Summary
It is now generally accepted that many of the physiological effects of alcohol consumption are a direct result of binding to specific sites in neuronal proteins such as ion channels or other components of neuronal signaling cascades. Binding to these targets generally occurs in water filled pockets and leads to alterations in protein structure and dynamics. However the precise interactions required to confer alcohol sensitivity to a particular protein remains undefined.
Using information from the previously solved crystal structures of the Drosophila melanogaster protein LUSH in complexes with short chain alcohols, we have designed and tested the effects of specific amino acid substitutions on alcohol binding. These effects of these substitutions, specifically S52A, T57S and T57A were examined using a combination of molecular dynamics, X-ray crystallography, fluorescence spectroscopy and thermal unfolding. These studies reveal that the binding of ethanol is highly sensitive to small changes in the composition of the alcohol binding site. We find that T57 is the most critical reside for binding alcohols, the T57A substitution completely abolishes binding, while the T57S substitution differentially affects ethanol binding compared to longer chain alcohols. The additional requirement for a potential hydrogen bond acceptor at position 52 suggests that both the presence of multiple hydrogen bonding groups and the identity of the hydrogen bonding residue are critical for defining an ethanol binding site. These results provide new insight into the detailed chemistry of alcohol’s interactions with proteins.
Keywords: Alcohol, Hydrogen Bonding, Binding affinities, LUSH, X-ray crystallography
Introduction
Alcohol exerts its effects predominantly through its actions on the activity of number of neuronal proteins. These include ligand-gated ion channels (LGICs) 1–5adenylyl cyclases (AC) 6, protein kinases, (PKC) 7–9 and tyrosine kinases Fyn, and phospholipase D (PLD) 10. Changes in the function, sub-cellular localization, and trafficking of these proteins is believed to contribute to changes in learning, memory and motor function that are associated with alcohol intoxication, dependency, tolerance, seizures during withdrawal, 1–5 and fetal alcohol syndrome11.
Many molecular targets of alcohol appear to contain specific binding sites that confer alcohol sensitivity. For example, different LGICs are sensitive to alcohols with different alkyl chain lengths 12–16 and domain swap experiments and/or specific amino acid substitutions can alter the chain length sensitivity or even abolish alcohol sensitivity 15 suggesting that there are sterically defined alcohol-binding pockets of different sizes in these channels17. Further, reaction of specific cysteine substitutions in the glycine α1 and GABA ρ1 receptors with alkyl methanethiosulfonate reagents mimics permanent occupation of an alcohol-binding site that is located in a water filled pocket within the transmembrane domains of these receptors 18–22.
Studies of the voltage gated Drosophila Shaw2 potassium channel suggest that a butanol binding site depends on the formation of well ordered structures in one loop of the receptor23–25. While the effects of alcohols on PKC depends on both the chain length and the mechanism of activation 26–30. Activation of PKCα in the absence of membranes depends on both the alcohol chain length and stereochemistry, and can be completely antagonized by short-chain alcohols 30. While for PKCδ, butanol and octanol can enhance binding of phorbol ester to the C1B domain31 and can directly quench intrinsic tryptophan fluorescence 32. Miller and co-workers have used photo-labile alcohol analogues to identify interactions with residues in the C1A32 and C1B31 domains that are in close proximity to the phorbol-ester binding site, which supports the hypothesis that alcohols may function by binding to discrete sites that lead to changes in protein structure and dynamics.
Alcohols bind to proteins with very low affinities with dissociation constants in the high micro to millimolar range. Natural and industrial fermentation of fruits and other sugars can easily produce alcohol concentrations between 3–15% v/v, which corresponds to 500 mM - 2.5 M ethanol. For humans, pharmacologically relevant concentrations of alcohol are in the range of 5–50 mM. For example, the legal limiting for driving in the UK and the USA is a blood alcohol concentration of 0.0 8% or 17 mM ethanol. These weak binding affinities, combined with the membrane bound nature of many of the molecular targets of alcohol, present enormous challenges in understanding the nature of alcohol binding sites in proteins.
We are using the odorant binding protein LUSH from Drosophila melanogaster as a model for alcohol binding sites in proteins. LUSH is a non-enzymatic protein required for both behavioral and electrophysiological responses of olfactory neurons to short chain n-alcohols 33; 34. The crystal structures of LUSH-alcohol complexes solved to high resolution in the presence of 30–50 mM alcohol 8 revealed a single alcohol-binding site 8 (Figure 1), suggesting this site has a significantly increased affinity for alcohol compared to other potential binding sites in the protein. The alcohols bind in a water filled hydrophobic cavity ~ 390 Å3 in volume, between a set of alpha-helices (Figure 1a) and potentially forms hydrogen bonds with two polar residues, S52 and T57 at one end of this cavity (Figure 1b). Based on measurements of the O-O distances we infer that the hydrogen bonds between the alcohol O and the T57 Oγ significantly shorter (dO-O = 2.51 Å, s.d. = 0.21 Å), than the hydrogen bonds between the alcohol O and the S52 Oγ (dO-O = 3.18 Å, s.d. = 0.14 Å), suggesting that T57 is forming a stronger hydrogen bond with the alcohol than S52.
Figure 1. Location and structure of the alcohol binding site in LUSH.
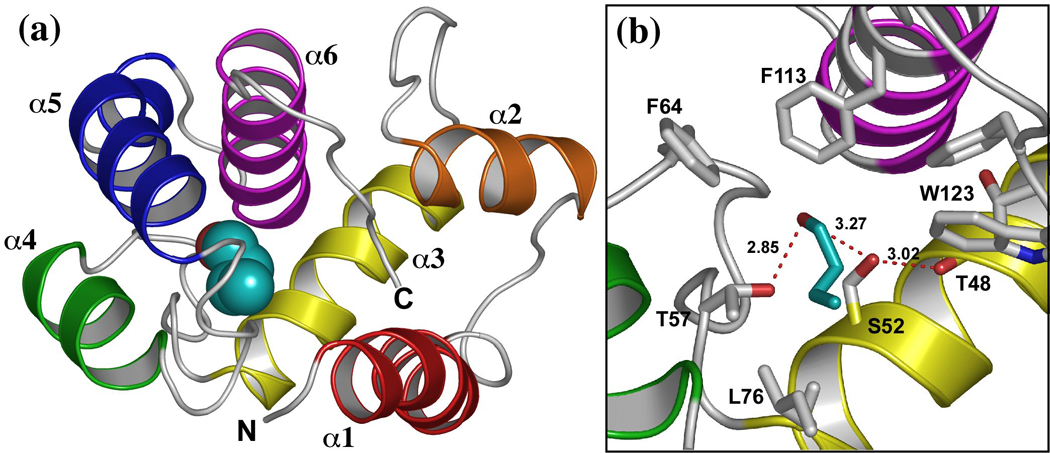
(a) Structure of the LUSH-butanol complex showing the position of butanol (cyan). (b) close up view of the alcohol binding site in the LUSH-butanol complex. The oxygen of alcohols forms potential hydrogen bonds with S52 and T57, while the alkyl chain of butanol forms contacts with residues F64, L76, F113, and W123. Figure based on PDB ID 1OOH. Ethanol binds to the same site. Distances between the T57 Oγ and alcohol O are 2.51 Å while the distance between the S52 Oγ and alcohol O is 3.18 Å.
The alcohol binding site contains a number of aromatic residues F113, W123, and F64 and this may be related to the ability of alcohols to stabilize the structure of the protein in a chain length dependent manner where each methylene group contributes approximately 1 kcal mol−1 to the overall stability of the complex 35. Previous studies have suggested that alcohols bind between two alpha helical segments 15; 36–39, and that binding of alcohol in a water filled cavity can stabilize specific conformations of alcohol sensitive receptors 40. Therefore LUSH appears to be a good model system for the study of alcohol binding to proteins.
To investigate the role of specific hydrogen bonding interactions in defining a “high-affinity” binding site for alcohol in LUSH we created specific amino acid substitutions, S52 to alanine (S52A), T57 to alanine (T57A), and T57 to serine (T57S) and tested the effect they had on alcohol binding. We show that ethanol is highly sensitive to even small changes in the binding site and these changes are much better tolerated with butanol. These results have important implications in understanding the nature of ethanol binding sites in proteins.
Results
Molecular Dynamics Simulations Suggest T57 Is Critical For Binding Alcohols
We postulated that the specific hydrogen bonding interactions between alcohols and LUSH may be important for formation of a high affinity binding site. Efforts to define these interactions using either atomic resolution X-ray crystallography or neutron diffraction have proven unsuccessful as per-deuterated crystals fail to diffract neutrons even though they diffract X-rays well. Therefore, we used molecular dynamics simulations of LUSH-alcohol complexes to gain insight into the nature of these interactions using the previously solved wild-type crystal structures of each complex as the initial starting coordinates. Simulation for a particular LUSH-alcohol complex were repeated at least three times over time frames of 200 ps to 1 ns, each time using a different initial random seed for directions and velocities of the atomic motions. As a result, the details of the atomic trajectories for each simulation performed with the same alcohol differ from each other, however the trends for simulations performed with same alcohol are similar.
In three of four separate simulations of the LUSH-ethanol complex, we found that the ethanol molecule is only transiently bound to the polar residues at T57 and S52, and moves out of hydrogen bonding distance in under 50–100ps (Figure 2 a and b). In the other simulation, it remains within hydrogen-bonding distance over the entire time period. When ethanol is within hydrogen-bonding distance of S52 and T57, it appears that T57 acts preferentially as an H-bond donor to the alcohol hydroxyl group (Figure 2a and b). In contrast, S52 rarely acts as an H-bond donor to the alcohol. Instead, S52 forms a hydrogen bond to the main-chain carbonyl group of T48 for almost the entire duration of all calculations performed. The interactions between ethanol and S52 are less well defined. At times, ethanol acts as a hydrogen bond donor to S52, while at other times the interaction of ethanol with S52 is bridged by a water molecule. In these latter cases, the position of the ethanol is shifted closer to T57 and away from S52 compared to its position observed in the crystal structures (Figure 3). In this conformation, the net residue dipole from T57 (4.36 D), is aligned head to tail with the net dipole of ethanol (2.35 D), whereas the net residue dipole from S52 (4.61 D), is orthogonal to the ethanol molecule by virtue of its position within helix 3. This may explain why T57 appears to be the most critical residue in forming interactions with the alcohols in these calculations.
Figure 2. Hydrogen Bonding Patterns in MD Calculations of LUSH-alcohol complexes.
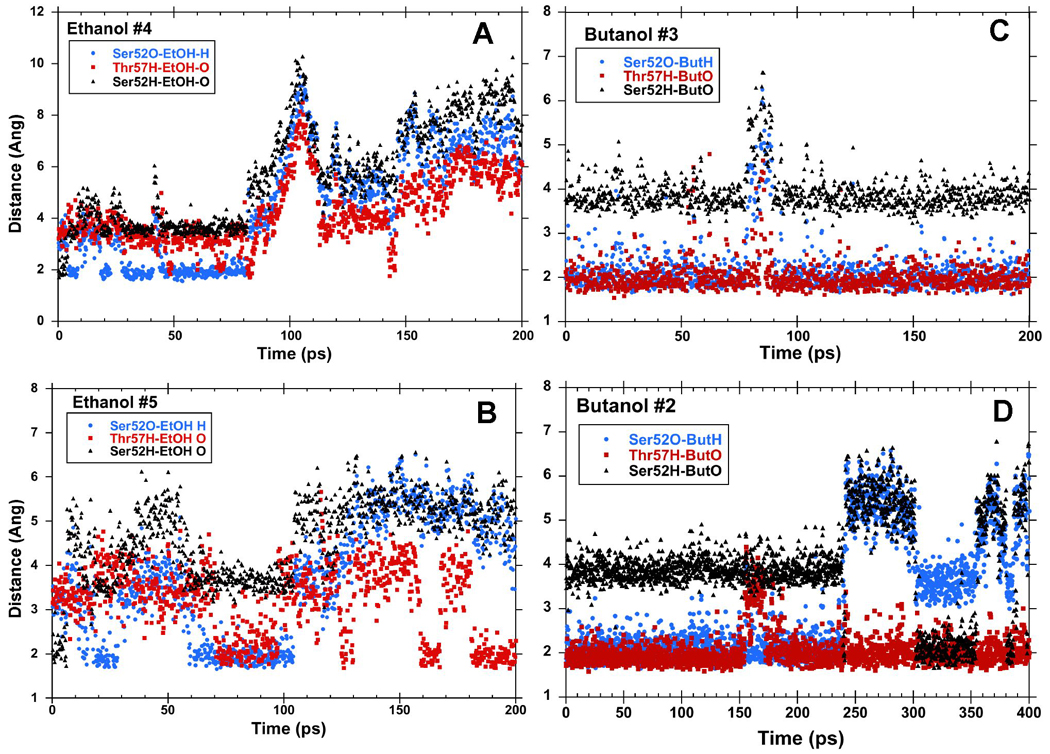
Hydrogen bond distances between alcohols and S57 and T57 in representative MD simulations of LUSH-ethanol complexes (a and b) and LUSH-butanol complexes (c and d). Butanol binds in the site for a longer period of time than ethanol. When bound, generally, alcohols act as hydrogen bond donors to S52 and accept a hydrogen bond from T57.
