Abstract
The significance of active vs. inactive flavin-containing monooxygenase 2 (FMO2) for human drug and xenobiotic metabolism and sensitivity is unknown, but the underlying ethnic polymorphism is well documented. We used quantitative real-time PCR to measure message levels of Fmo1, Fmo2, Fmo3 and Fmo5 in lung and liver from eight strains of eight week old female mice to determine if a strain could be identified that predominately expressed Fmo2 in lung, recapitulating the human FMO expression profile and being the ideal strain for Fmo2 knockout studies. We also characterized enzyme activity of baculovirus expressed mouse Fmo1, Fmo2 and Fmo3 to identify a substrate or incubation conditions capable of discriminating Fmo2 from Fmo mixtures. Fmo transcript expression patterns were similar for all strains. In lung, 59% of total FMO message was Fmo2, but Fmo1 levels were also high, averaging 34%, whereas Fmo3 and Fmo5 levels were 2 and 5%, respectively. In liver, Fmo1, Fmo2, Fmo3 and Fmo5 contributed 16, 1, 7 and 76% respectively, of detected message. Peak activity varied by isoform and was pH- and substrate-dependent. Fmo3 oxidation of methyl p-tolyl sulfide was negligible at pH 9.5, but Fmo3 oxidation of methimazole was comparable to Fmo1 and Fmo2. Heating microsomes at 50 °C for 10 min eliminated most Fmo1 and Fmo3 activity, while 94% of Fmo2 activity remained. Measurement of activity in heated and unheated lung and liver microsomes verified relative transcript abundance. Our results show that dual Fmo1/2 knockouts will be required to model the human lung FMO profile.
Keywords: quantitative real-time PCR, in vitro expression, flavin-containing monooxygenase, mouse, pulmonary expression, hepatic expression
1. Introduction
Mammalian flavin-containing monooxygenases (FMO, EC 1.14.13.8) are microsomal phase I enzymes that incorporate an atom of molecular oxygen into a wide range of nitrogen- and sulfur-containing drugs and xenobiotics (reviewed in Krueger and Williams 2005 [1]). Although oxygenation usually represents a detoxication reaction, some FMO-oxygenated sulfur substrates, such as thioureas, produce reactive sulfenic or sulfinic acids as major metabolites. Sulfenic acid metabolites can react with GSH inducing oxidative stress through a futile redox cycle [2-5].
Humans have five FMO genes (FMO1-5) and six pseudogenes (FMO6P-11P) [6-8], while mice have nine FMO genes (Fmo1-6, 9, 12 and 13) [7]. FMO1, FMO2 and FMO3 are the major mammalian drug and xenobiotic metabolizing isoforms from all species studied to date and are expressed in a tissue-, species-, sex-, and developmental-specific manner. In human and most other species including rabbit, monkey and guinea pig, FMO2 is expressed at high levels in lung and is the predominant drug metabolizing isoform in this organ [9-14]. FMO2 is co-expressed with other isoforms in kidney, intestine and nasal mucosa but is not the predominant FMO in these tissues.
A genetic polymorphism (g.23,238C>T; dbSNP#rs6661174) in human FMO2 converts a glutamine to a stop codon, p.Q472X [15]. Protein encoded by this allele (FMO2*2) does not bind FAD and is inactive [15,16]. This polymorphism is present in all Caucasians and Asians genotyped to date [15,17]. The FMO2*1 allele encoding full-length active protein is estimated to occur in 13-26% of individuals of African descent [17,18] and 2-7% of Hispanic origin [19,20]. Other polymorphisms of FMO2 have been documented [15,17,18], but are expected to be of minor impact as most are found exclusively or primarily as mutations secondary to the FMO2*2 allele [15,17,21].
Expressed human FMO2.1 efficiently catalyzes the oxidation of thioethers [22] and thioureas [5]. Ethnically- and racially-dependent differences may exist for metabolism upon exposure to these or other classes of compounds in the form of drugs (e.g. thioureas) and insecticides (e.g. thioethers), thus we are working to develop an animal model to test the relevance of these alterations. The FMO2 allele from laboratory rat strains (Rattus norvegicus) encodes truncated, inactive protein [23] but in a wild population from this species there is segregation for the wildtype and mutant FMO2 alleles [24]. Wild rats are not ideal to work with given that they were found to produce enzymatically active FMO1 in addition to FMO2 (in rats with at least one wildtype allele) [24], and any animals entering animal housing would need to undergo extensive cleanup to free them of infectious organisms that could threaten the health of other laboratory rodents.
