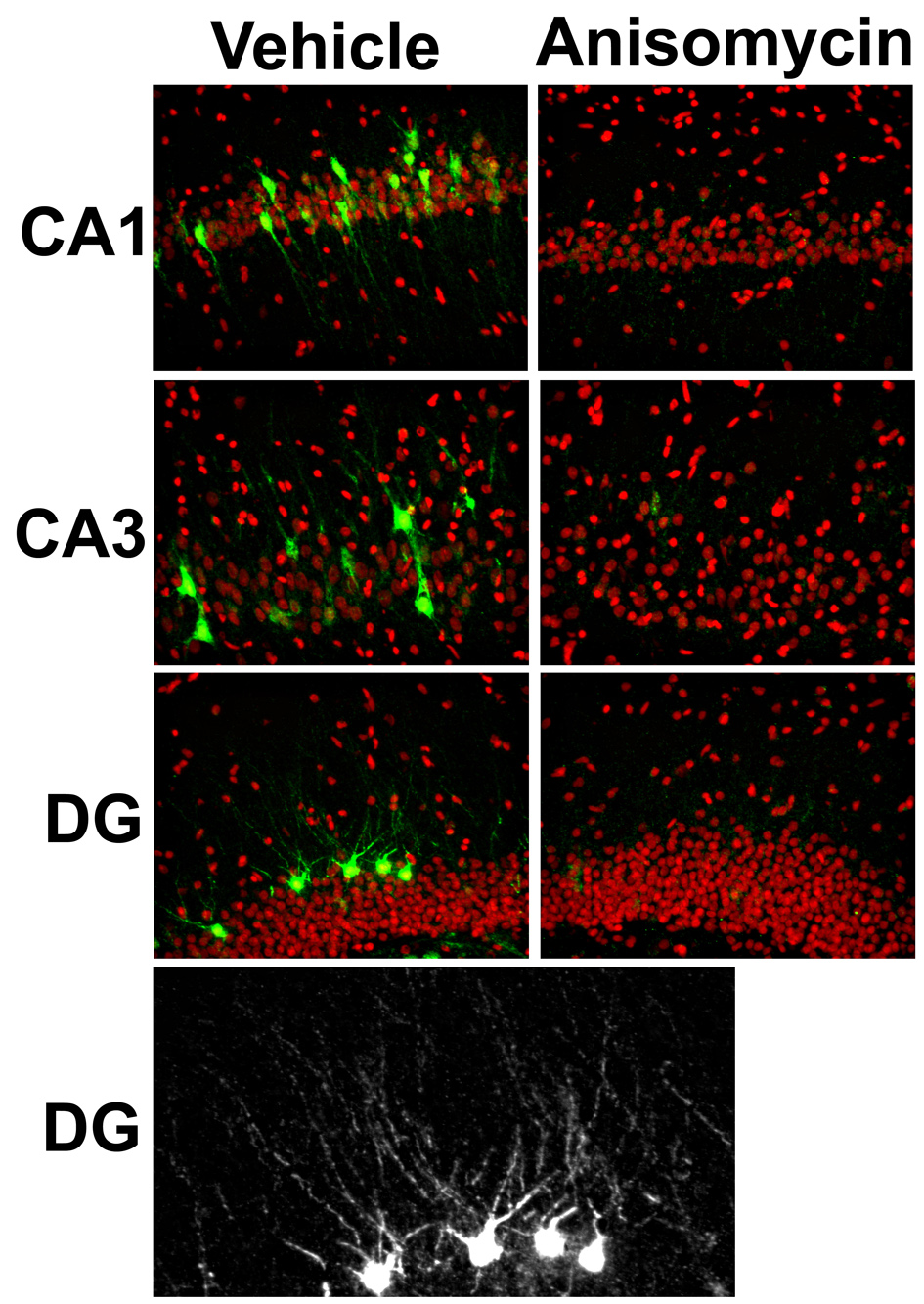
Rats were given 4 sessions (of 5 min each) of training on a rectangular track over a 90 min period, and then sacrificed. Ten minutes before the start of the behavioral procedure, the rats were given an i.p. injection of vehicle (left panels) or the protein synthesis inhibitor anisomycin (right panels; 210 mg / kg). As seen with Arc mRNA (Fig. 2), immunofluorescent detection of Arc protein (green) shows discrete labeling in a subset of the total population of cells (DAPI, here shown in red) in CA1, CA3, and DG regions of the hippocampus of vehicle treated rats (left panels). By contrast, the virtually absent staining in the sections from the anisomycin treated rats (right panels) is even lower than seen in caged controls (not shown), providing further evidence of the short half-life of Arc protein (see also Ramirez-Amaya et al., 2005). The lower grayscale panel is a crop of Arc protein staining for the DG of a vehicle treated rat. Please note the digital nature of Arc protein expression and its punctuate labeling within the dendritic arbors of specific granule cells.
Functional studies of Arc in synaptic plasticity and memory consolidation
Given its dynamic regulation by neural activity in vivo and localization to activated regions of the dendritic arbor (Moga et al., 2004; Steward et al., 1998; Steward & Worley, 2001a; Steward & Worley, 2001b), Arc is an excellent candidate for a critical agent in synaptic plasticity and long-term memory formation. The functional role of Arc in synaptic plasticity has been examined by inhibiting Arc protein synthesis using intrahippocampal infusions of antisense oligodeoxynucleotides (ODNs) to reversibly reduce expression by 60% relative to control scrambled ODNs (Guzowski et al., 2000). Additionally, knockout mice have been generated to assess the role of Arc in synaptic plasticity and memory. Antisense knockdown or transgenic knockout approaches provide greater specificity for investigating the role of gene expression as compared to the use of PSIs or transcription inhibitors (such as actinomycin D), which act globally and indiscriminately.
Antisense studies of Arc function
Ninety minutes before induction of perforant path / dentate gyrus LTP, Arc antisense ODNs were delivered to one hippocampus and scrambled ODNs to the contralateral one. The same LTP-inducing protocol was applied to both hemispheres. Whereas LTP induction (early-LTP) was equivalent in both hippocampi, its maintenance (late-LTP) in the Arc antisense ODN-treated hemispheres decayed more rapidly than in scrambled ODN-treated hemisphere, with a significant difference detected within hours after induction (Guzowski et al., 2000).
Using the same approach, the effects of inhibiting hippocampal Arc protein expression on retention of spatial memory were assessed in the reference version of the water maze (Guzowski et al., 2000). Arc antisense or scrambled control ODNs were infused bilaterally into the hippocampus 3 hr before training sessions. Neither infusion affected acquisition of this task, but the Arc antisense ODNs significantly impaired spatial memory in a probe test, given 2 days later. In a separate experiment, Arc antisense or scrambled control ODNs were infused into the hippocampus immediately after two training sessions in the spatial water maze task, with the two sessions separated by just 2 min. In addition, another group was given post-training infusion of the Arc antisense ODNs 8 hours after training. In this study, the animals given the scrambled ODNs immediately after training and those given the Arc antisense ODNs 8 hours after training performed indistinguishably in the 48 hour retention probe test, showing strong spatial bias for the training location. By contrast, the rats given the Arc antisense ODNs immediately after the training were strongly impaired relative to the other two groups. These results show that Arc expression in the hippocampus, immediately after training, is critical for consolidation of long-term spatial memory. In addition, pre-training intrahippocampal administration of Arc antisense ODNs also significantly impaired retention performance in inhibitory avoidance 48 hrs after training (McIntyre et al., 2005).
Arc knock-out mice
Recently, Kuhl and colleagues described the effect of germline knockout of the Arc/Arg3.1 gene in mice (Plath et al., 2006). These mice are congenitally completely deficient of the Arc open reading frame, but due to a lack of homology with any other genes, the possibility of compensatory alterations is low. The mice are viable with normal histological and electrophysiological phenotypic features. Nonetheless, Arc knockout mice were profoundly deficient in long-term plasticity. In both in vivo perforant path / dentate gyrus LTP and in vitro Schaffer collateral / CA1 LTP, Arc knockout mice (or hippocampal slices derived from them) showed an enhanced early- and deficient late- LTP. LTP in both regions returned to baseline levels within 60 to 90 min, coinciding with the time course of Arc protein expression. Furthermore, Arc knockouts showed impairments both in the initial induction as well as in the maintenance of Schaffer collateral / CA1 long-term depression (LTD). Thus, Arc knockout mice were incapable of stable modifications of synaptic weights in hippocampal networks.