Figure 3. Hydrogen Bonding in LUSH-alcohol complexes from MD Calculations.
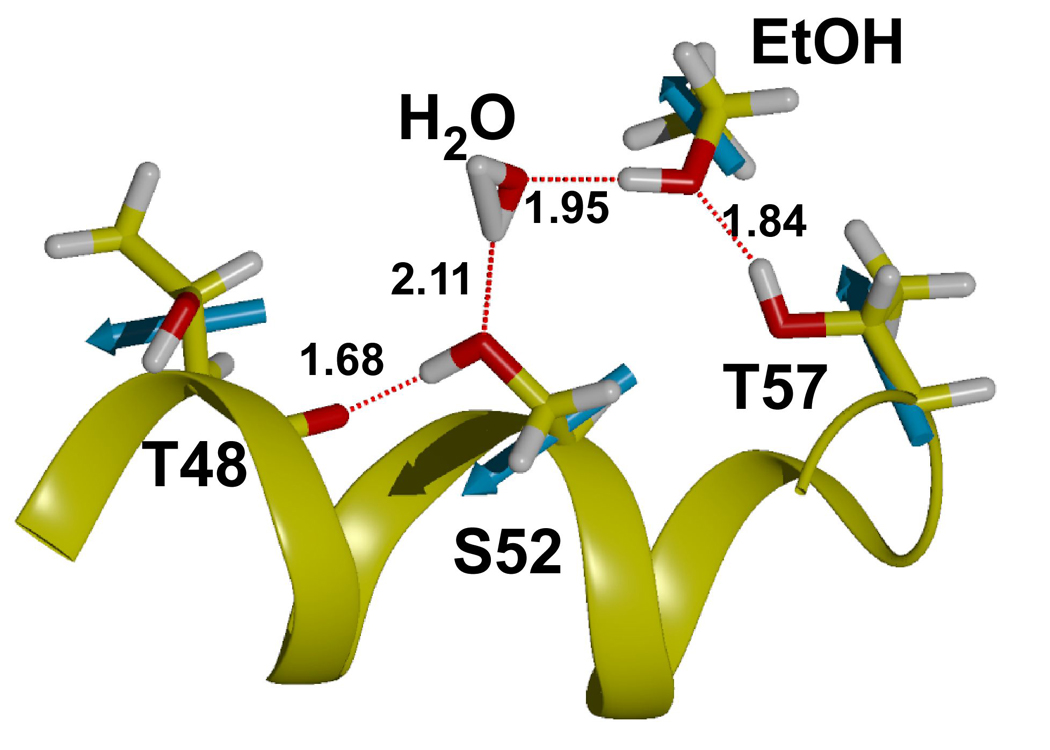
Snapshot of calculation shown in Figure 2b taken at ~200 ps. MD simulations predict that T57 acts preferentially as a donor to alcohols. S52 donates a hydrogen to the main chain of Thr48 and acts as an acceptor for a hydrogen bond from the alcohol or water molecules. Net residue dipoles for the key residues in the binding site and ethanol are shown in cyan. The dipole of T57 is oriented in a head to tail arrangement with the net dipole of ethanol and this may aid in increasing binding affinity at this site.
A total of 4 calculations were performed for the LUSH-butanol complex. A major contrast between these calculations and those of the LUSH-ethanol complex calculations is that the butanol appears to have a much longer residency time in the vicinity of S52/T57 (Figure 2c and d). In two calculations performed over a 200 ps time frame, the butanol remains hydrogen bonded to both S52 and T57 throughout the simulation. When these calculations were extended to 1 ns, it was found that this hydrogen bonding arrangement persisted for ~500 ps in one calculation, while in a fourth calculation a conformational shift occurs that disrupts the bond to S52 but maintains interactions with T57. In all of these calculations with butanol, S52 again forms a hydrogen-bond almost exclusively with the main-chain carbonyl of T48 while T57 is acting as a hydrogen bond donor to the alcohol and the butanol is donating an hydrogen bond to the side chain oxygen of S52.
These MD dynamics calculations predict that T57 is likely to be the major contributor to alcohol binding in LUSH. This is consistent with the potentially shorter hydrogen bonds we observe between T57 and the alcohol in the crystal structures of these complexes 8. In order to validate these predictions, we tested the effects of specific amino acid substitutions in LUSH designed to disrupt these potential hydrogen bonding interactions.
Crystal Structures of Substituted LUSH-Alcohol Complexes
We solved the crystal structures of wild-type apo LUSH (PDB 1T14), S52A with ethanol (PDB 3B7A) and butanol (PDB 3B6X), T57A in the presence of ethanol (PDB 3B88) and butanol (PDB 3B87), and T57S with ethanol (PDB 3B86). In all cases the concentration of alcohol used was ~30 mM. The structures of all these substituted proteins are very similar to previously solved LUSH-alcohol complexes 8 and show no significant conformational changes in structure. The proteins crystallized in different space groups, but each had two monomers in the asymmetric unit. These are arbitrarily labeled monomer A or B to distinguish them. Table 3 lists the backbone root mean squared deviations (RMSDs) for each monomer of each substituted protein compared to the wild-type LUSH-butanol structure (PDB 1OOH) 8.
Table 3.
RMSD values for substituted LUSH proteins compared to the wild-type protein. Monomer A of the substituted structure was superimposed on monomer A of the wild-type, and monomer B of the substituted structures was superimposed on monomer B of the wild-type protein.
| Main Chain | All Atoms | |||
|---|---|---|---|---|
| A | B | A | B | |
| S52A-butanol | 0.09 Å | 0.10 Å | 0.40 Å | 0.28 Å |
| S52A-ethanol | 0.17 Å | 0.16 Å | 0.41 Å | 0.33 Å |
| T57A-butanol | 0.47Å | 0.75 Å | 0.95 Å | 1.15 Å |
| T57A-ethanol | 0.21 Å | 0.27 Å | 0.51 Å | 0.64 Å |
| T57S-ethanol | 0.24 Å | 0.24 Å | 0.64 Å | 0.59 Å |
The details of each individual structure are described below, but in summary these structures again provide evidence that T57 is critical for alcohol binding. Substitution of this residue with alanine completely disrupts the binding of alcohol and water. In contrast, the T57S substitution had little or no effect upon binding of ethanol. All of these structure were solved to a resolution of 1.8–2.0 Å. At this level of resolution the positions of the hydrogen atoms are not observable and so the arrangement and strength of hydrogen bonds has to be inferred from a knowledge of the chemistry of the interacting groups and the distances between the oxygen atoms in the donor and acceptor groups. Although O-O distances are not the best predictor of hydrogen bond strength. Further, all the distances presented in the discussion below are subject to the experimental uncertainties in the coordinate positions given in Table 4.
Table 4.
Binding affinities of wild-type and substituted LUSH proteins for alcohols.
| Wild-type | T57S | S52A | T57A | |
|---|---|---|---|---|
| Ethanol | 107 ± 13 mM | * | * | * |
| Butanol | 11 ± 1 mM | 16 ± 2 mM | 158 ± 54 mM | >> 200mM |
| Pentanol | 2 ± 0.3 mM | 2 ± 0.3 mM | 6 ± 0.9 mM | |
| Hexanol | 0.20 ± 0.02 µM |
Indicates that no alcohol could compete for binding of ANS at any of the concentrations tested
A water molecule substitutes for alcohols in apo-LUSH
The structure of the apo-form of LUSH is essentially identical to the previously determined structures of LUSH solved in the presence of short chain n-alcohols 8, and the average RMSD between the individual monomers is 0.15 Å for main chain atoms and 0.56 Å for all heavy atoms. The notable difference between apo-LUSH and the LUSH-alcohol complexes is that the bound alcohol has been replaced with a well-ordered water molecule (not shown). This water molecule is located in essentially the same position as the hydroxyl group of the alcohols and makes the same hydrogen bonds with S52 and T57 as seen in the alcohol bound structures. Again hydrogen bonds formed between the water and T57 appear to be shorter (dO-O of 2.77 Å and 2.80 Å) than the potential hydrogen bonds to S52 (dO-O of 3.27 Å and 3.32 Å), suggesting that Thr57 forms the primary hydrogen bonding interaction with the ligand. The only other significant conformational difference between the alcohol bound complexes and the apo-protein is the positioning of F113. In both monomers of the asymmetric unit, F113 adopts two different conformations which reflects the open nature of the hydrophobic binding cavity.
The S52A substitution disrupts binding of ethanol but not of butanol
In the S52A-ethanol structure, extended electron density for ethanol is only observed in one monomer (A) of the asymmetric unit (Figure 4a). This ethanol is within hydrogen bonding distance of the Oγ of T57 (2.83 Å). However, in monomer B, the electron density in the binding site refines well as a water molecule instead of ethanol (Figure 4b). This water is in the same position as the water molecule observed in the binding site of the apo wild-type structure. This suggests that the S52A substitution has potentially weakened binding of ethanol at this site, suggesting that a potential hydrogen bond between S52 and the alcohol is important for ethanol binding, even though this interaction is likely to be very weak.
Figure 4. Electron Density Maps of the Binding Site in Substituted LUSH proteins.