The laboratory mouse (Mus musculus) represents a potentially viable alternative model. Because all tested mice have alleles encoding intact, active Fmo2, a knockout would be required to recapitulate the common human allelic variant. Studies from mice have conflicting results regarding the presence and abundance of Fmo2 in mouse lung. A 1992 study [25] demonstrated the presence of Fmo1 and Fmo2 transcript and protein in lung. More recently, a study of FMO gene expression in mice identified Fmo1 not Fmo2 as the major lung isoform [26]. We conducted this study to determine whether we could identify a mouse strain that primarily produced Fmo2 in the lung, as Fmo2 substrates would likely also be substrates for Fmo1, and would thus confound studies of Fmo2. In addition, we performed enzyme assays with methimazole (MMI) and methyl p-toly sulfide (MTS) as substrate for expressed Fmo1, Fmo2 and Fmo3 in search of conditions that could distinguish contributions by individual FMO isoforms.
2. Materials and Methods
2.1 Experimental animals
Four eight- week old, female mice of six different inbred strains and one hybrid strain were purchased from The Jackson Laboratory (Bar Harbor, ME). One additional outbred strain was purchased from Harlan (Indianapolis, IN) (Table 1). Tissues for RNA extraction were recovered from mice upon their arrival at the Laboratory Animal Resource Center at Oregon State University. All procedures were conducted according to National Institutes of Health guidelines and were approved by the Oregon State University Animal Care and Use Committee.
Table 1.
Mouse strains and sources.
| Mouse Strain | Abbreviation | Strain Type | Source |
|---|---|---|---|
| A/J | A/J | Inbred | Jackson Lab |
| C57BL/6J | C57 | Inbred | Jackson Lab |
| DBA/2J | DBA | Inbred | Jackson Lab |
| FVB/NJ | FVB | Inbred | Jackson Lab |
| SWR/J | SWR | Inbred | Jackson Lab |
| 129S1/SvImJ | 129 | Inbred | Jackson Lab |
| Hsd:ICR (CD-1) | ICR | Outbred | Harlan Lab |
| B6129SF1/Ja | F1 Hyb | F1 Hybrid | Jackson Lab |
C57BL6J female × 129S1/SvImJ male.
Frozen lungs and livers from 8 to 10 week old female Swiss Webster mice were purchased (Pel-Freez, Rogers, AR) for the preparation of microsomes. Lungs and livers were not necessarily from the same mice. Upon receipt, tissue was stored at -80 °C until microsomes were prepared by differential centrifugation [27] from two pools each of 15 lung pairs and five livers. Protein concentrations were subsequently determined [28].
2.2 Tissue harvest and RNA extraction
Lungs and livers were harvested from mice euthanized with an overdose of CO2. Tissues were immediately placed in Trizol® reagent (Invitrogen, Carlsbad, CA), snap frozen in liquid nitrogen and stored at -80 °C until total RNA was extracted per manufacturer’s recommendation. RNA was further purified with RNeasy (Qiagen, Valencia, CA). RNA quality was measured on an Agilent 2100 Bioanalyzer (Santa Clara, CA) and quantitated with a NanoDrop ND-1000 UV-Vis spectrophotometer (Wilmington, DE).
2.3 DNA synthesis and real-time PCR
For each sample 2.0 μg RNA was treated with amplification grade DNase I (Invitrogen) and then reverse transcribed following Invitrogen’s recommendations using Superscript III and random hexamers (Invitrogen), as not all FMOs have an apparent poly-A tail. dNTPs were from Fermentas Inc. (Hanover, MD). Reactions were run on a PTC-100 ThermalCycler (MJ Research Inc., Waltham, MA), 25 °C for 5 min, 42 °C for 1 h, and 70 °C 15 min. cDNA reaction volumes were brought to 50 μl with nuclease free water and stored at 4 °C until quantitative PCR was completed for all of the genes of interest.