Consistent with the profound deficits in long-term synaptic plasticity, Arc knockout mice showed impaired long-term memory in several behavioral tasks, including spatial water maze task, cued and contextual fear conditioning, conditioned taste aversion, and object recognition (Plath et al., 2006). In the tasks where measures of short-term memory and acquisition were available, the Arc knockout mice did not differ from their littermate controls in those measures. These findings are consistent with the earlier studies using Arc antisense ODNs showing that Arc does not play a role in acquisition of a task or short-term memory, but is essential for long-term memory consolidation.
Transgenic mice that express GFP in place of Arc (Arc-GFP+) have also been used to study plasticity in the visual cortex in response to visual experience (Wang et al., 2006). In Arc+GFP+ heterozyogous mice, everyday changes in the orientation of visual stimuli resulted in tuning of neuronal ensembles that respond to stimulus orientation in the visual cortex. In contrast, experience-dependent plasticity in the size as well as specificity of neuronal ensembles responding to the stimuli orientation was absent in the Arc-GFP+ homozygous mice.
In summation, the use of physiological, behavioral, and molecular approaches all support the critical role of Arc expression in synaptic plasticity and the formation of long-term memory.
Arc in memory modulation
Considerable experimental evidence indicates that the basolateral complex of the amygdala (BLA) modulates memory storage by facilitating plastic changes in other brain regions, including the hippocampus (Guzowski & McGaugh, 1997). It has been shown that BLA neuronal activity can modulate hippocampal LTP (Ikegaya, Saito, & Abe, 1994; Ikegaya, Saito, & Abe, 1995) and performance in hippocampal dependent task (Hatfield & McGaugh, 1999; Liang, Juler, & McGaugh, 1986). Given that Arc plays a critical role in the maintenance of hippocampal LTP (Guzowski et al., 2000), McIntyre et al. (2005) tested the hypothesis whether BLA neuronal activity influences learning induced Arc gene expression in the hippocampus. Infusions of the β-adrenoreceptor agonist, clenbuterol, into the BLA immediately after training in an inhibitory avoidance task enhanced memory tested 48 h later. The same dose of clenbuterol significantly increased Arc protein levels in the dorsal hippocampus 45 min after training. Additionally, posttraining intra-BLA infusions of a memory-impairing dose of a local anesthetic lidocaine significantly reduced Arc protein levels in the dorsal hippocampus at the same time point. Interestingly, these changes in Arc protein levels, associated with intra-BLA clenbuterol or lidocaine, were not accompanied by similar alterations in Arc mRNA levels 30 min after training. These data are consistent with the notion that amygdala modulation of Arc protein expression in efferent brain regions occurs at a posttranscriptional level. This regulation of Arc protein expression may be mediated by mTOR pathway (Takei, Inamura, Kawamura, Namba, Hara, Yonezawa, & Nawa, 2004; Tsokas, Ma, Iyengar, Landau, & Blitzer, 2007). McIntyre et al. then used Arc antisense ODNs to directly block training induced Arc protein expression in the hippocampus. This manipulation significantly impaired 48 h retention performance in the inhibitory avoidance, supporting the hypothesis that the BLA influences long-term memory, at least in part, by modulating plasticity in the hippocampus. The above findings together suggest that Arc mRNA and protein are both required for and underlie memory consolidation, although memory can be modulated by regulation of Arc protein expression.
Reported cellular functions of Arc
The critical role for Arc in the maintenance of long-term plasticity and memory consolidation drew interest in determining the cellular function, or functions, of Arc. In studies of transfected primary neurons, Fujimoto et al., (2004) suggested a role for Arc in destabilizing the cytoskeleton through interactions with microtubule associate protein 2 (MAP2). Such interactions could be important for dendritic remodeling associated with long-lasting alterations of synaptic efficacy. Another in vitro study suggested that Arc interacts with calcium-calmodulin kinase II (CaMKII), and that this interaction promotes neurite outgrowth in cultured neuroblastoma cells (Donai, Sugiura, Ara, Yoshimura, Yamagata, & Yamauchi, 2003). In a recent study using immunofluorescence, Arc protein was shown to co-localize with GluR4 receptors and co-precipitate with actin and GluR4 in turtle brain (Mokin, Lindahl, & Keifer, 2006).
While the above studies are all consistent with Arc interacting with a number of different dendritic proteins (in NMDA receptor multiprotein complexes, with MAP2, with CaMKII, and with actin and GluR4), they are largely correlative or come from heterologous model systems. Moreover, these studies lacked a strong mechanistic functional dissection. Recently, Worley and colleagues identified a novel cellular function for Arc. Initially, yeast two-hybrid screens were used to identify potential protein interaction partners of Arc. Two partners were identified that implicate Arc in the neural endocytotic pathway: dynamin 2 and endophilin 3. In a series of well designed cell biological studies, Chowdury et al. (2006) provided strong evidence that Arc facilitates endocytosis of AMPA receptors through its interactions with dynamin 2 and endophilin 3. Shepherd et al. (2006) extended these findings by showing that Arc plays a critical role in homeostatic synaptic scaling of AMPA receptors in cultured hippocampal and cortical neurons. In normal neuronal cultures blocking neural activity with tetrodotoxin decreased Arc expression and led to upregulation of surface expression of AMPA receptors. By contrast, increasing neural activity in normal cultures by picrotoxin, a GABA A receptor antagonist, increased Arc expression and decreased surface expression of AMPA receptors. In neuronal cultures from Arc knockout mice the ability to modulate surface AMPA receptor expression was absent in both tetrodotoxin and picrotoxin containing media. However, transfection of wild type Arc into Arc knockout neurons rescued the synaptic scaling phenotype, whereas Arc mutants with deletion in the regions necessary for interaction with endophilin 3 did not result in rescue. The hypothesis that Arc regulates surface AMPA receptor equilibrium by enhancing endocytosis was also supported in a study using hippocampal organotypic slice cultures (Rial Verde et al., 2006).
Although the cell biological data implicating Arc in regulating AMPA receptor endocytosis is compelling, it is not easy to see how endocytosis of AMPA receptors by an Arc-dynamin-endophilin complex could be involved in stabilizing LTP, which is believed to be, at least in part, due to insertion of AMPA receptors (Malinow & Malenka, 2002). Rial Verde et al., (2006) addressed this issue by tentatively suggesting that whereas activity-dependent increase in synaptic efficacy occurs via insertion GluR1 subunit-containing AMPA receptors, the Arc-mediated endocytosis is specific for GluR2 receptor subunit. Recently potentiated synapses are then less affected by the Arc-mediated endocytosis because of their higher GluR1 content. This explanation, however, will require further testing in reconciling these cell biological data with studies on in vivo plasticity and memory in behaving animals (Tzingounis & Nicoll, 2006). Nevertheless, from the perspective of memory consolidation and storage, it is attractive to think of Arc as a critical molecule for maintaining synaptic homeostasis in hippocampal and neocortical networks. It is important to note that in this view, the homeostatic function is an integral part of synaptic plasticity, and its impairment results in rapid deterioration of the system, as documented by the studies using Arc knockout mice or Arc antisense ODNs.
REGULATION OF ARC IN BEHAVIORALLY RELEVANT NEURAL CIRCUTS
To date, there are at least 40 publications showing upregulation of Arc in response to learning, memory retrieval, or stimulus presentation in rodents. Of the studies examining Arc expression in learning and memory, perhaps the most compelling argument for a specific role for Arc in modifying discrete neural ensembles associated with information processing has come from the so-called “catFISH” studies (“cellular compartment analysis of temporal activity by fluorescence in situ hybridization”; Guzowski, Timlin, Roysam, McNaughton, Worley, & Barnes, 2005). catFISH exploits the precise timing of Arc transcription activation and mRNA processing (described in the section, “A Particularly Instructive IEG: Arc) to allow the experimenter to compare two cellular activity maps, for two discrete behavioral experiences, within the same brain. In such experiments, cytoplasmic Arc mRNA provides a marker of neurons active 30 min prior to death, and Arc intranuclear transcription foci identify neurons active immediately before death. In this way, catFISH provides both cellular and temporal resolution for monitoring not only the proportion of neurons activated by an experience, but also the degree of overlap of ensembles for a similar or different experience. catFISH was first validated by examining neuronal activation patterns in the CA1 region of the hippocampus of rats exposed sequentially to either the same environment twice or to two distinct environments (Guzowski et al., 1999). Exploration of the same environment twice induced Arc in highly similar ensembles of neurons, whereas exploration of different environments induces Arc in distinct ensembles. This and subsequent studies demonstrated that the activation of Arc transcription in hippocampal pyramidal neurons is information content specific (Guzowski et al., 1999; Guzowski et al., 2006; Vazdarjanova et al., 2002).