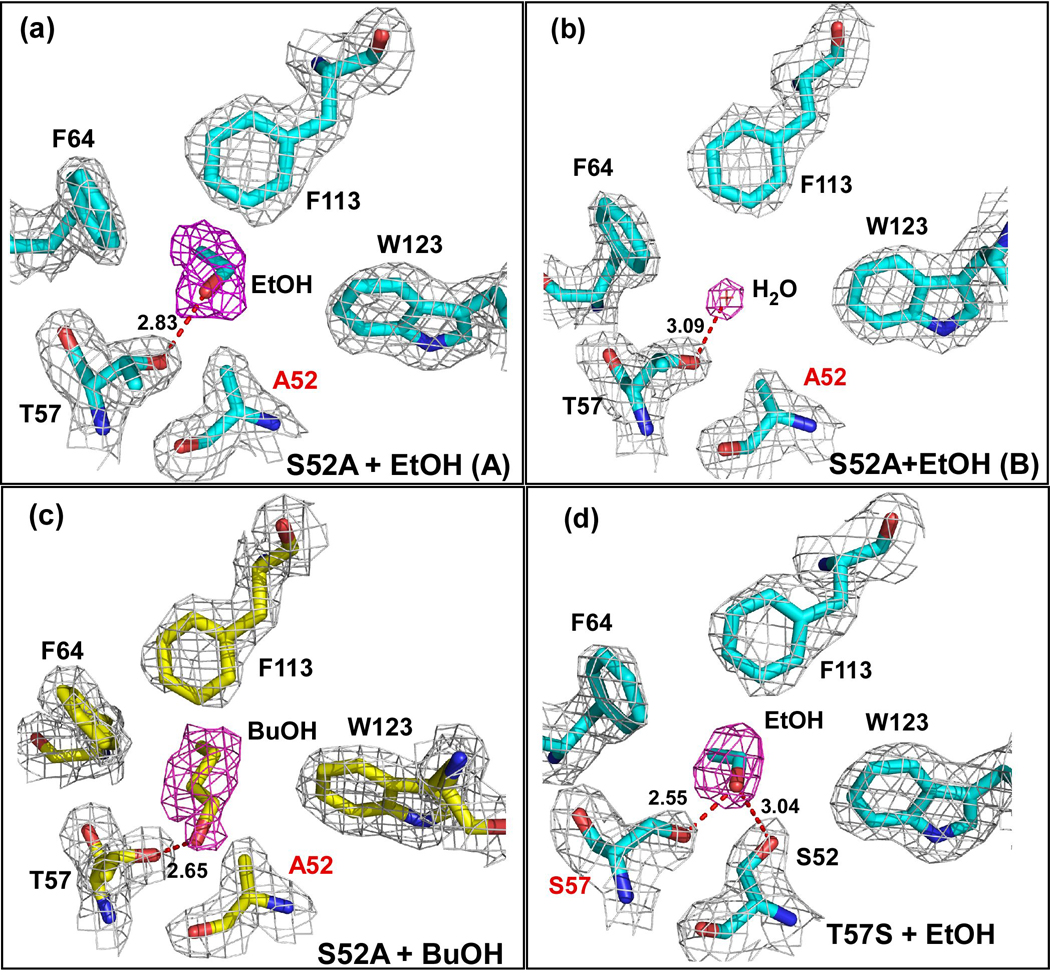
Electron density in the final normalized 2Fo-Fc maps contoured to 1.0 σ are shown in gray, while the electron density from Fo-Fc omit maps contoured at 2.5 σ are shown in magenta for (a) Monomer A of the S52A-ethanol structure. The ethanol molecule is in the same position as seen in the wild-type protein. The distance between the T57 Oγ and alcohol O is 2.83 Å; (b) Monomer B of the S52A-ethanol structure. A water molecule is observed in the place of the ethanol suggesting that this substitution might have affected ethanol binding. The distance between the T57 Oγ and water O is 3.09 Å. (c) S52A-butanol structure. Density for butanol is clear in both monomers of the asymmetric unit. The distances between the T57 Oγ and alcohol O are 2.65 Å in chain A and 2.82 Å in chain B. (d) The T57S-ethanol complex. Again density for an ethanol molecule is visible in both monomers of this structure, in the same position as seen in the wild-type protein. The distance between the S57 Oγ and alcohol O is 2.55 Å in both chains while the distance between the S52 Oγ and alcohol O is 3.04 Å in both chains. In all cases the proteins were crystallized in the presence of 30 mM of the corresponding alcohol. Each protein is in approximately the same orientation as shown in figure 1b.
In both monomers of the S52A-butanol structure, there is clear electron density for butanol in the alcohol-binding site (Figure 4c). The butanol is within hydrogen bonding distance of the Oγ atom of T57, 2.65Å in monomer A and 2.82 Å in monomer B. This compares to distances of 2.65 Å and 2.34 Å, observed in the wild-type butanol structure. The distance from the S52 Cβ to alcohol O is 3.58 Å in monomer A and 2.62 Å in monomer B. This compares to a distance of 3.58 Å and 3.52 Å in the wild-type complex respectively. Substitution of S52 with alanine generates a small cavity so that the alkyl chain of the butanol molecule is orientated in the opposite direction to that seen in the wild-type protein so that it makes van der Waals interactions with F64 and F113. We had previously observed that ethanol and propanol can both adopt this alternate binding conformation 8. However, this is the first time that we have seen butanol bind so that the alkyl chain does not extend out into the central cavity.
The T57S Substitution Has No Effect on Binding Alcohols in The Crystal
Substitution of T57 with serine had no apparent effect on the binding of ethanol to the protein in the crystal structure. Both monomers in the asymmetric unit of the complex show clear electron density for an ethanol molecule binding to S57 and S52 (Figure 4d). The distance from the O of the ethanol to the Oγ of Ser57 is 2.55 Å and to the Oγ of S52 is 3.04 Å in both monomers. These are essentially identical, within coordinate error, to the O-O distances seen in the wild-type protein, suggesting that this substitution has not significantly altered the chemistry in the binding site.
A feature of this structure is the presence of a portion of a polyethylene glycol (PEG) molecule within the central hydrophobic cavity (Figure 5) that originates from the crystallization buffers. We also see a PEG bound in the same place in both structures of the T57A substituted protein (see below). The PEG molecule partially fills the ligand binding cavity, however, it does not interfere with alcohol binding. PEG has also been observed in the structure of a closely related odorant binding protein from Anopheles gambiae (AgOBP-1) where it was the only ligand within the cavity and in fact was shared between the two monomers in the asymmetric unit 41.
Figure 5. A PEG molecule is present in both monomers of the T57S-ethanol structure.
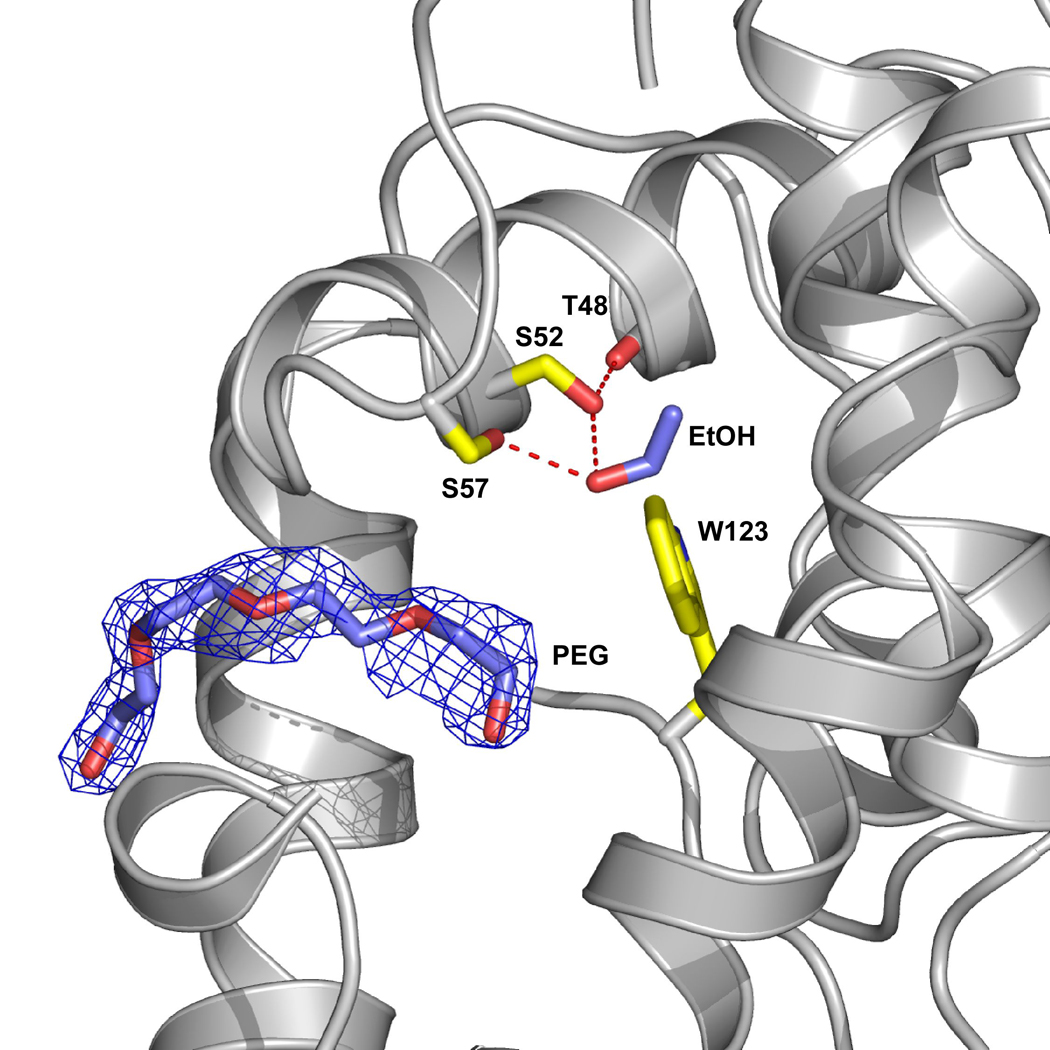
A region of a PEG molecule (blue) is observed to enter the site but does not interfere with alcohol binding. Yellow residues highlight the ethanol binding site and the ethanol is shown in blue. The protein was crystallized using 25–29% PEG4000, which contains ~ 90 repeating units, and the remainder of the PEG molecule, which is presumably outside of the binding site is not defined.
The T57A Substitution Disrupts Binding of All Alcohols
The crystal structures of the T57A substituted protein complexes reveal that this substitution has a dramatic effect on binding of alcohol. No electron density could be observed for any alcohol within the binding site, and we did not even observe density for a water molecule at this site despite the fact that there are several other ordered water molecules within the central cavity. None of these waters are within hydrogen bonding distance of S52 suggesting that the T57A substitution is unfavorable for hydrogen bonding interactions at this site. This contrasts with S52A substituted structures in which a water molecule is present in place of an ethanol molecule in one monomer and forms a hydrogen bond with T57. This suggests that a potential hydrogen bond donor, such as T57, is critical at this position for alcohol binding.
The structure of these substituted LUSH structures provide support for the importance of T57 as a key residue in contributing to a high affinity binding site. However it also appears that a hydrogen bond acceptor at position 52, albeit forming a weak interaction, is also important for ethanol binding. To further examine the effects of these substitutions we have determined binding affinities of alcohols for these proteins.
Differential Effects of Amino Acid Substitutions on Alcohol Dissociation Constants
The dissociation constants for alcohols to LUSH were determined using a competition binding experiment in which alcohols compete for binding of the fluorescent probe, 1-anilino-8-naphthalenesulfonic acid (ANS). ANS binds to hydrophobic patches of a protein, and binding is accompanied by an increase in ANS fluorescence 42; 43. We have previously shown that alcohols can compete for ANS binding to LUSH in a chain length dependent manner when present at the same concentration 35.
The alcohol-binding site of LUSH contains a single tryptophan (W123) residue in close proximity to T57 and S52 (Figure 1). When ANS is titrated into samples of the apo-protein there is an increase in ANS fluorescence that is correlated with a quenching of endogenous tryptophan fluorescence (Figure 6). This suggests that ANS binds in close proximity to this tryptophan. Furthermore, substitutions in LUSH designed to disrupt alcohol binding also change the binding affinity of the apo protein for ANS, providing further support that ANS is most likely binding to the same site as the alcohols. In addition, the crystal structure of an OBP from the cockroach L. madrae was solved with a single molecule of ANS in the hydrophobic binding pocket of the protein 44. It appears that ANS may bind to LUSH in an analogous fashion. In these experiments we are determining the inhibition constants for alcohols inhibiting ANS binding to LUSH. Analysis using Scatchard or Eadie-Hoftsee plots indicates that binding is competitive, therefore we are equating these to be equivalent to the dissociation constants for alcohol binding to the protein. However, it is possible that binding of alcohols and ANS does not occur at exactly the same site and so is not truly competitive. If this is the case, those alcohols that overlap the most with the ANS binding site will appear to have an increased binding affinity compared to the binding of other alcohols. However we cannot distinguish these possibilities with the present structural data.
Figure 6. Quenching of intrinsic tryptophan fluorescence by ANS.