Gene-specific primer pairs (Table 2) were synthesized (Invitrogen) and used to produce DNA amplicons for real-time PCR calibration curves, and for amplification reactions with test samples. In addition to mouse Fmo1-3, and Fmo5, four housekeeping genes including cytoplasmic β-actin (Actb), hypoxanthine guanine phosphoribosyl transferase 1 (Hprt1), large ribosomal protein 13a (Rpl13a), and TATA box binding protein (Tbp) were quantitated from lungs and livers from all mice.
Table 2.
Primers used for quantitative PCR of mouse FMO and housekeeping genes.
| Gene | Primer Sequence (5’ to 3’) | Exon | Amplicon Size (bp) | Annealing Temperature | ENSEMBLE Transcript1 | Reference |
|---|---|---|---|---|---|---|
| Fmo1 | 5’-TGTCTCTGGACAGTGGGAAGT | 4 | 217 | 56 | ENSMUST00000046049 | [26] |
| 5’-CATTCCAACTACAAGGACTCG | 5 | |||||
| Fmo2 | 5’-AGGCTCCATCTTCCCAACCGTA | 7 | 382 | 56 | ENSMUST00000045902 | |
| 5’-CCGGGTCTTTAAGGGTTTCAGG | 9 | |||||
| Fmo3 | 5’-CACCACTGAAAAGCACGGTA | 4 | 445 | 58 | ENSMUST00000028010 | [26] |
| 5’-GTTTAAAGGCACCAAACCATAG | 6 | |||||
| Fmo5 | 5’-ATCACACGGATGCTCACCTG | 4 | 235 | 58 | ENSMUST00000029729 | [26] |
| 5’-GCTTGCCTACACGGTTCAAG | 6 | |||||
| Hprt1 | 5’-AGCTACTGTAATGATCAGTCAAC | 3/4 | 180 | 53 | ENSMUST00000026723 | RT Primer DB ID:502 |
| 5’-AGAGGTCCTTTTCACCAGCA | 7 | |||||
| Rpl13a | 5’-GCTCTCAAGGTTGTTCGGCTGA | 6 | 157 | 53 | ENSMUST00000102669 | |
| 5’-AGATCTGCTTCTTCTTCCGATA | 7 | |||||
| Tbp | 5’-GGCCTCTCAGAAGCATCACT | 2/3 | 167 | 53 | ENSMUST00000039079 | RT Primer DB ID:492 |
| 5’-GCCAAGCCCTGAGCATAA | 3 | |||||
| Actb | 5’-CTACAGCTTCACCACCACAG | 4 | 137 | 62 | ENSMUST00000031564 | |
| 5’-GCAGCTCATAGCTCTTCTCC | 4 |
Gene-specific standards were constructed to quantitate each gene target. Lung and liver cDNA from eight mice was pooled and PCR products generated using the conditions for real-time PCR reactions described below, except that melt curve analysis was not performed. PCR products were electrophoresed on agarose gels then purified with a Zymoclean™ Gel DNA Recovery kit (Zymo Research Corp., Orange, CA), quantitated with Quant-iT™ Picogreen® (Molecular Probes, Eugene, OR) and diluted to cover a 106-fold range for calibration curves, using reactions containing 0.0001 pg to 100 pg gel-purified DNA.
Real-time PCR reactions contained 1.0 μl cDNA template (or appropriately diluted DNA standard), 0.4 μM forward primer, 0.4 μM reverse primer, 10 μl 2X DyNAmo™ SYBR® green master mix (New England Biolabs, Ipswich, MA), and water to 20 μl. PCR was performed in an MJ Research (BioRad Research, Hercules, CA) Opticon 2 thermal cycler using Opticon Monitor version 3.1 software (BioRad Research). Reactions were initially denatured at 95 °C for 10 min followed by 35 cycles of 10 s denaturing at 94 °C, 20 s annealing (see Table 2 for specific temperatures), 18 s extension at 72 °C, an extension step for 7 min at 72 °C, melt curve at 50-93 °C at 0.2 °C intervals, and a final extension for 10 min at 72 °C. In order to determine whether primer pairs amplified a single product of the correct size we used both Opticon Monitor software’s melt curve analysis and gel electrophoresis. Products were run on 6% TBE gels (Invitrogen) and visualized by ethidium bromide staining. Results from each of the four housekeeping genes were considered separately and as the geometric mean of all four, to determine the most appropriate gene for normalizing real-time reactions for Fmo1-3, and Fmo5. Normalized data were expressed as a ratio of the calculated values (copies Fmo isoform/μg total RNA : copies Hprt1/μg total RNA) for each sample.