A variant of catFISH imaging (Arc/Homer1a catFISH) uses the different temporal profiles of expression of two IEGs, Arc and Homer 1a, to provide an even more robust readout of neural activity maps for two experiences (Guzowski, Timlin, Roysam, McNaughton, Worley, & Barnes, 2005; Vazdarjanova & Guzowski, 2004). Like Arc catFISH, neurons active just prior to death are detected by Arc intranuclear transcription foci, whereas neurons active 30 min before death are marked by Homer 1a intranuclear transcription foci using a specific riboprobe to the 3’ untranslated region of Homer 1a. Arc/Homer1a catFISH has been used to probe the dynamics of information processing within the hippocampus, by comparing the ensemble responses of CA3 and CA1 neurons to defined perturbations of the environment (Vazdarjanova & Guzowski, 2004). These studies of Arc and Homer 1a activation in CA1 and CA3 are consistent, both qualitatively and quantitatively, with single unit recording studies of complex spiking activity of hippocampal neurons (Guzowski, Knierim, & Moser, 2004; Guzowski et al., 2006). Our current working model is that the expression of a hippocampal firing field, characterized by bursts of action potentials, is sufficient to activate transcription of Arc and probably other similarly sensitive IEGs such as zif268 and Homer 1a.
Assessment of Arc transcription using FISH and confocal microscopy has been a powerful method to assay neuronal activity engaged during distinct behavioral epochs in different brain regions [e.g., (Burke, Chawla, Penner, Crowell, Worley, Barnes, & McNaughton, 2005; Chawla, Guzowski, Ramirez-Amaya, Lipa, Hoffman, Marriott, Worley, McNaughton, & Barnes, 2005; Guzowski, Setlow, Wagner, & McGaugh, 2001; Han, Kushner, Yiu, Cole, Matynia, Brown, Neve, Guzowski, Silva, & Josselyn, 2007; Ramirez-Amaya et al., 2005; Zhang, Guzowski, & Thomas, 2005; Zou & Buck, 2006)]. In addition to the studies in hippocampus, Arc catFISH has been used to investigate neuronal ensemble responses in parietal (Burke et al., 2005) and olfactory (Zou & Buck, 2006) cortices. Lastly, Petrovich and colleagues combined Arc/Homer 1a catFISH with tract tracing methods to study the influence of amygdalar and prefrontal cortical inputs to the lateral hypothalamus in driving conditioned potentiation of feeding (Petrovich, Holland, & Gallagher, 2005). The catFISH approaches to brain activity mapping have been very informative at a “systems” level of analysis in memory formation, revealing neuronal network organization and activation patterns during encoding and retrieval in vivo. More germane to the current article, these various studies have shown that IEG induction is induced in neural circuits in an information content specific manner, providing further evidence of an instructive (and specific) role for activity-regulated gene expression to memory consolidation processes.
WHAT ARC STUDIES TELL US ABOUT MEMORY CONSOLIDATION
As discussed above, Arc mRNA and protein are dynamically regulated in hippocampal neural ensembles associated with information processing, and in a dramatic all-or-none fashion (Figure 2 and Figure 3; Guzowski et al., 1999; Ramirez-Amaya et al.,, 2005, Vazdarjanova and Guzowski, 2004). In addition, Arc mRNA and protein are targeted to active dendrites (Moga et al., 2004; Steward & Worley, 2001a; Steward & Worley, 2001b), and can be locally translated in synaptoneurosomes by well defined mechanisms such as the BDNF and mTOR pathways (Takei et al., 2004; Yin, Edelman, & Vanderklish, 2002). Lastly, Arc gene expression is critical to the maintenance of changes in synaptic efficacy and in long-term memory consolidation (Guzowski et al., 2000; Plath et al., 2006). Therefore, Arc studies argue against hypotheses that downplay the importance of de novo protein synthesis to the formation of long-term memory.
The cumulative data on Arc are hard to reconcile with the strict interpretation of the “PTM” model of Routtenberg and Reckart (2005). Specifically, the only stated role for new protein synthesis is in the service of passive “replenishment” of proteins depleted in earlier plastic events. As demonstrated repeatedly, there is no constitutive Arc protein present to replenish in the basal state of neurons (Guzowski et al., 1999, Vazdarjanova et al., 2002, Ramirez-Amaya, et al., 2005; Guzowski et al., 2006; Figure 2 and Figure 3). Instead, the dynamic regulation of Arc transcription and translation can more readily be interpreted as “instructive” to synaptic plasticity. Moreover, the aforementioned dendritic expression of Arc addresses perceived deficiencies of synapse specificity in protein synthetic plasticity mechanisms as described by Routtenberg and Reckart (2005). To be clear, however, there was a second and larger component of the PTM model, which is very much in line with our view (see Figure 1). This second element in the PTM model (p. 16, 2nd paragraph in the original publication) describes the metastability of memory networks and suggests “that change underlying memory storage is never stable at the level of an individual synapse”. This view is similar to ours (Figure 1) and that of others in the field, who suggest a greater degree of dynamics in the synaptic nature of memory traces (Abraham & Robins, 2005; Gold, 2006; Wittenberg, Sullivan, & Tsien, 2002).
The other alternative hypothesis suggests that de novo protein synthesis is not a critical substrate of memory formation, but it takes part in memory modulation (Gold, 2006). This claim is based on findings that amnesia induced by PSIs or inhibition of a specific substrate such as RTF CREB can be “rescued” by pharmacological manipulations. Indeed, McIntyre et al., (2005) demonstrated that Arc protein expression can be “modulated”. However, modulation does not exclude a direct role in consolidation. Mechanisms that are involved in consolidation may be defined as essential in memory, but the same mechanisms can be enhanced or suppressed. The reviewed data suggest that Arc is certainly a part of memory consolidation, but also plays a part in modulation.
Our view of the role of gene expression in memory is not a “genocentric” one, where gene expression drives a deterministic “molecular cascade” to long-term memory. Rather, we suggest an integrated model in which experience-dependent gene expression plays a necessary, but not sufficient, role in the formation of lasting memories (Figure 1). Central to this view, is the idea that memory is not a function of single genes, but rather of neural circuits. The pattern of gene expression within a given neuron, at a given time, is one factor in dictating the range of responses of that neuron, which influence its recruitment to, and stabilization in, neural circuits. In this sense, experience-dependent gene expression plays a role in enabling plastic states important for memory, just as intrinsic rhythms (such as hippocampal theta) do.
NETWORKS OF NEURONS, NETWORKS OF GENES: A SYSTEMS BIOLOGY VIEW OF THE ROLE OF EXPERIENCE-DEPENDENT GENE EXPRESSION TO NEURAL FUNCTION AND MEMORY
As described in the preceding sections, Arc plays a critical role in the maintenance of synaptic change and in consolidation of long-term memory. Furthermore, Arc gene expression is tightly linked to distinct patterns of neural activity in the behaving rodent. Thus, Arc is induced in defined neural ensembles associated with encoding of new information, and its expression is essential for consolidating this information. These data fit with the idea that new learning recruits plastic processes, such as those requiring alterations in genomic or proteomic processes. But what about retrieval of memory from an established and stable circuit—are the same or different activity-dependent molecular processes engaged? It is sometimes assumed that neural activity during retrieval is distinct from that during initial encoding and does not engage the same plasticity mechanisms. Our studies of Arc expression, however, fail to show disengagement of Arc induction in conditions of familiarity or memory retrieval. Specifically, the activation of Arc transcription in hippocampal neurons does not habituate with repeated exposures to the same environment (Guzowski et al., 2006), with overtraining in the spatial water maze task (Guzowski et al, 2001; Miyashita and Guzowski, unpublished observation), or with days of repeated exposure on a minimal track environment (Miyashita, Kubik, and Guzowski; unpublished observation). Moreover, we continue to see elevated Arc protein expression in rats overtrained in the spatial water maze task (Miyashita and Guzowski, unpublished observation). Thus, Arc gene expression remains coupled to neural activation, and does not distinguish between neural activity associated with new learning or memory retrieval.