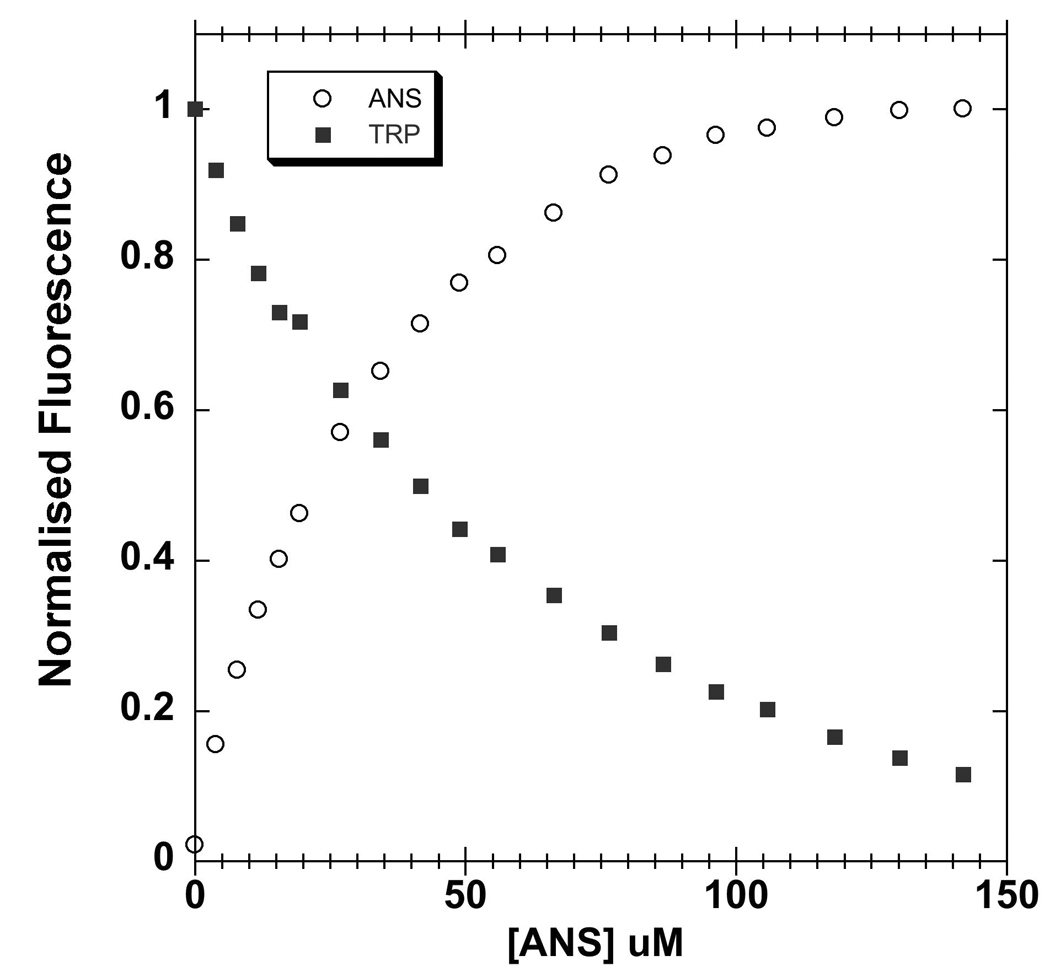
Increasing concentrations of ANS (circles) quenches intrinsic tryptophan fluorescence (squares), suggesting that ANS binds in proximity to W123 in the vicinity of the alcohol binding site. The fluorescence signal was normalized as a fraction of maximal signal. The protein concentration was 5 µM. Intrinsic tryptophan fluorescence was excited at 295 nm and recorded at 342 nm. ANS fluoresence was excited at 370 nm and recorded at 457 nn
The results of ANS titrations performed with LUSH-alcohol complexes are shown in Figure 7 and Figure 8. A summary of the calculated KD values for the alcohol for each complex are given in Table 4. To obtain these values each series of experiments with a single alcohol were globally fit with an equation that accounts for ligand depletion of the ANS, which is present at concentrations of the same order as the protein concentration and the dissociation constants. Concentrations and dissociation constants of alcohol were much higher (100–5000x) than the concentrations of protein used in each titration, so the free alcohol concentration was assumed to be equal to the total concentration of alcohol.
Figure 7. Short chain n-alcohols compete for binding of ANS to wild-type LUSH.
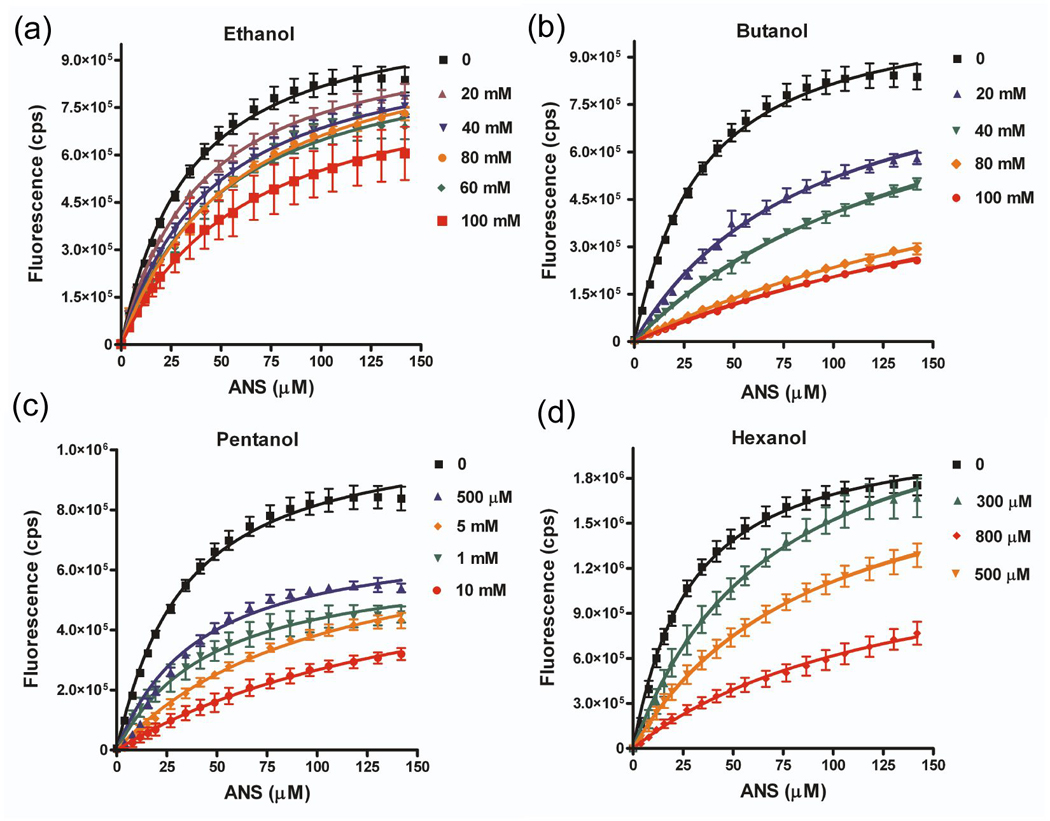
Graphs show titrations of LUSH-alcohol complexes with ANS as a function of increasing alcohol concentrations for (a) ethanol, (b) butanol, (c) pentanol, and (d) hexanol. The fitted curves are the results of a global fit using a competitive binding isotherm (see Materials and Methods). All alcohols can compete for binding to the wild-type protein.
Figure 8. Effect of aT57S and S52A amino acid substitutions on ability of alcohol to compete for ANS binding.
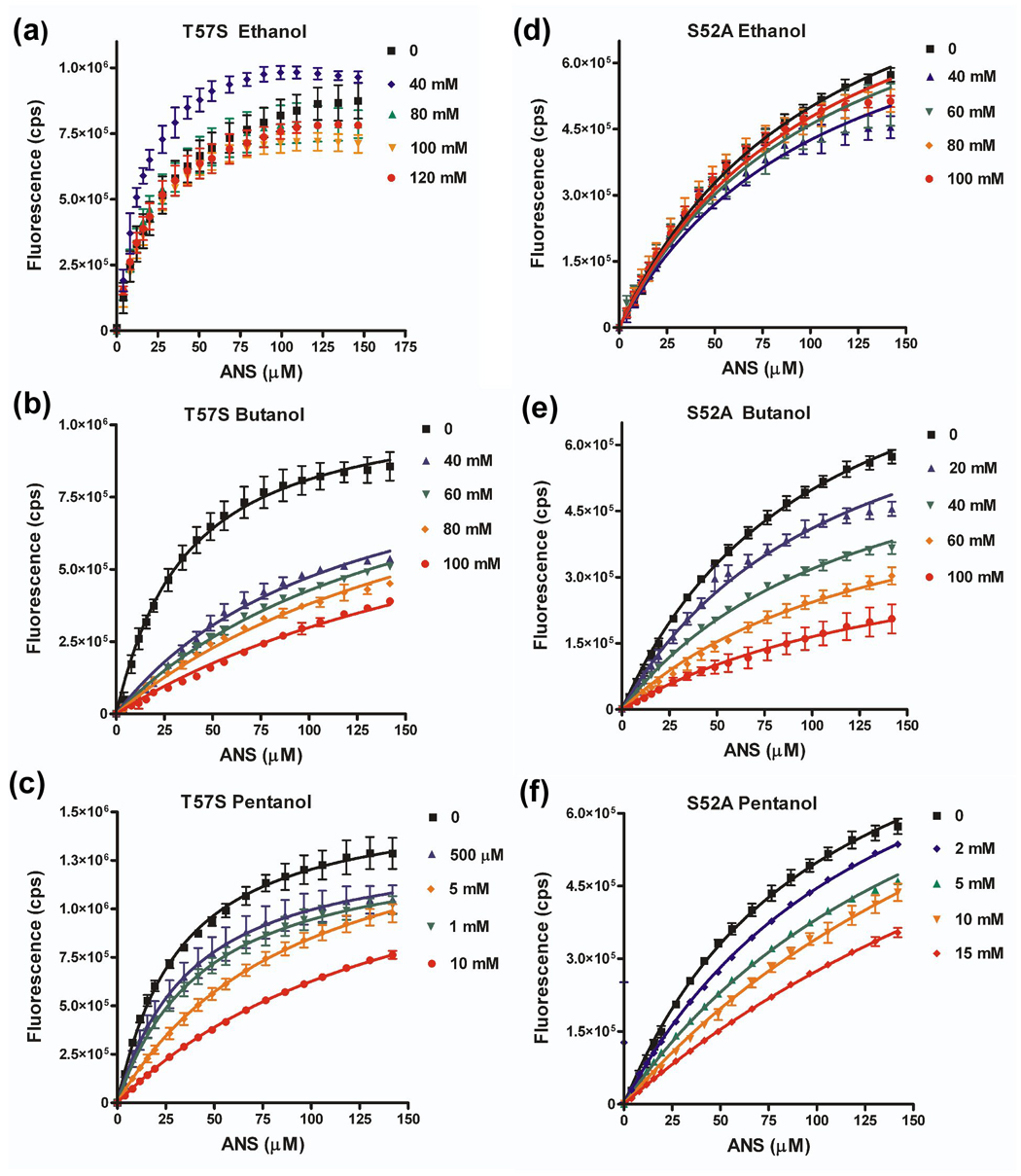
Binding isotherms for LUSH-alcohol complexes with ANS as a function of increasing alcohol concentrations for (a) T57S-ethanol, (b) T57S-butanol, (c) T57S-pentanol, (d) S52A-ethanol, (e) S52A-butanol, and (f) S52A-pentanol. Curves were fit in Graphpad Prism to a competitive binding isotherm as described in materials and methods. Ethanol could not compete for binding of ANS with either the T57S or S52A proteins. The concentration of alcohol is indicated
The detailed results of these experiments are discussed below, but in summary, amino acid substitutions within the alcohol-binding site appear to differentially affect ethanol binding compared to that of longer chain alcohols. Substitutions that remove a hydrogen bonding interaction, S52A and T57A, affect both ethanol and butanol binding, while the T57S substitution showed an effect on ethanol but not butanol binding. Therefore, we conclude that subtle changes in the chemistry of the binding site can lead to differential changes in the binding affinities for alcohols.