Statistical analysis was done using InStat software (GraphPad Inc., SanDiego, CA). Bartlett’s test was used to determine whether groups exhibited similar variance. Those groups with significant difference in variance were ln transformed (Y=lnY) before analyzing. Strain differences for each FMO isoform and levels of expression within each strain were compared with one-way ANOVA followed by a Tukey’s multiple comparisons test when significant differences (p <0.05) of means occurred.
2.4 cDNA synthesis, cloning and protein expression
First strand cDNA synthesis was performed on RNA (2.0 μg) isolated from a female F1 Hyb mouse liver (Fmo1 and Fmo3) and lung (Fmo2) using SuperScript III (Invitrogen) according to Invitrogen’s recommendations. The final reaction volume was 40 μl. Fmo-specific reverse primers (Table 3) were used in the reaction at a final concentration of 0.1 μM (2 pmol/μg RNA). First strand PCR conditions were the same as described above for cDNA used in quantitative PCR. First strand synthesis product was diluted to 100 μl with water; 4 μl was subsequently used as template in 20 μl PCR reactions with 1 unit Expand High Fidelity polymerase (Roche, Indianapolis, IN) and Fmo-specific forward and reverse primers (Table 3) at 0.2 μM, each. Robocycler (Stratagene, La Jolla, CA) conditions were: 4 cycles of 45 s at 94 °C, 45 s at 56 °C, 120 s at 72 °C; 30 cycles of 30 s at 94 °C, 30 s at 56 °C, 90 s at 72 °C; the extension time was extended by 10 min in the final cycle.
Table 3.
Primers used to clone mouse Fmo1, Fmo2 and Fmo3.
| Isoform | Primer Type | Primer Sequence 5’ to 3’a |
|---|---|---|
| Fmo1 | Forward | 5’-CACC ATG GCA AAG CGA GTG GCA AT |
| Fmo1 | Reverse | 5’-TTA CAG GAA AAT CAA GAA GAC AGC T |
| Fmo2 | Forward | 5’-CACC ATG GCA AAG AAG GTT G |
| Fmo2 | Reverse | 5’-TTA AAA CCC CTG AAG TTG GA |
| Fmo3 | Forward | 5’-CACC ATG AAG AAG AAA GTG GCC ATC |
| Fmo3 | Reverse | 5’-TCA GAT CAA CAC AAG GAA CAA AGC |
Italicized nucleotides were added for vector compatibility. The sequences corresponding to start and stop codons are underlined.
Full-length cDNA products were cloned into pENTR/SD/D-TOPO (Invitrogen) and expressed in Sf9 cells. Plasmid DNA was isolated (Qiagen) and cDNA sequenced (Center for Genome Research and Biocomputing, Oregon State University, Corvallis, OR), to confirm that translated sequences matched the corresponding NCBI reference sequences NP_034361, NP_061369 and NP_032056 for Fmo1, Fmo2 and Fmo3, respectively. Vector DNA from each of the clones was recombined with BaculoDirect linear DNA (Invitrogen) to yield recombinant baculovirus DNA for transfection of Sf9 cells to generate primary virus. Recombinant protein was produced for each clone as described previously [21,22] with the addition of FAD to the media (10 μg/ml) of batches that microsomes were isolated from. Microsomes were isolated [16] from cells harvested 96 h post-infection and were resuspended in storage buffer (10 mM potassium phosphate pH 7.4, 20% glycerol, 1 mM EDTA, 100 mM phenylmethylsulfonyl fluoride). Protein concentration was determined by the Bradford method [28]. FMO content was calculated from HPLC determination of FAD content [22].