To investigate whether other genes show differential expression at distinct learning stages, we examined the hippocampal gene expression profiles in rats during various stages of learning. In this experiment, rats were trained to learn the location of a submerged platform in the standard spatial water maze task using past protocols (Guzowski et al., 2001). Each training session consisted of 5 trials, each of which ended as soon as the rat located the platform or after a maximum of 60s elapsed. The location of the platform remained constant during these sessions, but the release locations varied across trials. Groups of 6 rats were sacrificed at various times during the course of the experiment according to the following: (1) D1-30m - rats were sacrificed 30 min after an initial training session (5 trials), (2) D1-3h – rats were sacrificed 3h after the initial training session, (3) D5-30m – rats were sacrificed 30 min after training on the 5th day, with the platform in the same location as the previous 4 days, (4) D5R-30m - rats were sacrificed 30 min after a single reversal training session on Day 5, and (5) Caged Control (CC) - rats were sacrificed directly from the home cage to establish basal gene expression levels. The D5-30m and D5R-30m groups were given 4 days of training, with 2 sessions of 5 trials given on each of these 4 days. The stages of learning & memory represented by these groups were: (1) acquisition / new learning (D1 groups), (2) stable reference memory retrieval (D5-30m), and (3) memory extinction coincident with new learning (D5R-30m). At the appropriate time, rats were sacrificed and dorsal hippocampi were dissected. Hippocampal RNA was prepared for each rat and an equal amount of RNA from each of 3 rats (per group) was combined to form a replicate pool. In this way, there were 10 pooled samples—two for each behavioral group.
The behavioral performance of all groups is shown in Figure 4.1 and demonstrates that the rats learned the platform location and achieved an asymptotic performance by the end of day 3 (Panel A). Panel B shows the performance levels for all groups in their final session before sacrifice. The short swim latencies of the D5-30m rats indicate that the rats had formed and were retrieving a memory of the platform location. Notably, the D1 and D5R-30m groups had similar performances. These results demonstrate that both groups of rats were acquiring new platform location information. However, in addition to acquiring the new platform location, the D5R group also had to extinguish their memory of the previous platform location.
Figure 4. Distinct hippocampal gene expression profiles are associated with distinct stages of learning & memory.
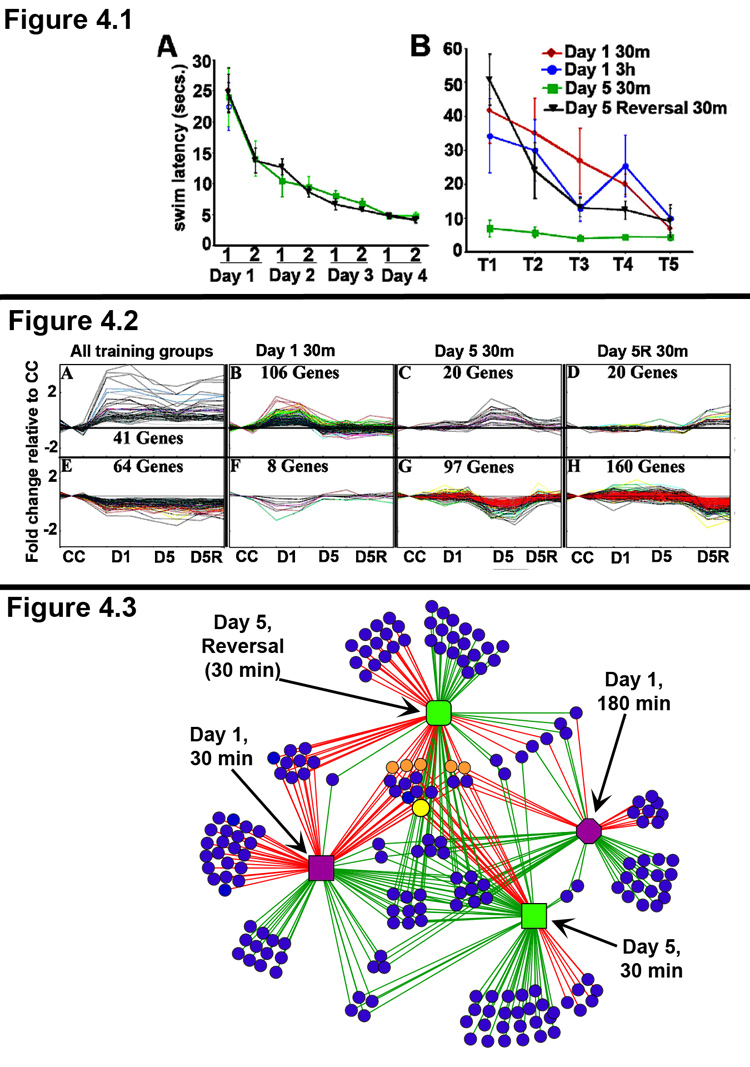
(Panel 4.1) Behavioral performance in the spatial water maze task. Different groups of rats were trained in the spatial water maze task, as described in the text. N = 6 rats per group. Panel A: The rats given multiple days of training learned the task well and showed an asymptotic performance achieved by end of day 3. Panel B: The behavioral performance of each group in the last training session before being sacrificed is shown. Note, that the Day 5 reversal rats were required to extinguish their response to the old platform location, in order to locate the new platform location. Accordingly, the behavioral performance of the Day 5 Reversal rats is similar to that of the Day 1 rats.
(Panel 4.2) Pattern template matching of microarray expression data reveals gene expression profiles associated with distinct stages of learning. RNA was isolated from the dorsal hippocampi of 6 rats from each behavioral group, and RNA from 3 rats each was used to generate replicate pools for each behavioral condition. Genome-wide gene expression analysis was done using Affymetrix® GeneChip® Rat230_2 arrays for each replicate pool. The Affymetrix PLIER algorithm was used to generate normalized gene expression values. TIGR MeV was used to define a set of ~400 significant genes with an ANOVA value of <0.05 and a fold change of < 1.5 or >1.5 relative to the caged control value. The gene expression profiles shown here were generated using a pattern template matching clustering algorithm. Gene expression levels are normalized to caged controls (CC), and the group labels are listed at the bottom of the panel (CC, D1, D5, and D5R). The data for each replicate chip is shown as a single data point, resulting in the “mountains” (panels B, C, and D) or “valleys” (panels F, G, and H) for genes differentially expressed in only one behavioral condition. Panels A & E show several genes that are induced (A) and repressed (E) in all behavior groups at 30m post-behavior. Panels B–D and F–H show distinct clusters of genes that are regulated only by initial learning (B &F), overtraining (reference memory retrieval; C &G), or reversal learning (reference memory extinction, coincident with new information acquisition; D &H). Notably, although the behavioral performance of D1 and D5R rats were similar, the gene expression profiles are distinct. Additionally, several genes in the D5 30m expression profile are associated with axonal outgrowth and new synapse formation, suggesting that overtraining might recruit additional plasticity mechanisms which play a role in the long-term stability of highly learned information (Reckart et al., 2007).
(Panel 4.3) Complex network analysis of gene expression networks regulated by distinct stages of learning & memory. Network of gene expression regulated by behavior in the spatial water maze task at D1 30m (purple square), D1 3h (purple octagon), D5 30m (green square), D5R 30m (green rounded-square). The lines connect behavior groups with the genes (blue circle nodes) that are differentially regulated by that behavior. Red & Green edges indicate up- and down- regulation of gene expression, respectively. Note that the several nodes in the center of the network represent genes that are regulated across multiple stages of learning and memory. This is also demonstrated by the high connectivity of these nodes in the network. Of these “core” genes several common IEGs are indicated as orange circle nodes and Arc is shown as a larger yellow circle node. In contrast, low connectivity genes, represented by blue circles connected to only one behavior group, are regulated only by a single behavior (i.e., in a distinct state of learning and memory). The degree of similarity or difference of the gene expression networks between any two of the behavior groups (stages of learning and memory) can be culled from the number of shared and distinct regulated genes. For example, D1 30m and D5R 30m exclusively share 9 up-regulated and 1 down regulated genes (left side of figure).
We examined the hippocampal gene expression profiles of each behavioral group using standard microarray methods (Figure 4.2) and complex network analysis (Figure 4.3). Although validation experiments are in progress for many of the identified genes, the microarray values for Arc, zif268, and c-fos were essentially identical to those achieved in an earlier study using the same behavioral groups and RNase protection assays (Guzowski et al., 2001). These results encourage strong confidence in the microarray data. Moreover, the purpose of presenting this data is not for a detailed discussion of the specific gene networks, but rather to show that distinct stages of learning differentially influence mRNA expression patterns in a complex fashion. Using a template pattern matching algorithm (TIGR Multiple Experiment Viewer), we identified distinct gene expression profiles. Interestingly, there were ~40 genes that, similar to Arc, were upregulated in all of the 30m behavioral groups (Figure 4.2, Panel A). Among the commonly upregulated genes were the IEGs Arc, Homer 1a, zif268, Egr-4, jun-B, and c-fos. These IEGs could be considered the “usual suspects” of many behavioral studies of brain gene expression. Moreover, there were ~60 genes down-regulated in all of the 30m behavioral groups (Figure 4.2, Panel B). These data indicate that there is a core neural gene expression network that is engaged by basic information processing. Additionally, and of considerable interest, is the finding that there were other distinct elements of coordinately regulated gene expression uniquely associated with each behavior group (i.e. stage of learning & memory). As shown in Figure 4.2, there were genes differentially regulated only in the D1-30m group (Panels B & F), or in the D5-30m group (Panels C & G), or in the D5R-30m group (Panels D & H). Again, genes were both down- and up-regulated relative to the caged controls.