For the wild-type protein, we find a chain length dependence of the alcohol binding affinity. There is a 9.7 fold difference in the KD values for ethanol and butanol, a 5.5 fold difference between butanol and pentanol, and a 10 fold difference between pentanol and hexanol. This increase in binding affinity as a function of chain length is not unexpected, and can be attributed to the additional interactions formed by the longer alkyl chains. Similar dissociation constants have been observed for the binding of ethanol (232 mM), butanol (13 mM), and hexanol, (1.5 mM) in the Shaw2 Drosophila potassium channel 23.
Substitution of T57 with serine has only a limited effect on the binding affinities of butanol and pentanol compared to the wild-type protein. In contrast, ethanol could not compete for binding with ANS to the LUSHT57S substituted protein over the concentration range tested. There is no detectable decrease in fluorescence upon increasing ethanol concentrations. We did not test higher concentrations of ethanol as we are concerned about non-specific interactions or potential protein unfolding that starts to occur at concentrations of alcohol > 1% v/v (170 mM). Even so, we can still conclude that a threonine at position 57 appears critical to achieve the highest binding affinity of ethanol, but binding of butanol can tolerate a threonine or serine at this position.
Substitution of the S52 with alanine leads to a 14 fold decrease in the binding affinity for butanol and a three fold decrease in the binding affinity of pentanol compared to the wild-type protein. This corresponds to a change in the free energy of binding for butanol of 1.6 kcal mol−1 by deleting this one potential hydrogen bond, while there is a 0.55 kcal mol−1 change in binding energy for binding of pentanol. These results demonstrate that the relative importance of hydrogen bonding interactions to the overall binding energy are reduced as the chain length increases and the contributions of van der Waals interactions and/or hydrophobic effects begin to dominate the interaction.
Again ethanol cannot compete for ANS binding to the S52A substituted protein. This is consistent with the crystal structure of the S52A-ethanol complex in which ethanol is only observed to bind to one monomer of the asymmetric unit. Together we conclude that a potential hydrogen bond acceptor appears to required for the highest binding affinity of ethanol at this site.
Substitution of T57 with alanine has a significant effect on both ethanol and butanol binding. This substitution also weakens binding of ANS (128 µM compared to 30 µM for wild-type). This reduced affinity for ANS could potentially enhance the ability of alcohol to compete for binding. However, there is no observable change in ANS fluorescence upon increasing concentrations of ethanol or butanol. This result is consistent with our crystal structures, in which neither alcohol nor water is bound in any monomer of either structure as well as our molecular dynamics simulations, which suggest that T57 is the most important residue for binding alcohols.
Changes in Binding Affinity are not due to Changes in Protein Stability
LUSH is a highly dynamic protein, with significant regions undergoing conformational exchange on the micro-milliseconds timescale in solution 35. We postulate that this highly dynamic state regulates LUSH’s ability to activate olfactory signaling in insects 34. Binding of ligands such as alcohols leads to a shift to a more ordered state, and the degree of ordering depends on the chain length of the alcohol 35. Therefore it is possible that changes in the binding affinity of alcohols in the substituted LUSH proteins is due to stabilization of a preferred binding site. Therefore, we examined the effects the substitutions on the overall stability of the protein by comparing changes in the melting temperature (TM) of the substituted and wild-type proteins in the apo and butanol bound states.
Far UV circular dichroism spectra of the wild-type and substituted proteins in both the apo form and in the presence of 80 mM butanol were recorded as a function of temperature over the range of 5°C to 95° while the molar ellipticity was monitored at 222 nm (Figure 9a–d). Data were then processed using the method of John and Weeks 45 to obtain values of the TM and van't Hoff enthalpies of unfolding (ΔVH) for each complex (Figure 9e–h). The results of this analysis are given in Table 5.
Figure 9. Effect of amino acids substitutions on thermal unfolding of wild-type and substituted LUSH proteins.
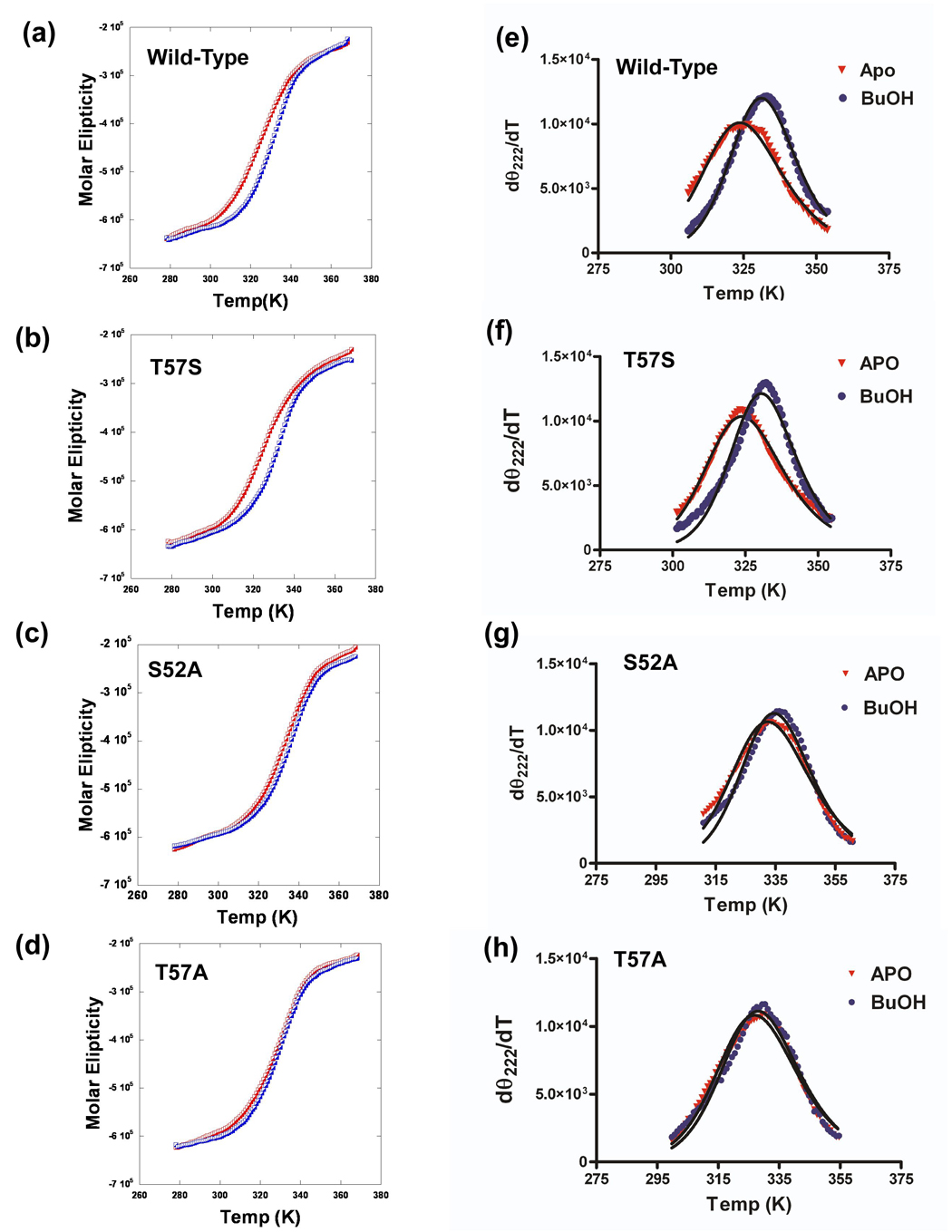
(Left) Thermal unfolding of LUSH proteins in the apo state (red) and in the presence of 80mM butanol (blue) monitored using CD at 222 nM and (Right) differential unfolding curves derived from the data in the left panel to obtain values of Tm and ΔVH 45 for (a & e) Wild-type, (b & f) T57S, (c & g) S52A, and (d & h) T57A proteins.
Table 5.
Summary of melting temperatures of WT and substituted LUSH proteins determined by circular dichroism. In all cases the concentration of butanol was 80 mM.
| TM (°C) Apo | TM (°C) + butanol | Δ TM (°C) | |
|---|---|---|---|
| Wild-Type | 49.0 ± 0.2 | 56.0 ± 1.8 | 7.0 |
| T57S | 49.1 ± 1.9 | 56.4 ± 1.2 | 7.3 |
| S52A | 58.2 ± 1.1 | 61.7 ±1.9 | 3.5 |
| T57A | 53.4 ± 2.3 | 54.2 ± 2.4 | 0.8 |
Apo-LUSH has a TM of 49.0 °C. The T57S protein had a similar melting temperature of 48.1 °C, while the S52A and T57A proteins had higher TM values of 58.2 °C and 53.4 °C. This suggests that the T57S substitution had no effect on the overall stability of the protein while replacement of S52 or T57 with alanine both result in an increase in the stability of the protein to thermal denaturation, although to differing degrees. When these experiments are repeated in the presence of 80 mM butanol, we find that the ability of the alcohol to stabilize the protein against thermal unfolding is correlated with the binding affinity for the alcohol. For example, the wild-type protein showed an increase in TM of 7 °C in the presence of butanol. The T57S protein, which has a similar binding affinity for butanol as the wild-type protein, shows the same increase in TM of 7 °C in the presence of butanol. In contrast, the S52A substitution, which reduces the binding affinity for butanol, shows only a 3°C increase in TM in the presence of butanol. Finally, the T57A substituted protein, which does not bind butanol, shows no significant change in its TM in the presence of butanol compared to the wild-type protein. However, butanol does appear to slightly stabilize a pre-unfolding transition (Figure 9d).
These studies also reveal that a change in the overall stability due to a specific substitution does not correlate with a change in the binding affinity for alcohols. For example, the S52A substitution results in an increase in the TM of the apo protein by 9°C. If this substitution led to stabilization of an alcohol-binding site we may expect that it would have an increased binding affinity for alcohol. Instead, the S52A substituted protein shows a significant decrease in binding affinities for alcohols when compared to the wild-type protein. We observe a similar effect for the T57A substituted protein which exhibits an increase in TM of 4.5 °C but this substitution completely disrupts alcohol binding.
Discussion
Previous work has established that in D. melanogaster LUSH is required for both behavioral 33 and electrophysiological responses of olfactory neurons 34 to alcohols. Further, X-ray crystallography and NMR spectroscopy8 clearly show that short chain n-alcohols bind to a single site in LUSH and this binding leads to a conformational shift from a highly dynamic structure to a well-ordered structure 35. Work by others 46 however, has suggested that alcohols are not ligands for LUSH, as alcohols failed to displace fluorescent dyes or chemical plasticizers from the binding site of LUSH, whereas a plasticizer, dibutyl phthalate can displace these compounds from the binding site. Additionally flies exhibit an avoidance response to dibutyl phthalate 46. Our present data suggest that the concentrations of alcohols used in those experiments were too low to observe any effect on the binding of these artificial ligands. It may appear that alcohol concentrations in the millimolar range are physiologically irrelevant, however, alcohol concentrations in nature can easily reach levels of ~4–15% v/v or 0.7–2.4 molar. The involvement of LUSH in the avoidance response to dibutyl phthalate also has not been established, as the responses to phthalates in lush null flies has not been tested. Indeed a number of different olfactory receptor neurons that do not express LUSH are known to respond to aromatic compounds47; 48, and so it is entirely possible that responses to dibutyl phthalate are mediated by a completely different subset of olfactory neurons. In contrast, there is strong evidence for the involvement of LUSH in responses to alcohols. lush null flies show altered behavioral and electrophysiological responses to alcohols and this wild-type behavior can be recovered in flies that contain a lush rescue mutation 33; 49. Therefore, while it is now clear that LUSH is also required for the responses to the Drosophila pheromone 11 cis-vaccneyl acetate in a different subset of olfactory neurons49, behavioral, electrophysiological, high resolution structural and biophysical data all support the observation that LUSH binds to alcohols.