Using complex network analysis (CNA), we visualized the gene regulation network associated with- and interconnecting- each behavior group (i.e. stage of learning & memory). CNA is a way to visualize and mine information from large data sets. In CNA, data is visualized as a graphical network of nodes that are interconnected by edges (lines). In a single-mode gene expression network, nodes representing differentially regulated genes are connected by edges. The edges define the relationship between the genes. We used Cytoscape 2.4 and a 2-mode variation of CNA to examine our gene expression data. In our 2-mode network, circle nodes represent genes and square, octagon, or rounded-square nodes represent behavioral groups (Figure 4.3). Each behavioral group is connected to a corresponding group of genes that are significantly regulated. The edge connecting a behavioral group to a gene is red or green, indicating that the gene is up- or down- regulated by the behavior training respectively. In figure 4.3, genes that are regulated in multiple stages of learning and memory are positioned in the center of the network and are highly connected. For example, the node representing Arc is the yellow node in the center of the network and is connected to all of the 30m behavioral training groups by red lines. Likewise, the other IEGs Homer 1a, zif268, Egr4, jun-B, and c-fos (orange circle nodes) are also densely connected and central to the network. Interestingly, most of the genes that are regulated in multiple behavior groups are regulated in the same direction. Conversely, gene nodes (circles) that are connected by a single line to a single behavior group are clearly regulated uniquely by that stage of learning and memory. The observation of distinct gene expression networks during the progression of learning, underscores the complexity and importance of gene expression involved in long-term memory.
In reconciling our focused and array studies of experience-dependent gene expression, we hypothesize that there are “core” plasticity genes (e.g., Arc, Homer 1a, c-fos, zif268, etc) and “state-specific” plasticity genes (ie., those that are learning stage dependent). These distinct patterns may reflect an interaction between glutamatergic and neuromodulatory systems (e.g., cholinergic, noradrenergic, etc.) that are engaged differentially by the stage of learning. Many of the core plasticity genes have already been implicated in neural plasticity and memory (reviewed in Guzowski et al., 2002; Tischmeyer and Grimm, 1999). Additionally, the state-specific genes include many neural proteins involved in processes such as cell adhesion, neurotransmitter function (both at receptor and neurotransmitter exocytosis steps), ion channel function, regulation of synaptic structure, intracellular signaling, etc. The combination of “core” and “state-specific” plasticity genes may enable distinct plastic processes to subserve memory.
One of the central guiding principles of modern biology is that cell function is dictated by the network of interacting proteins within a cell. Regulation of the protein interaction network can be achieved at multiple levels including (i) gene transcription regulation, (ii) mRNA processing and trafficking, (iii) control of mRNA stability / degradation, (iv) translation regulation, (v) post-translational modification of existing proteins, and (vi) protein trafficking and localization. A distinctive feature of neurons is to integrate a myriad of extracellular signals (neurotransmitters, neuromodulators, hormones, growth factors, cytokines, etc) into an orchestrated change in genomic expression. While there has been a focus on transcriptional induction / repression in the regulation of synaptic plasticity, it is appreciated that there is significant post-transcriptional regulation of mRNA (Bentley, 2002; Proudfoot, Furger, & Dye, 2002). An additional level of complexity is further suggested by recent findings that microRNAs can be regulated by cAMP and neural activity (Klein, Impey, & Goodman, 2005; Vo, Klein, Varlamova, Keller, Yamamoto, Goodman, & Impey, 2005). Moreover, RNAi mediated mechanisms play a role in synaptic plasticity and memory (Ashraf, McLoon, Sclarsic, & Kunes, 2006). Beyond controlling protein synthesis, there is extensive evidence that regulation of existing proteins via spatial localization and post-translational modification (PTM) also control neuronal plasticity. With regard to the central focus of this review, we suggest that IEGs provide a powerful way to modify the plasticity protein interaction network—the plasticity “interactome”. In this view (Figure 5), IEGs represent protein nodes that exert their function by being present or absent. There is evidence that IEGs, such as Arc, enable new protein-protein interactions to alter neuronal function in many ways (Figure 5, IEG 1) (Chowdhury, Shepherd, Okuno, Lyford, Petralia, Plath, Kuhl, Huganir, & Worley, 2006; Xu, Hopf, Reddy, Cho, Guo, Lanahan, Petralia, Wenthold, O'Brien, & Worley, 2003). Additionally, other IEGs, such as Homer 1a, act in a dominant negative fashion to alter neuronal signaling (Figure 5, IEG 2) (Kammermeier & Worley, 2007; Tu, Xiao, Yuan, Lanahan, Leoffert, Li, Linden, & Worley, 1998). Of course, IEGs provide just one of the means to modify the plasticity interactome, with post-translational modification (PTM; Routtenberg and Reckart, 2005) providing a powerful means to rapidly modify protein network function (Figure 5, PO4 node). Nonetheless, the accumulating evidence on IEGs such as Arc suggests that PTM of preexisting synaptic proteins is necessary, but not sufficient, for the establishment of lasting memories.
Figure 5. Modification of the Plasticity Interactome by IEG Expression.
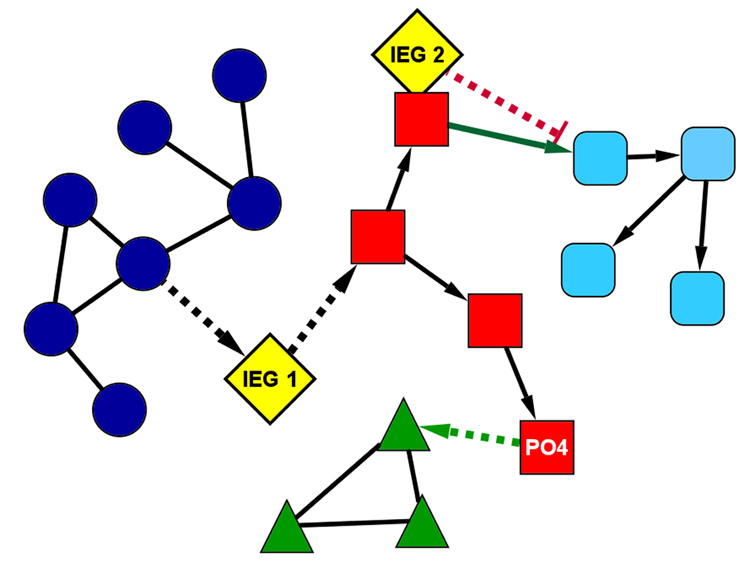
A simplified model of a protein interaction network containing 4 distinct subnetworks (dark blue circles, red squares, green triangles, and aqua rounded-squares). Solid lines between protein nodes represent constitutive interactions and dotted lines represent conditional interactions. Directionality of interactions is noted with arrowheads. Prior to patterned synaptic input, the two IEGs (IEG1 and IEG2, yellow diamonds) are absent and the protein in the red subnetwork is not phosphorylated. Under these basal conditions, the green and blue subnetworks function in isolation, and the red and aqua subnetworks interact. Following patterned synaptic activity, IEGs 1 and 2 are induced and the protein in the red network is phosphorylated (indicated by “PO4”), activating the conditional interactions. With these conditional interactions now active, the dialog between the subnetworks changes dramatically. IEG1 couples the blue and red subnetworks, and IEG2 functions as a dominant negative gene to block the interaction between red and aqua subnetworks. The phosphorylation in the red subnetwork now enables interaction with the green subnetwork. These 3 changes (induction of the two IEGs and the phosphorylation in the red subnetwork) markedly change the behavior of the entire network, with now a directed interaction between blue, red, and green networks, which did not occur in the basal state. Please note the powerful capacity of IEGs to modify the dialog of the plasticity interactome, and by regulation of a relatively limited number of IEGs.
THE DYNAMIC MEMORY TRACE?
Findings from our laboratory indicate that Arc transcription is induced in rats after repeated exposures to the same environment separated by 24 hrs (Guzowski et al., 2006) or even during overtraining in the spatial water maze task (Miyashita et al., unpublished observation; Guzowski et al, 2001). The findings seem counterintuitive at first; if Arc enables plasticity, then why is it induced at times when behavioral learning is no longer occurring? The discrepancy stems from the tacit assumption of the memory consolidation hypothesis, that once synaptic modifications that represent a memory trace are encoded and consolidated, the memory remains in an immutable stable state permanently. In that case, plasticity should no longer occur when memory is fully consolidated. However, our findings of continued Arc induction in a familiar environment or during overtraining suggests that plasticity may occur constantly and continuously, coupled with a rat’s experience. In a way, rats never stop learning, even if no changes in behavior are detected. Ongoing plasticity as indicated by Arc induction may signify incremental changes within a network ensemble representing a memory trace to be fine-tuned and maintained, even during overtraining.
The idea that plastic changes of synapses are completed and maintained permanently imply a kind of “synaptic phrenology” (Abraham & Robins, 2005) assuming that information is encoded and stored in a fixed configuration of synaptic weights in specific neural circuits. Such organization would allow a limited range of configurations and available connections and would saturate very quickly. Instead, continued updating of synaptic weights within a dynamic network would greatly increase its capacity to process and store information. Beyond possible advantages in capacity, studies using artificial neural networks have shown that ongoing adjustments in connection weights within network ensembles are necessary to maximize maintenance of previously stored information (Abraham and Robins, 2005). Activity at the time of initial encoding must be repeated to maintain changes in connections. The same study also suggested that such iterative processing of synaptic weights could take place during quiet periods or sleep. Evidence of such offline updating processes are derived from studies of neural reactivation during quiet awake periods and sleep (Foster & Wilson, 2006; Louie & Wilson, 2001; Wilson & McNaughton, 1994). Reverberation of neuronal ensemble at these time points may be necessary for triggering changes in the plasticity inteactome, updating and reorganizing of synaptic weights, and culminating in refinement and maintenance of memory traces [Figure 1]. Accordingly, different profiles of gene activation are found in different stages of sleep (Cirelli, 2005; Ribeiro, Mello, Velho, Gardner, Jarvis, & Pavlides, 2002), indicative of intrinsic reverberation during sleep eliciting changes in gene expression and plasticity within networked elements. Moreover, studies using mice with conditional knockout of NR1 subunit of NMDA receptors further support the theory that continuous reverberation (mediated by NMDA receptor activation) is required for maintenance of memory, even as old as 9 month retention of contextual fear conditioning (Cui, Wang, Tan, Zaia, Zhang, & Tsien, 2004).
The recent studies suggesting a role for Arc in homeostatic synaptic scaling of AMPA receptors (Chowdhury, et al., 2006; Rial Verde, et al., 2006; Shepherd et al., 2006) have come as a surprise to many in the field, but may speak to the nature of network dynamics in the storage of memories. Given the robust Arc induction we see by behavior, it is attractive to think that Arc could act as a critical molecule for maintaining synaptic homeostasis in hippocampal and neocortical networks. By virtue of its tight coupling to neural activity and its ability to provide synapse specific targeting, Arc would be well suited to continually refine synaptic weights within the network to allow capacity within the network for new memories, while maintaining and updating earlier memory traces.
In conclusion, studies of Arc and other IEGs support a role for de novo protein synthesis in long-term memory. These studies show that IEG expression remains tightly coupled to neural activity across behaviors and stages of memory. Despite this, we do not view IEG expression as isomorphic to memory, but instead suggest that IEG expression can enable specific plastic states or provide critical homeostatic functions to maintain the viability of network function. To better understand the complex relationship between neural activity, experience-dependent gene expression, and memory, we will need to continue to define the functions of individual genes and the higher order function of gene interaction networks associated with distinct behaviors. In this view, terms such as “permissive” and “instructive” do not speak to defined biological mechanisms, and are of limited use to understanding the molecular, cellular, and systems bases of memory. Memory is necessarily complex involving numerous interactions amongst many levels of biological organization—we might never find THE substrate of memory because no one substrate exists. Framing memory in the context of false dichotomies will not help us understand the many subtle, but important, roles played by a myriad of biological mechanisms in the brain supporting memory.
Acknowledgments
This research was supported by NIH grant MH060123 (JFG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham WC, Mason SE, Demmer J, Williams JM, Richardson CL, Tate WP, Lawlor PA, Dragunow M. Correlations between immediate early gene induction and the persistence of long-term potentiation. Neuroscience. 1993;56:717–727. doi: 10.1016/0306-4522(93)90369-q. [DOI] [PubMed] [Google Scholar]
- Abraham WC, Robins A. Memory retention--the synaptic stability versus plasticity dilemma. Trends in Neurosciences. 2005;28:73–78. doi: 10.1016/j.tins.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Adams JP, Dudek SM. Late-phase long-term potentiation: getting to the nucleus. Nature Reviews Neuroscience. 2005;6:737–743. doi: 10.1038/nrn1749. [DOI] [PubMed] [Google Scholar]
- Agranoff BW, Davis RE, Brink JJ. Memory fixation in the goldfish. Proceedings of the National Academy of Sciences of the United States of America. 1965;54:788–793. doi: 10.1073/pnas.54.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Bentley D. The mRNA assembly line: transcription and processing machines in the same factory. Current Opinion in Cell Biolology. 2002;14:336–342. doi: 10.1016/s0955-0674(02)00333-2. [DOI] [PubMed] [Google Scholar]
- Berridge MJ. Neuronal calcium signaling. Neuron. 1998;21:13–26. doi: 10.1016/s0896-6273(00)80510-3. [DOI] [PubMed] [Google Scholar]
- Burke SN, Chawla MK, Penner MR, Crowell BE, Worley PF, Barnes CA, McNaughton BL. Differential encoding of behavior and spatial context in deep and superficial layers of the neocortex. Neuron. 2005;45:667–674. doi: 10.1016/j.neuron.2005.01.042. [DOI] [PubMed] [Google Scholar]
- Cammarota M, Bevilaqua LR, Ardenghi P, Paratcha G, Levi de Stein M, Izquierdo I, Medina JH. Learning-associated activation of nuclear MAPK, CREB and Elk-1, along with Fos production, in the rat hippocampus after a one-trial avoidance learning: abolition by NMDA receptor blockade. Brain Research Molecular Brain Research. 2000;76:36–46. doi: 10.1016/s0169-328x(99)00329-0. [DOI] [PubMed] [Google Scholar]
- Chawla MK, Guzowski JF, Ramirez-Amaya V, Lipa P, Hoffman KL, Marriott LK, Worley PF, McNaughton BL, Barnes CA. Sparse, environmentally selective expression of Arc RNA in the upper blade of the rodent fascia dentata by brief spatial experience. Hippocampus. 2005;15:579–586. doi: 10.1002/hipo.20091. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C. A molecular window on sleep: changes in gene expression between sleep and wakefulness. Neuroscientist. 2005;11:63–74. doi: 10.1177/1073858404270900. [DOI] [PubMed] [Google Scholar]
- Clayton DF. The genomic action potential. Neurobiology of Learning and Memory. 2000;74:185–216. doi: 10.1006/nlme.2000.3967. [DOI] [PubMed] [Google Scholar]
- Cui Z, Wang H, Tan Y, Zaia KA, Zhang S, Tsien JZ. Inducible and reversible NR1 knockout reveals crucial role of the NMDA receptor in preserving remote memories in the brain. Neuron. 2004;41:781–793. doi: 10.1016/s0896-6273(04)00072-8. [DOI] [PubMed] [Google Scholar]
- Davis HP, Squire LR. Protein synthesis and memory: a review. Psychological Bulletin. 1984;96:518–559. [PubMed] [Google Scholar]