The alcohol binding site in LUSH has a unique chemistry that leads to an increased binding affinity for alcohols at this site compared to other potential binding sites within the protein. In addition to a number of hydrophobic residues, this binding site contains a set of polar residues that form a network of hydrogen bonds with alcohols and water. Small changes in the nature of the polar residues in this site significantly disrupt binding of alcohols. Our results reveal that T57 is the most critical residue within this network. Making a T57S substitution has little or no effect on binding of longer chain alcohols such as butanol or pentanol but substantially reduces the binding affinity for ethanol. Deletion of this hydrogen bond by the T57A substitution dramatically reduces the binding affinity for all short chain alcohols. In addition, we find that binding of ethanol also requires the ability to form a hydrogen bond with S52. Hydrogen bonding interactions formed with this residue are likely to be very weak based on the observed O-O distances of the donor and acceptor groups in the crystal structures of the wild-type protein (3.18 Å s.d. 0.14 Å). However, removal of this potential hydrogen bonding interaction leads to a 14 fold reduction in the binding affinities for butanol and disrupts ethanol binding to the point where it cannot bind at concentrations under 150 mM. This indicates that optimal binding affinity for ethanol is only achieved by having both of these hydrogen bonds, or at least the potential to form an additional dipolar interaction.
The relative importance of the potential hydrogen bonding interactions to the overall free energy of binding appears to diminish with increasing alcohol chain length. The overall binding affinity of the alcohol is dependent on chain length and each methylene groups contributes 0.9 Kcal mol−1 to the binding. Similar values of 0.8 Kcal mol−1 per methylene group have been found in studies of the Drosophila Shaw2 K+ voltage sensitive channel for alcohols contain 2 to 6 carbons 24. For longer-chain alcohols such as pentanol and hexanol, the additional interactions formed by the alkyl chain can compensate for the loss of high energy hydrogen bonds. In contrast, binding of ethanol appears to be dominated by the formation of hydrogen bonds or dipolar interactions with polar side groups, such as serine and threonine residues.
Molecular dynamics simulations suggest that there is a preferred arrangement of hydrogen bonds between LUSH and the alcohol. The preferred arrangement has T57 donating a hydrogen bond to the alcohol, which in turn donates a hydrogen bond to S52, while S52 acts as hydrogen-bond donor to the main-chain carbonyl group. It is known that hydrogen bonding groups have altered abilities to donate or accept hydrogen bonds depending on if they act preferentially as donors or acceptors 50. If S52 acts as a preferential donor to T48 and makes this interaction in the absence of alcohols, this may prime this group as a better hydrogen bond acceptor for a hydrogen bond from the alcohol, and this may act to increase the strength of this interaction.
We have established that for LUSH, the specific nature of the hydrogen bonding interactions are critical for defining a high affinity ethanol binding site. We hypothesize that such sites may also exist in other alcohol sensitive proteins, and indeed, many ion channels contain numerous clusters of threonine and serine residues at the interfaces of the transmembrane domains that could provide high affinity binding sites. It is clear that there are a number of other factors that could potentially enhance alcohol binding to a protein. These include, but are not limited to, the presence of ordered waters within the binding site, the polarizability of alcohols, and changes in the local dielectric constant.
Release of ordered water molecules into the bulk solvent can enhance ligand binding, and it has been estimated that this can contribute ~1.9 Kcal mol−1 to the binding energy 51–53. The alcohol-binding site of LUSH exists in a water filled cavity, and a water molecule is bound when no alcohol is present. An ethanol molecule could displace the equivalent of two water molecules, leading to a favorable entropy for binding of alcohol in this site, while longer chain alcohols could potentially displace even more waters.
The effects of alcohols on protein structure and the ability to promote folding or unfolding have been attributed to both the polar or non-polar nature of the molecule and changes in the dielectric constant of the solvent 54–57 or by disrupting normal protein solvent interactions. However, almost all of these effects of alcohol occur at concentrations greater than 1% v/v alcohol. Changes in the local dielectric can also potentially have large impacts on electrostatic interactions between ligands and proteins 58. Displacement of water by alcohol in the LUSH binding could potentially lead to a decrease in the local microscopic dielectric and enhance the strength of dipolar interactions in the vicinity of the binding site. As the nature of the dipolar interactions appear to be critical for binding of ethanol, differences in the overall polarizability of ethanol compared to water could lead to the induction of larger dipoles and therefore stronger interactions within the binding site for ethanol compared to water. Estimates for the polarizability of ethanol range from 5.11 Å3 59 to 5.26 Å3 60, compared to 1.49 Å3 for water 59. Differences in polarizability have been recognized for some time as potentially important determinants of anesthetic potency 61–63 and recent computational studies suggest that the polarization of anesthetics by the protein could potentially enhance binding by 1.2 Kcal mol−1 compared to the non-polarized state 64. A similar effect may occur for alcohols. It is notable that in our MD simulations, the net residue dipole of T57 is oriented so it forms a favorable interaction with the dipole of ethanol. This may explain why T57 appears to be so critical for binding of all alcohols.
While multiple factors contribute to the formation of a high affinity alcohol binding site in LUSH and other proteins, our present results reveal that the binding of ethanol appears highly sensitive to the specific chemistry of the ligand binding site. The requirement of hydrogen bonding groups and hydrophobic residues can be easily predicted, however the requirement for a threonine over a serine residue was not readily predicted. Further, we found that these factors are critical for ethanol binding but appear less so for longer chain alcohols. These findings may be important in understanding the nature of alcohol binding sites in other proteins such as ligand gated ion channels which are molecular targets for alcohol.
Materials and Methods
Site Directed Mutagenesis, Protein Expression and Purification
DNA primers (Operon) for polymerase chain reaction of 25–35 base pairs were designed to create T57S, T57A, and S52A mutations in the lush gene. The forward and reverse primer sequence are, T57S 5′ GTGTCTTTGATGGCGGGCTCTGTGAACAAAAAGGGGGAG 3′ and 5′ CACAGAAACTACCGCCCGAGACACTTGTTTTTCCCCCCTC 5′. S52A 5′ TGCTACACAAAGTGTGTGGCGTTGATGGCGGGCACTGTG 3′ and 5′ CACAGTGCCCGCCATCAACGCCACACACTTTGTGTAGCA 3′ T57A 5′ GTGTCTTTGATGGCGGGCGCGGTGAACAAAAGGGGGAG 3′ and 5′ CTCCCCCTTTTTGTTCACCGCGCCCGCCATCAAAGACAC 3′
The primers were purified using size exclusion chromatography using a Superdex 75 column (Amersham Pharmacia Biotech). Mutagenesis was performed as recommended in the Stratagene QuikChange site-directed mutagenesis kit, and after sequence verification plasmid DNA with the desired mutation was transformed into E. Coli BL21 (DE3) cells (Novagen) for protein expression and purification as previously described 35.
Crystallization and data collection
Crystals of apo-LUSH and substituted LUSH proteins were grown in sodium acetate 100 mM, pH 4.0, 25–30% (w/v) PEG 4000, and 30 mM alcohol at 18 °C and flash frozen in liquid nitrogen. Data for the apo-LUSH and LUSHS52A with butanol were collected in the UCHSC crystallography facility using a Rigaku MSC Ru-H3R generator with an Raxis IV++ area detector and was processed using HKL2000 65. Data for the LUSHT57S-ethanol, LUSHS52A-ethanol, and LUSHT57A-ethanol complexes were collected at the Molecular Biology Consortium beamline 4.2.2 at the Advanced Light Source at Lawrence Berkeley National Laboratory, Berkeley California. Data were processed using D*trek 66.
Structure Determination and Refinement
All structures of the substituted LUSH proteins were solved by molecular replacement using the wild-type LUSH structure (PDB entry 1OOH) as a model in CNS 67 or CCP4 68 programs. Model building was performed in O 69. The LUSH-S52A structures refined in the P43 spacegroup, the LUSH-T57S and LUSH-T57A-butanol structures were refined in the P212121 spacegroup, and the LUSH-T57A-ethanol structure was refined in the C2221 spacegroup using REFMAC in CCP4. The RMSD values were calculated using the program Superpose 70.
In all cases structures were initially refined without any alcohol present in the ligand binding pocket. Any electron density observed in the binding site in either 2Fo-Fc (contoured at 1 σ) or Fo-Fc maps (contoured at 2.5 σ) was initially modeled as a water and only replaced with alcohol if the refinement indicated the presence of additional density that was not satisfied by the water molecule. No additional restraints were placed on the alcohols, however in all cases no significant deviations from the ideal bond lengths and bond angles were observed in the refined structures. Inclusion of ethanol at these sites was reinforced by the observation that butanol also bound in the same site, and this has clearly elongated density compared to a water molecule.
Molecular Dynamics Simulations
Calculations were performed in the UCHSC Computational Facility on a Dell Poweredge cluster with 24 1 GHz Pentium III processors using the program NAMD 71 and the Charmm22 force field 71; 72. Each simulation used the coordinates of a single LUSH-alcohol complex from the crystal structure and any crystallographic waters within 4 Å of the protein surface. Hydrogen atoms were added using XPLOR 73 and bulk solvent was generated using the SOLVATE plug in of VMD 74 and truncated to a box of 56 Å length along each side. Sodium and chloride ions were added at random positions to maintain charge neutrality. Periodic boundary conditions were used to maintain the geometry of the system and molecules were allowed to wrap throughout the system. Electrostatic interaction were treated using a Particle Mesh Ewald approximation with a cut-off distance of 12.0 Å and a pair-list distance of 13.5 Å. A timestep integrator of 1 fs was used in all calculations and the full electrostatic potential was recalculated every 4 fs.
Each complex was energy minimized and equilibrated prior to simulation runs. Initially the positions of waters and ions only were minimized for 5 ps using conjugate gradient energy minimization. Second, all atoms were subjected to 5 ps of free dynamics at 140 K, followed by equilibration from 140 K to 298 K over a period of 5 ps. The resulting coordinate sets were used for multiple simulations repeated with initial random number seeds for atomic velocities and directions. The equilibrium of the system during calculations was evaluated by monitoring the temperature and overall energies of the system. The resulting trajectories were analyzed using Xplor 73 and VMD 74.
Fluorescence Spectroscopy
Samples of wild-type and substituted proteins prepared as above and diluted to a final protein concentration of 5 µM, and incubated with ethanol and butanol at concentrations of 0, 20, 40, 60, 80, and 100 mM and pentanol at 500 µM, 1 mM, 5 mM, and 10 mM. Wild-type protein was also incubated with hexanol concentrations of 0, 300, 500, and 800 µM. Samples were equilibrated with stirring for 30 minutes at 25 °C. A solution containing1-anilino-8-naphthalenesulfonic acid (ANS) (Molecular Probes) at a concentration of 1mM, in the same buffer used to make the protein solutions, was titrated into the equilibrated sample to a final concentration of 147 µM using a Gilson F-3005/6 titrator that was controlled by the instrument software. The concentration of stock solutions of ANS was calculated using an extinction coefficient of 7800 cm−1 M−1 at 372 nm (Molecular Probes Handbook).
Fluorescence experiments were performed at 25 °C on a Jobin-Yvon Fluorolog3 spectrometer with an excitation wavelength of 370 nm, and the emission detected over the range of 390–600 nm at 1 nm intervals with a 0.5 s integration time and a slit width of 2 nm for both the excitation and emission monochromators. Data from two scans were averaged for each experiment, and three independent experiments were performed for each complex. Data were corrected by subtraction of the spectrum of buffer containing the appropriate alcohol, correction for the dilution factor during titration, subtraction of ANS in buffer, and a correction for the inner filter effect of the ANS using an extinction coefficient of 7800 cm−1 M−1 for the excitation and 0 cm−1M−1 for the emission wavelengths. The correction for the inner filter effect was verified by titration of ANS into buffer alone. After this correction, a plot of ANS fluorescence as a function of concentration was linear.