- Davis S, Laroche S. Mitogen-activated protein kinase/extracellular regulated kinase signalling and memory stabilization: a review. Genes Brain and Behavior. 2006;5 Suppl 2:61–72. doi: 10.1111/j.1601-183X.2006.00230.x. [DOI] [PubMed] [Google Scholar]
- Davis S, Vanhoutte P, Pages C, Caboche J, Laroche S. The MAPK/ERK cascade targets both Elk-1 and cAMP response element-binding protein to control long-term potentiation-dependent gene expression in the dentate gyrus in vivo. Journal of Neuroscience. 2000;20:4563–4572. doi: 10.1523/JNEUROSCI.20-12-04563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Heist EK, Tsien RW. Translocation of calmodulin to the nucleus supports CREB phosphorylation in hippocampal neurons. Nature. 1998;392:198–202. doi: 10.1038/32448. [DOI] [PubMed] [Google Scholar]
- Delghandi MP, Johannessen M, Moens U. The cAMP signalling pathway activates CREB through PKA, p38 and MSK1 in NIH 3T3 cells. Cell Signaling. 2005;17:1343–1351. doi: 10.1016/j.cellsig.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Donai H, Sugiura H, Ara D, Yoshimura Y, Yamagata K, Yamauchi T. Interaction of Arc with CaM kinase II and stimulation of neurite extension by Arc in neuroblastoma cells expressing CaM kinase II. Neuroscience Research. 2003;47:399–408. doi: 10.1016/j.neures.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Dudai Y. The neurobiology of consolidations, or, how stable is the engram? Annual Reviews of Psychology. 2004;55:51–86. doi: 10.1146/annurev.psych.55.090902.142050. [DOI] [PubMed] [Google Scholar]
- Duncan CP. The Retroactive effect of electroshock on learning. Journal of Comparative Physiology and Psychology. 1949;42:32–44. doi: 10.1037/h0058173. [DOI] [PubMed] [Google Scholar]
- Finkbeiner S, Greenberg ME. Ca2+ channel-regulated neuronal gene expression. Journal of Neurobiology. 1998;37:171–189. [PubMed] [Google Scholar]
- Flexner JB, Flexner LB, Stellar E. Memory in mice as affected by intracerebral puromycin. Science. 1963;141:57–59. doi: 10.1126/science.141.3575.57. [DOI] [PubMed] [Google Scholar]
- Foster DJ, Wilson MA. Reverse replay of behavioural sequences in hippocampal place cells during the awake state. Nature. 2006;440:680–683. doi: 10.1038/nature04587. [DOI] [PubMed] [Google Scholar]
- Frankland PW, Bontempi B. The organization of recent and remote memories. Nature Reviews Neuroscience. 2005;6:119–130. doi: 10.1038/nrn1607. [DOI] [PubMed] [Google Scholar]
- Fujimoto T, Tanaka H, Kumamaru E, Okamura K, Miki N. Arc interacts with microtubules/microtubule-associated protein 2 and attenuates microtubule-associated protein 2 immunoreactivity in the dendrites. Journal of Neuroscience Research. 2004;76:51–63. doi: 10.1002/jnr.20056. [DOI] [PubMed] [Google Scholar]
- Gerard RW. Physiology and Psychiatry. American Journal of Psychiatry. 1949;106:161–173. doi: 10.1176/ajp.106.3.161. [DOI] [PubMed] [Google Scholar]
- Ginty DD. Calcium regulation of gene expression: isn't that spatial? Neuron. 1997;18:183–186. doi: 10.1016/s0896-6273(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Goelet P, Castellucci VF, Schacher S, Kandel ER. The long and the short of long-term memory--a molecular framework. Nature. 1986;322:419–422. doi: 10.1038/322419a0. [DOI] [PubMed] [Google Scholar]
- Gold PE. The many faces of amnesia. Learning and Memory. 2006;13:506–514. doi: 10.1101/lm.277406. [DOI] [PubMed] [Google Scholar]
- Greenberg ME, Ziff EB, Greene LA. Stimulation of neuronal acetylcholine receptors induces rapid gene transcription. Science. 1986;234:80–83. doi: 10.1126/science.3749894. [DOI] [PubMed] [Google Scholar]
- Guzowski JF. Insights into immediate-early gene function in hippocampal memory consolidation using antisense oligonucleotide and fluorescent imaging approaches. Hippocampus. 2002;12:86–104. doi: 10.1002/hipo.10010. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Knierim JJ, Moser EI. Ensemble dynamics of hippocampal regions CA3 and CA1. Neuron. 2004;44:581–584. doi: 10.1016/j.neuron.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. Journal of Neuroscience. 2000;20:3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McGaugh JL. Interaction of neuromodulatory systems regulating memory storage. In: Decker M, Brioni JD, editors. Alzheimer's Disease: Molecular Aspects and Pharmacological Treatments. Wiley-Liss; 1997. pp. 37–61. [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nature Neuroscience. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Miyashita T, Chawla MK, Sanderson J, Maes LI, Houston FP, Lipa P, McNaughton BL, Worley PF, Barnes CA. Recent behavioral history modifies coupling between cell activity and Arc gene transcription in hippocampal CA1 neurons. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1077–1082. doi: 10.1073/pnas.0505519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, Setlow B, Wagner EK, McGaugh JL. Experience-dependent gene expression in the rat hippocampus after spatial learning: A comparison of the immediate-early genes Arc, c-fos, and zif268. J Neurosci. 2001;21:5089–5098. doi: 10.1523/JNEUROSCI.21-14-05089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, Timlin JA, Roysam B, McNaughton BL, Worley PF, Barnes CA. Mapping behaviorally relevant neural circuits with immediate-early gene expression. Current Opinion in Neurobiology. 2005;15:599–606. doi: 10.1016/j.conb.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Han JH, Kushner SA, Yiu AP, Cole CJ, Matynia A, Brown RA, Neve RL, Guzowski JF, Silva AJ, Josselyn SA. Neuronal competition and selection during memory formation. Science. 2007;316:457–460. doi: 10.1126/science.1139438. [DOI] [PubMed] [Google Scholar]
- Hatfield T, McGaugh JL. Norepinephrine infused into the basolateral amygdala posttraining enhances retention in a spatial water maze task. Neurobiology of Learning and Memory. 1999;71:232–239. doi: 10.1006/nlme.1998.3875. [DOI] [PubMed] [Google Scholar]
- Ikegaya Y, Saito H, Abe K. Attenuated hippocampal long-term potentiation in basolateral amygdala-lesioned rats. Brain Research. 1994;656:157–164. doi: 10.1016/0006-8993(94)91377-3. [DOI] [PubMed] [Google Scholar]
- Ikegaya Y, Saito H, Abe K. Requirement of basolateral amygdala neuron activity for the induction of long-term potentiation in the dentate gyrus in vivo. Brain Research. 1995;671:351–354. doi: 10.1016/0006-8993(94)01403-5. [DOI] [PubMed] [Google Scholar]
- Kaczmarek L. Gene expression in learning processes. Acta Neurobiologiae Experimentalis. 2000;60:419–424. doi: 10.55782/ane-2000-1361. [DOI] [PubMed] [Google Scholar]
- Kammermeier PJ, Worley PF. Homer 1a uncouples metabotropic glutamate receptor 5 from postsynaptic effectors. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6055–6060. doi: 10.1073/pnas.0608991104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein ME, Impey S, Goodman RH. Role reversal: the regulation of neuronal gene expression by microRNAs. Current Opinion in Neurobiology. 2005;15:507–513. doi: 10.1016/j.conb.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Lanahan A, Worley P. Immediate-early genes and synaptic function. Neurobiology of Learning and Memory. 1998;70:37–43. doi: 10.1006/nlme.1998.3836. [DOI] [PubMed] [Google Scholar]
- Liang KC, Juler RG, McGaugh JL. Modulating effects of posttraining epinephrine on memory: involvement of the amygdala noradrenergic system. Brain Research. 1986;368:125–133. doi: 10.1016/0006-8993(86)91049-8. [DOI] [PubMed] [Google Scholar]
- Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl D. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:5734–5738. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie K, Wilson MA. Temporally structured replay of awake hippocampal ensemble activity during rapid eye movement sleep. Neuron. 2001;29:145–156. doi: 10.1016/s0896-6273(01)00186-6. [DOI] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annual Reviews of Neuroscience. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- McGaugh JL. Memory--a century of consolidation. Science. 2000;287:248–251. doi: 10.1126/science.287.5451.248. [DOI] [PubMed] [Google Scholar]
- McIntyre CK, Miyashita T, Setlow B, Marjon KD, Steward O, Guzowski JF, McGaugh JL. Memory-influencing intra-basolateral amygdala drug infusions modulate expression of Arc protein in the hippocampus. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(30):10718–10723. doi: 10.1073/pnas.0504436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moga DE, Calhoun ME, Chowdhury A, Worley P, Morrison JH, Shapiro ML. Activity-regulated cytoskeletal-associated protein is localized to recently activated excitatory synapses. Neuroscience. 2004;125:7–11. doi: 10.1016/j.neuroscience.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Mokin M, Lindahl JS, Keifer J. Immediate-early gene-encoded protein Arc is associated with synaptic delivery of GluR4-containing AMPA receptors during in vitro classical conditioning. Journal of Neurophysiology. 2006;95:215–224. doi: 10.1152/jn.00737.2005. [DOI] [PubMed] [Google Scholar]