The fluorescence emission at 457 nM as a function of ANS concentration was fit to equation 1 to account for ligand depletion of the ANS 75; 76 in GraphPad Prism version 4.00, GraphPad Software, San Diego California USA.
| (Eqn 1) |
| (Eqn 2) |
In this equation, F is the measured fluorescence, Fmax is maximal fluorescence at the end of each experiment, [P] is the protein concentration, [ANS] is the total concentration of ANS, [I] is the concentration of alcohol, KD is the dissociation constant of ANS, and KI is the inhibition constant of the alcohol, which in this case is equivalent to the dissociation constant for the alcohol. The KD for ANS was determined separately for each individually substituted protein and was fixed in the subsequent analysis. The alcohol concentration, and the protein concentration were fixed. All data collected for a single protein with one alcohol were fit globally over the range of alcohol concentrations assuming the value of KI is the same for each alcohol concentration. The value of Fmax was allowed to vary for each experiment to allow for experimental differences in alcohol, protein and ANS concentrations.
Circular Dichroism Studies
Samples of wild-type and substituted protein were prepared and purified as above and diluted to 100 µM stock solutions. These concentrations were confirmed by amino acid analysis and then diluted to a final concentration of 20 µM. CD experiments were performed on a Jasco-810 spectropolarimeter in the UCHSC biophysics core facility. Thermal denaturation was performed over the range of 5°C to 95°C while the ellipticity was monitored at 222 nm. Each experiment was repeated three times. In order to determine the melting temperature (TM), the derivative of the temperature in Kelvin versus molar ellipticity was calculated and fit in Graphpad Prism to obtain values of and TM□and Van't Hoff enthalpies using the method of John and Weeks 45.
Table 1.
X-ray data collection statistics for LUSH-alcohol complexes
| WT apo | S52A ethanol | S52A butanol | T57S ethanol | T57A ethanol | T57A butanol | |
|---|---|---|---|---|---|---|
| Resolution (Å) | 15.0−1.9 | 30.0−1.9 | 30.0−2.0 | 33.0−2.0 | 40.0−2.0 | 40.0−2.0 |
| Completenessa (%) | 96.7 (99.2) | 98.9 (99.8) | 98.0(99.6) | 99.9 (99.9) | 99.6 (99.9) | 100 (99.9) |
| Rmerge b(%) | 0.06 (0.28) | 0.07 (0.46) | 0.05 (0.21) | 0.07 (0.39) | 0.07 (0.34) | 0.12 (0.44) |
| I/σ | 28.6 (4.6) | 8.4 (2.3) | 8.8 (2.7) | 10.0 (2.8) | 15.0 (5.0) | 8.0 (3.1) |
| Total Reflections | 89,172 | 86668 | 111721 | 58680 | 115397 | 126477 |
| Unique Reflections | 19,577 | 25896 | 15963 | 16219 | 16772 | 18362 |
| Space Group | P43 | P43 | P43 | P43 | C2221 | P212121 |
| Cell Dimensions | ||||||
| a (Å) | 46.94 | 46.81 | 46.87 | 46.82 | 65.12 | 42.66 |
| b (Å) | 46.94 | 46.81 | 46.87 | 46.82 | 67.28 | 45.64 |
| c (Å) | 111.36 | 111.20 | 111.32 | 111.24 | 110.53 | 114.50 |
Numbers in parenthesis represent values for the highest resolution shell.
Rmerge = Σ | I − <I> | / ΣI, where I is the intensity of a given reflection.
Table 2.
X-ray Structure Refinement Statistics
| WT apo | S52A ethanol | S52A butanol | T57S ethanol | T57A ethanol | T57A butanol | |
|---|---|---|---|---|---|---|
| Resolution (Å) | 15.0 −1.9 | 30.0−1.9 | 30.0−2.0 | 33.0−2.0 | 40.0−2.0 | 40.0−2.0 |
| Rfree (%) | 25.5 | 23.0 | 22.4 | 25.8 | 23.5 | 22.9 |
| Rcryst (%) | 18.7 | 19.6 | 16.3 | 21.2 | 19.3 | 18.6 |
| Number of Atoms | 2236 | 2,289 | 2,353 | 2,226 | 2,201 | 2,212 |
| Protein | 2021 | 2,097 | 2,067 | 2,093 | 2,018 | 2,053 |
| Ligand | - | 6 | 10 | 6 | - | - |
| Water | 207 | 179 | 278 | 113 | 179 | 222 |
| PEG | - | - | - | 21 | 28 | 25 |
| Acetate | 8 | 20 | 20 | 16 | 4 | 12 |
| Avg. B Factor (Å2) | 33.8 | 29.4 | 21.7 | 39.0 | 28.7 | 20.1 |
| Protein | 33.6 | 28.5 | 20.3 | 41.7 | 28.7 | 19.6 |
| Ligand | - | 42.7(EtOH) | 36.6(BuOH) | 53.8(EtOH) | 53.8(PEG) | 39.2(PEG) |
| RMSD | ||||||
| Bond Angle (°) | 1.56 | 1.09 | 1.05 | 1.38 | 1.13 | 1.17 |
| Bond Length (Å) | 0.02 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 |
| Ramachandran | ||||||
| Most Favored (%) | 95 | 98.5 | 95 | 95 | 95 | 95 |
| Allowed (%) | 100 | 100 | 100 | 100 | 100 | 100 |
| Coordinate Error | ||||||
| Luzzati | 0.21 | .21 | .19 | .25 | .23 | .22 |
| DPI | 0.20 | .16 | .17 | .19 | .19 | .17 |
Acknowledgments
We thank Dr Robert Hodges and Dr Brian Tripet for amino acid analysis, Dr. Stan Kwok for assistance with circular dichroism studies, and Dr Mair Churchill for assistance with X-ray crystallography. The X-ray crystallography facilities at UCDSOM were purchased with assistance from the Howard Hughes Medical Institute and their operation is supported by the Program in Biomolecular Structure and the University of Colorado Cancer Center. This work was funded by a grant from NIH NIAAA R01 AA013618 to DNMJ.
Coordinates: The coordinates for wild-type apo LUSH and all the substituted LUSH-alcohol complexes have been deposited in the Protein Data Bank with the following accession numbers: 1T14, 3B7A, 3B6X, 3B88, 3B87, 3B86.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tabakoff B, Hoffman PL. Alcohol addiction: an enigma among us. Neuron. 1996;16:909–912. doi: 10.1016/s0896-6273(00)80113-0. [DOI] [PubMed] [Google Scholar]
- 2.Diamond I, Gordon AS. Cellular and molecular neuroscience of alcoholism. Physiol. Rev. 1997;77:1–20. doi: 10.1152/physrev.1997.77.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Lovinger DM. Alcohols and neurotransmitter gated ion channels: past, present and future. Naunyn. Schmiedebergs Arch. Pharmacol. 1997;356:267–282. doi: 10.1007/pl00005051. [DOI] [PubMed] [Google Scholar]
- 4.Faingold CL, N'Gouemo P, Riaz A. Ethanol and neurotransmitter interactions--from molecular to integrative effects. Prog. Neurobiol. 1998;55:509–535. doi: 10.1016/s0301-0082(98)00027-6. [DOI] [PubMed] [Google Scholar]
- 5.Harris RA. Ethanol actions on multiple ion channels: which are important? Alcohol. Clin. Exp. Res. 1999;23:1563–1570. [PubMed] [Google Scholar]
- 6.Kou J, Yoshimura M. Isoform-specific enhancement of adenylyl cyclase activity by n-alkanols. Alcohol Clin. Exp. Res. 2007;31:1467–1472. doi: 10.1111/j.1530-0277.2007.00455.x. [DOI] [PubMed] [Google Scholar]
- 7.Stubbs CD, Slater SJ. Ethanol and protein kinase C. Alcohol Clin. Exp. Res. 1999;23:1552–1560. [PubMed] [Google Scholar]
- 8.Kruse SW, Zhao R, Smith DP, Jones DN. Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster. Nat. Struct. Biol. 2003;10:694–700. doi: 10.1038/nsb960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace MJ, Newton PM, Oyasu M, McMahon T, Chou WH, Connolly J, Messing RO. Acute functional tolerance to ethanol mediated by protein kinase C epsilon. Neuropsychopharmacol. 2007;32:127–136. doi: 10.1038/sj.npp.1301059. [DOI] [PubMed] [Google Scholar]
- 10.Newton PM, Messing RO. Intracellular signaling pathways that regulate behavioral responses to ethanol. Pharmacol. Ther. 2006;109:227–237. doi: 10.1016/j.pharmthera.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Ikonomidou C, Bittigau P, Ishimaru MJ, Wozniak DF, Koch C, Genz K, Price MT, Stefovska V, Horster F, Tenkova T, Dikranian K, Olney JW. Ethanol-induced apoptotic neurodegeneration and fetal alcohol syndrome. Science. 2000;287:1056–1060. doi: 10.1126/science.287.5455.1056. [DOI] [PubMed] [Google Scholar]
- 12.McCreery MJ, Hunt WA. Physico-chemical correlates of alcohol intoxication. Neuropharmacology. 1978;17:451–461. doi: 10.1016/0028-3908(78)90050-3. [DOI] [PubMed] [Google Scholar]
- 13.Lyon RC, McComb JA, Schreurs J, Goldstein DB. A relationship between alcohol intoxication and the disordering of brain membranes by a series of short-chain alcohols. J. Pharmacol. Exp. Ther. 1981;218:669–675. [PubMed] [Google Scholar]
- 14.Alifimoff JK, Firestone LL, Miller KW. Anaesthetic potencies of primary alkanols: implications for the molecular dimensions of the anaesthetic site. Br. J. Pharmacol. 1989;96:9–16. doi: 10.1111/j.1476-5381.1989.tb11777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wick MJ, Mihic SJ, Ueno S, Mascia MP, Trudell JR, Brozowski SJ, Ye Q, Harrison NL, Harris RA. Mutations of gamma-aminobutyric acid and glycine receptors change alcohol cutoff: evidence for an alcohol receptor? Proc. Natl. Acad. Sc.i USA. 1998;95:6504–6509. doi: 10.1073/pnas.95.11.6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peoples RW, Stewart RR. Alcohols inhibit N-methyl-D-aspartate receptors via a site exposed to the extracellular environment. Neuropharmacology. 2000;39:1681–1691. doi: 10.1016/s0028-3908(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 17.Mihic SJ, Ye Q, Wick MJ, Koltchine VV, Krasowski MD, Finn SE, Mascia MP, Valenzuela CF, Hanson KK, Greenblatt EP, Harris RA, Harrison NL. Sites of alcohol and volatile anaesthetic action on GABA(A) and glycine receptors. Nature. 1997;389:385–389. doi: 10.1038/38738. [DOI] [PubMed] [Google Scholar]
- 18.Mascia MP, Trudell JR, Harris RA. Specific binding sites for alcohols and anesthetics on ligand-gated ion channels. Proc. Natl. Acad. Sci. USA. 2000;97:9305–9310. doi: 10.1073/pnas.160128797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trudell JR, Bertaccini E. Comparative modeling of a GABAA alpha1 receptor using three crystal structures as templates. J. Mol. Graph. Model. 2004;23:39–49. doi: 10.1016/j.jmgm.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Bertaccini EJ, Shapiro J, Brutlag DL, Trudell JR. Homology modeling of a human glycine alpha 1 receptor reveals a plausible anesthetic binding site. J. Chem. Inf. Model. 2005;45:128–135. doi: 10.1021/ci0497399. [DOI] [PubMed] [Google Scholar]
- 21.Jung S, Harris RA. Sites in TM2 and 3 are critical for alcohol-induced conformational changes in GABA receptors. J. Neurochem. 2006;96:885–892. doi: 10.1111/j.1471-4159.2005.03617.x. [DOI] [PubMed] [Google Scholar]
- 22.Crawford DK, Trudell JR, Bertaccini EJ, Li K, Davies DL, Alkana RL. Evidence that ethanol acts on a target in Loop 2 of the extracellular domain of alpha1 glycine receptors. J. Neurochem. 2007;102:2097–2109. doi: 10.1111/j.1471-4159.2007.04680.x. [DOI] [PubMed] [Google Scholar]
- 23.Covarrubias M, Vyas TB, Escobar L, Wei A. Alcohols inhibit a cloned potassium channel at a discrete saturable site. Insights into the molecular basis of general anesthesia. J. Biol. Chem. 1995;270:19408–19416. doi: 10.1074/jbc.270.33.19408. [DOI] [PubMed] [Google Scholar]
- 24.Shahidullah M, Harris T, Germann MW, Covarrubias M. Molecular features of an alcohol binding site in a neuronal potassium channel. Biochemistry. 2003;42:11243–11252. doi: 10.1021/bi034738f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattacharji A, Kaplan B, Harris T, Qu X, Germann MW, Covarrubias M. The concerted contribution of the S4–S5 linker and the S6 segment to the modulation of a Kv channel by 1-alkanols. Mol. Pharmacol. 2006;70:1542–1554. doi: 10.1124/mol.106.026187. [DOI] [PubMed] [Google Scholar]
- 26.Slater SJ, Ho C, Kelly MB, Larkin JD, Taddeo FJ, Yeager MD, Stubbs CD. Protein kinase Calpha contains two activator binding sites that bind phorbol esters and diacylglycerols with opposite affinities. J. Biol. Chem. 1996;271:4627–4631. doi: 10.1074/jbc.271.9.4627. [DOI] [PubMed] [Google Scholar]
- 27.Slater SJ, Kelly MB, Larkin JD, Ho C, Mazurek A, Taddeo FJ, Yeager MD, Stubbs CD. Interaction of alcohols and anesthetics with protein kinase Calpha. J. Biol. Chem. 1997;272:6167–6173. doi: 10.1074/jbc.272.10.6167. [DOI] [PubMed] [Google Scholar]
- 28.Shen YM, Chertihin OI, Biltonen RL, Sando JJ. Lipid-dependent activation of protein kinase C-alpha by normal alcohols. J. Biol. Chem. 1999;274:34036–34044. doi: 10.1074/jbc.274.48.34036. [DOI] [PubMed] [Google Scholar]
- 29.Slater SJ, Cook AC, Seiz JL, Malinowski SA, Stagliano BA, Stubbs CD. Effects of ethanol on protein kinase C alpha activity induced by association with Rho GTPases. Biochemistry. 2003;42:12105–12114. doi: 10.1021/bi034860e. [DOI] [PubMed] [Google Scholar]
- 30.Slater SJ, Malinowski SA, Stubbs CD. The nature of the hydrophobic n-alkanol binding site within the C1 domains of protein kinase Calpha. Biochemistry. 2004;43:7601–7609. doi: 10.1021/bi049755z. [DOI] [PubMed] [Google Scholar]
- 31.Das J, Addona GH, Sandberg WS, Husain SS, Stehle T, Miller KW. Identification of a general anesthetic binding site in the diacylglycerol-binding domain of protein kinase Cdelta. J. Biol. Chem. 2004;279:37964–37972. doi: 10.1074/jbc.M405137200. [DOI] [PubMed] [Google Scholar]
- 32.Das J, Zhou X, Miller KW. Identification of an alcohol binding site in the first cysteine-rich domain of protein kinase Cdelta. Protein Sci. 2006;15:2107–2119. doi: 10.1110/ps.062237606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim MS, Repp A, Smith DP. LUSH odorant-binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster. Genetics. 1998;150:711–721. doi: 10.1093/genetics/150.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu P, Atkinson R, Jones DNM, Smith DP. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron. 2005;45:193–200. doi: 10.1016/j.neuron.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 35.Bucci BK, Kruse SW, Thode AB, Alvarado SM, Jones DNM. Effect of n-alcohols on the structure and stability of the Drosophila odorant binding protein LUSH. Biochemistry. 2006;45:1693–1701. doi: 10.1021/bi0516576. [DOI] [PubMed] [Google Scholar]
- 36.Jenkins A, Greenblatt EP, Faulkner HJ, Bertaccini E, Light A, Lin A, Andreasen A, Viner A, Trudell JR, Harrison NL. Evidence for a common binding cavity for three general anesthetics within the GABAA receptor. J. Neurosci. 2001;21:RC136. doi: 10.1523/JNEUROSCI.21-06-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamakura T, Bertaccini E, Trudell JR, Harris RA. Anesthetics and ion channels: molecular models and sites of action. Annu. Rev. Pharmacol. Toxicol. 2001;41:23–51. doi: 10.1146/annurev.pharmtox.41.1.23. [DOI] [PubMed] [Google Scholar]
- 38.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 39.Ye S, Strzalka J, Churbanova IY, Zheng S, Johansson JS, Blasie JK. A model membrane protein for binding volatile anesthetics. Biophys. J. 2004;87:4065–4074. doi: 10.1529/biophysj.104.051045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lobo IA, Mascia MP, Trudell JR, Harris RA. Channel gating of the glycine receptor changes accessibility to residues implicated in receptor potentiation by alcohols and anesthetics. J. Biol. Chem. 2004;279:33919–33927. doi: 10.1074/jbc.M313941200. [DOI] [PubMed] [Google Scholar]
- 41.Wogulis M, Morgan T, Ishida Y, Leal WS, Wilson DK. The crystal structure of an odorant binding protein from Anopheles gambiae: evidence for a common ligand release mechanism. Biochem. Biophys. Res. Commun. 2006;339:157–164. doi: 10.1016/j.bbrc.2005.10.191. [DOI] [PubMed] [Google Scholar]
- 42.Semisotnov GV, Rodionova NA, Razgulyaev OI, Uversky VN, Gripas AF, Gilmanshin RI. Study of the "molten globule" intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers. 1991;31:119–128. doi: 10.1002/bip.360310111. [DOI] [PubMed] [Google Scholar]
- 43.Tcherkasskaya O, Uversky VN. Denatured collapsed states in protein folding: example of apomyoglobin. Proteins. 2001;44:244–254. doi: 10.1002/prot.1089. [DOI] [PubMed] [Google Scholar]
- 44.Lartigue A, Gruez A, Spinelli S, Riviere S, Brossut R, Tegoni M, Cambillau C. The crystal structure of a cockroach pheromone-binding protein suggests a new ligand binding and release mechanism. J. Biol. Chem. 2003;278:30213–30218. doi: 10.1074/jbc.M304688200. [DOI] [PubMed] [Google Scholar]
- 45.John DM, Weeks KM. van't Hoff enthalpies without baselines. Protein Sci. 2000;9:1416–1419. doi: 10.1110/ps.9.7.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou JJ, Zhang GA, Huang W, Birkett MA, Field LM, Pickett JA, Pelosi P. Revisiting the odorant-binding protein LUSH of Drosophila melanogaster: evidence for odour recognition and discrimination. FEBS Lett. 2004;558:23–26. doi: 10.1016/S0014-5793(03)01521-7. [DOI] [PubMed] [Google Scholar]
- 47.Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 48.Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 49.Xu P, Atkinson R, Jones DNM, Smith DP. Drosophila OBP LUSH is Required for Activity of Pheromone-Sensitive Neurons. Neuron. 2005;45:193–200. doi: 10.1016/j.neuron.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 50.Laurence CBM. Observations on the strength of hydrogen bonding. Perspect. Drug Discov. Design. 2000;18:39–60. [Google Scholar]
- 51.Dunitz J. The entropic cost of bound water in crystal structures and biomolecules. Science. 1994;264:670. doi: 10.1126/science.264.5159.670. [DOI] [PubMed] [Google Scholar]
- 52.Ringe D. What makes a binding site a binding site? Curr. Opin. Struct. Biol. 1995;5:825–829. doi: 10.1016/0959-440x(95)80017-4. [DOI] [PubMed] [Google Scholar]
- 53.Trudell JR, Harris RA. Are sobriety and consciousness determined by water in protein cavities? Alcohol Clin. Exp. Res. 2004;28:1–3. doi: 10.1097/01.ALC.0000108648.32241.BD. [DOI] [PubMed] [Google Scholar]
- 54.Wilkinson KD, Mayer AN. Alcohol-induced conformational changes of ubiquitin. Arch. Biochem. Biophys. 1986;250:390–399. doi: 10.1016/0003-9861(86)90741-1. [DOI] [PubMed] [Google Scholar]
- 55.Dufour E, Bertrand-Harb C, Haertle T. Reversible effects of medium dielectric constant on structural transformation of beta-lactoglobulin and its retinol binding. Biopolymers. 1993;33:589–598. doi: 10.1002/bip.360330408. [DOI] [PubMed] [Google Scholar]
- 56.Bychkova VE, Dujsekina AE, Klenin SI, Tiktopulo EI, Uversky VN, Ptitsyn OB. Molten globule-like state of cytochrome c under conditions simulating those near the membrane surface. Biochemistry. 1996;35:6058–6063. doi: 10.1021/bi9522460. [DOI] [PubMed] [Google Scholar]
- 57.Perham M, Liao J, Wittung-Stafshede P. Differential effects of alcohols on conformational switchovers in alpha-helical and beta-sheet protein models. Biochemistry. 2006;45:7740–7749. doi: 10.1021/bi060464v. [DOI] [PubMed] [Google Scholar]
- 58.Huang N, Kalyanaraman C, Bernacki K, Jacobson MP. Molecular mechanics methods for predicting protein-ligand binding. Phys. Chem. Chem. Phys. 2006;8:5166–5177. [Google Scholar]
- 59.Applequist JEA. An Atom Dipole Interaction Model for molecular Polarizability. Application to Polyatomic Molecules and Determination of Atom Polarizabilities. J. . Amer. Chem. Soc. 1971;94:2952–2960. [Google Scholar]
- 60.Wang S, Cann NM. Polarizable and flexible model for ethanol. J. Chem. Phys. 2007;126:214502. doi: 10.1063/1.2730837. [DOI] [PubMed] [Google Scholar]
- 61.Eckenhoff RG, Johansson JS. Molecular interactions between inhaled anesthetics and proteins. Pharmacol. Rev. 1997;49:343–367. [PubMed] [Google Scholar]
- 62.Trudell JR. Contributions of dipole moments, quadrupole moments, and molecular polarizabilities to the anesthetic potency of fluorobenzenes. Biophys. Chem. 1998;73:7–11. doi: 10.1016/s0301-4622(98)00161-6. [DOI] [PubMed] [Google Scholar]
- 63.Trudell JR, Koblin DD, Eger EI., 2nd A molecular description of how noble gases and nitrogen bind to a model site of anesthetic action. Anesth. Analg. 1998;87:411–418. doi: 10.1097/00000539-199808000-00034. [DOI] [PubMed] [Google Scholar]
- 64.Bertaccini EJ, Trudell JR, Franks NP. The common chemical motifs within anesthetic binding sites. Anesth. Analg. 2007;104:318–324. doi: 10.1213/01.ane.0000253029.67331.8d. [DOI] [PubMed] [Google Scholar]
- 65.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta. Crystallog.r D Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 66.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta. Crystallogr. D Biol. Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 67.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta. Crystallogr. D Biol. Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 68.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta. Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 69.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta. Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 70.Maiti R, Van Domselaar GH, Zhang H, Wishart DS. SuperPose: a simple server for sophisticated structural superposition. Nucl. Acids Res. 2004;32:W590–W594. doi: 10.1093/nar/gkh477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nelson M, Humphrey W, Gursoy A, Dalke A, Kale LV, Skeel RD, Schulten K. NAMD- A parallel, object-oriented molecular dynamics program. Int. J. of Supercomp. Appl. High Perf. Comput. 1996;10:251–268. [Google Scholar]
- 72.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Chem. Phys. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 73.Brunger AT. A system for X-ray crystallography and NMR. X-PLOR version 3.1. New Haven, CT: Yale University Press; 1992. [Google Scholar]
- 74.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 75.Swillens S. Interpretation of binding burves obtained with high receptor roncentrations: A practical aid for computer analysis. Mol. Pharmacol. 1995;47:1197–1203. [PubMed] [Google Scholar]
- 76.Stein RA, Wilkinson JC, Guyer CA, Staros JV. An analytical approach to the measurement of equilibrium binding constants: application to EGF binding to EGF receptors in intact cells measured by flow cytometry. Biochemistry. 2001;40:6142–6154. doi: 10.1021/bi002817a. [DOI] [PubMed] [Google Scholar]