- Morgan JI, Curran T. Stimulus-transcription coupling in the nervous system: involvement of the inducible proto-oncogenes fos and jun. Annual Reviews of Neuroscience. 1991;14:421–451. doi: 10.1146/annurev.ne.14.030191.002225. [DOI] [PubMed] [Google Scholar]
- Müller GE, Pilzecker A. Experimentelle Beiträge zur Lehre vom Gedächtnis" (Experimental Contributions to the Science of Memory) Z Psychology. 1900;1:1–288. [Google Scholar]
- Nedivi E, Hevroni D, Naot D, Israeli D, Citri Y. Numerous candidate plasticity-related genes revealed by differential cDNA cloning. Nature. 1993;363:718–722. doi: 10.1038/363718a0. [DOI] [PubMed] [Google Scholar]
- Petrovich GD, Holland PC, Gallagher M. Amygdalar and prefrontal pathways to the lateral hypothalamus are activated by a learned cue that stimulates eating. Journal of Neuroscience. 2005;25:8295–8302. doi: 10.1523/JNEUROSCI.2480-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, Kobalz U, Stawrakakis A, Fernandez E, Waltereit R, Bick-Sander A, Therstappen E, Cooke SF, Blanquet V, Wurst W, Salmen B, Bosl MR, Lipp HP, Grant SG, Bliss TV, Wolfer DP, Kuhl D. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Ramirez-Amaya V, Vazdarjanova A, Mikhael D, Rosi S, Worley PF, Barnes CA. Spatial exploration-induced Arc mRNA and protein expression: evidence for selective, network-specific reactivation. Journal of Neuroscience. 2005;25:1761–1768. doi: 10.1523/JNEUROSCI.4342-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reckart JL, Sandoval CJ, Bermudez-Rattoni F, Routtenberg A. Remodeling of hippocampal mossy fibers is selectively induced seven days after the acquisition of a spatial but not a cued reference memory task. Learning & Memory. 2007;14(6):416–421. doi: 10.1101/lm.516507. [DOI] [PubMed] [Google Scholar]
- Rial Verde EM, Lee-Osbourne J, Worley PF, Malinow R, Cline HT. Increased expression of the immediate-early gene arc/arg3.1 reduces AMPA receptor-mediated synaptic transmission. Neuron. 2006;52:461–474. doi: 10.1016/j.neuron.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro S, Mello CV, Velho T, Gardner TJ, Jarvis ED, Pavlides C. Induction of Hippocampal Long-Term Potentiation during Waking Leads to Increased Extrahippocampal zif-268 Expression during Ensuing Rapid-Eye-Movement Sleep. Journal of Neuroscience. 2002;22:10914–10923. doi: 10.1523/JNEUROSCI.22-24-10914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routtenberg A, Rekart JL. Post-translational protein modification as the substrate for long-lasting memory. Trends in Neurosciences. 2005;28:12–19. doi: 10.1016/j.tins.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Rudy JW, Biedenkapp JC, Moineau J, Bolding K. Anisomycin and the reconsolidation hypothesis. Learning and Memory. 2006;13:1–3. doi: 10.1101/lm.157806. [DOI] [PubMed] [Google Scholar]
- Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annual Reviews of Biochemistry. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderling TR, Chang B, Brickey D. Cellular signaling through multifunctional Ca2+/calmodulin-dependent protein kinase II. Journal of Biological Chemistry. 2001;276:3719–3722. doi: 10.1074/jbc.R000013200. [DOI] [PubMed] [Google Scholar]
- Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- Steward O, Worley PF. A cellular mechanism for targeting newly synthesized mRNAs to synaptic sites on dendrites. Proceedings of the National Academy of Sciences of the United States of America. 2001a;98:7062–7068. doi: 10.1073/pnas.131146398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Worley PF. Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron. 2001b;30:227–240. doi: 10.1016/s0896-6273(01)00275-6. [DOI] [PubMed] [Google Scholar]
- Sweatt JD. The neuronal MAP kinase cascade: a biochemical signal integration system subserving synaptic plasticity and memory. Journal of Neurochemistry. 2001;76:1–10. doi: 10.1046/j.1471-4159.2001.00054.x. [DOI] [PubMed] [Google Scholar]
- Takei N, Inamura N, Kawamura M, Namba H, Hara K, Yonezawa K, Nawa H. Brain-Derived Neurotrophic Factor Induces Mammalian Target of Rapamycin-Dependent Local Activation of Translation Machinery and Protein Synthesis in Neuronal Dendrites. Journal of Neuroscience. 2004;24:9760–9769. doi: 10.1523/JNEUROSCI.1427-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischmeyer W, Grimm R. Activation of immediate early genes and memory formation. Cellular and Molecular Life Sciences. 1999;55:564–574. doi: 10.1007/s000180050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokas P, Ma T, Iyengar R, Landau EM, Blitzer RD. Mitogen-activated protein kinase upregulates the dendritic translation machinery in long-term potentiation by controlling the mammalian target of rapamycin pathway. Journal of Neuroscience. 2007;27:5885–5894. doi: 10.1523/JNEUROSCI.4548-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21:717–726. doi: 10.1016/s0896-6273(00)80589-9. [DOI] [PubMed] [Google Scholar]
- Tzingounis AV, Nicoll RA. Arc/Arg3.1: linking gene expression to synaptic plasticity and memory. Neuron. 2006;52:403–407. doi: 10.1016/j.neuron.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Vazdarjanova A, Guzowski JF. Differences in hippocampal neuronal population responses to modifications of an environmental context: evidence for distinct, yet complementary, functions of CA3 and CA1 ensembles. Journal of Neuroscience. 2004;24:6489–6496. doi: 10.1523/JNEUROSCI.0350-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazdarjanova A, McNaughton BL, Barnes CA, Worley PF, Guzowski JF. Experience-dependent coincident expression of the effector immediate-early genes arc and Homer 1a in hippocampal and neocortical neuronal networks. Journal of Neuroscience. 2002;22:10067–10071. doi: 10.1523/JNEUROSCI.22-23-10067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazdarjanova A, Ramirez-Amaya V, Insel N, Plummer TK, Rosi S, Chowdhury S, Mikhael D, Worley PF, Guzowski JF, Barnes CA. Spatial exploration induces ARC, a plasticity-related immediate-early gene, only in calcium/calmodulin-dependent protein kinase II-positive principal excitatory and inhibitory neurons of the rat forebrain. Journal of Comparative Neurology. 2006;498:317–329. doi: 10.1002/cne.21003. [DOI] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. From The Cover: A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltereit R, Dammermann B, Wulff P, Scafidi J, Staubli U, Kauselmann G, Bundman M, Kuhl D. Arg3.1/Arc mRNA induction by Ca2+ and cAMP requires protein kinase A and mitogen-activated protein kinase/extracellular regulated kinase activation. Journal of Neuroscience. 2001;21:5484–5493. doi: 10.1523/JNEUROSCI.21-15-05484.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, McNaughton BL. Reactivation of hippocampal ensemble memories during sleep. Science. 1994;265:676–679. doi: 10.1126/science.8036517. [DOI] [PubMed] [Google Scholar]
- Wittenberg GM, Sullivan MR, Tsien JZ. Synaptic reentry reinforcement based network model for long-term memory consolidation. Hippocampus. 2002;12:637–647. doi: 10.1002/hipo.10102. [DOI] [PubMed] [Google Scholar]
- Worley PF, Bhat RV, Baraban JM, Erickson CA, McNaughton BL, Barnes CA. Thresholds for synaptic activation of transcription factors in hippocampus: correlation with long-term enhancement. Journal of Neuroscience. 1993;13:4776–4786. doi: 10.1523/JNEUROSCI.13-11-04776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GY, Deisseroth K, Tsien RW. Spaced stimuli stabilize MAPK pathway activation and its effects on dendritic morphology. Nature Neuroscience. 2001;4:151–158. doi: 10.1038/83976. [DOI] [PubMed] [Google Scholar]
- Xu D, Hopf C, Reddy R, Cho RW, Guo L, Lanahan A, Petralia RS, Wenthold RJ, O'Brien RJ, Worley P. Narp and NP1 form heterocomplexes that function in developmental and activity-dependent synaptic plasticity. Neuron. 2003;39:513–528. doi: 10.1016/s0896-6273(03)00463-x. [DOI] [PubMed] [Google Scholar]
- Yin Y, Edelman GM, Vanderklish PW. The brain-derived neurotrophic factor enhances synthesis of Arc in synaptoneurosomes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:2368–2373. doi: 10.1073/pnas.042693699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WP, Guzowski JF, Thomas SA. Mapping neuronal activation and the influence of adrenergic signaling during contextual memory retrieval. Learning and Memory. 2005;12:239–247. doi: 10.1101/lm.90005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Buck LB. Combinatorial effects of odorant mixes in olfactory cortex. Science. 2006;311:1477–1481. doi: 10.1126/science.1124755. [DOI] [PubMed] [Google Scholar]