2.5 Enzyme assays
Enzyme activity was estimated for expressed mouse Fmo1, Fmo2 and Fmo3 by following MMI-dependent oxidation of TNB [29] as detailed previously [16], except that MMI was from Lancaster Synthesis (Ward Hill, MA), or by following MTS oxidation, as detailed below. The limit of detection for the MMI assay in our laboratory was 0.035 nmol TNB oxidized min-1 (corresponding to 0.1, 0.5 and 0.3 pmol of Fmo1, Fmo2 and Fmo3, respectively, at pH 8.5). Benchmarking assays were performed daily to confirm that assay run variability did not exceed 10%. Study parameters included pH profiles (pH 7.5 to 9.5) performed with MTS (200 μM) and MMI (2 mM) as substrate; sodium cholate addition (0, 1 and 1.5%) with MMI (2 mM) at pH 8.5; and activity retention following heat treatment (all proteins were diluted to 3 mg/ml in storage buffer for the 10 min pre-incubation at 50 °C) at pH 8.5 using MMI (2 mM), or pH 9.5 using MTS (200 μM). Kinetic constants for MTS oxidation (2 to 20 μM substrate) were calculated for Fmo1 and Fmo2 at pH 9.5, from two independently expressed replicates of each isoform using Lineweaver-Burk and Eadie-Hofstee plots. A two-tailed unpaired t-test (InStat software, GraphPad Inc.) was used to test for significance of kinetic estimates; data were ln transformed when variances were not equal.
Oxidation of MTS was analyzed by the method of Rettie, et al. [30] with some modifications. MTS and methyl p-tolyl sulfoxide (MTSO, racemic mixture and individual enantiomers) were from Sigma-Aldrich (St. Louis, MO). Standard microsomal incubations contained 100 μg of expressed protein in a total reaction volume of 250 μl (0.1 M Tricine pH 9.5, 1 mM EDTA, 1 mM NADPH). After 3 min preincubation on ice, reactions were initiated by the addition of MTS and transferred to a Dubnoff shaking incubator at 37 °C. Reactions were stopped after 5 min (2.5 min for kinetics) by addition of 75 μl acetonitrile on ice. After centrifugation at 10,000 × g for 30 min at 4 °C, supernatants were analyzed by HPLC with a Waters 2695 system (Waters Corp., Milford, MA) equipped with a photodiode array detector. MTSO was separated from MTS on a Waters Novapak C18 (150 × 3.9 mm) column at 40 °C, a flow rate of 0.8 ml/min and detection at 237 nm. Mobile phase was 30% acetonitrile for 4 min; to 80% acetonitrile in 2 min; returned to 30% acetonitrile at 8 min, in 2 min. The retention times of racemic MTSO and MTS were 2.7 and 8.7 min, respectively. Standard curves (r2 = 0.9965) of racemic MTSO (0.5 to 5 nmol) were run each day of analysis and MTSO values calculated by linear regression with a detection limit of 50 pmols.
For determination of the (S)-(-)- and (R)-(+)-MTSO enantiomers, incubations were stopped by placing them on ice and extracting twice with 1.0 ml ethyl acetate. The combined extracts were dried under vacuum with a Savant speed-vac (Thermo Scientific, Waltham, MA) and dissolved in 50 μl hexane:isopropyl alcohol (90:10) for analysis by chiral HPLC. The MTSO enantiomers were separated on a Phenomenex (Torrance, CA) Chirex (R)-PGYL and DNB column (250 × 4.6 mm) at 35 °C with detection at 237 nm. The eluant was 90% hexane/10% isopropyl alcohol with 0.1% trifluoroacetic acid, pumped at 1.5 ml/min. Retention times of the (S)-(-)- and (R)-(+)-MTSO enantiomers were 23.2 and 25.2 min, respectively. Separate standard curves were run with chiral separation for each enantiomer with a linear range of standard (2.5 to 20 nmol). The detection limits for (R)-(+)-MTSO (r2 = 0.9975) and (S)-(-)- MTSO (r2 = 0.9990) were 0.2 and 0.1 nmol, respectively.
Following characterization of individual expressed FMOs, the FMO activity of female mouse lung and liver microsomes was assessed under limited conditions. Activity toward MMI was performed without pre-treating microsomes and following 10 min heat treatment at 50 °C, during which the protein concentration was 3 mg/ml in storage buffer, as already described. Assays were performed in duplicate for each microsomal pool at pH 8.5 with 2 mM MMI and 200 μg protein.
3. Results
3.1 Message levels
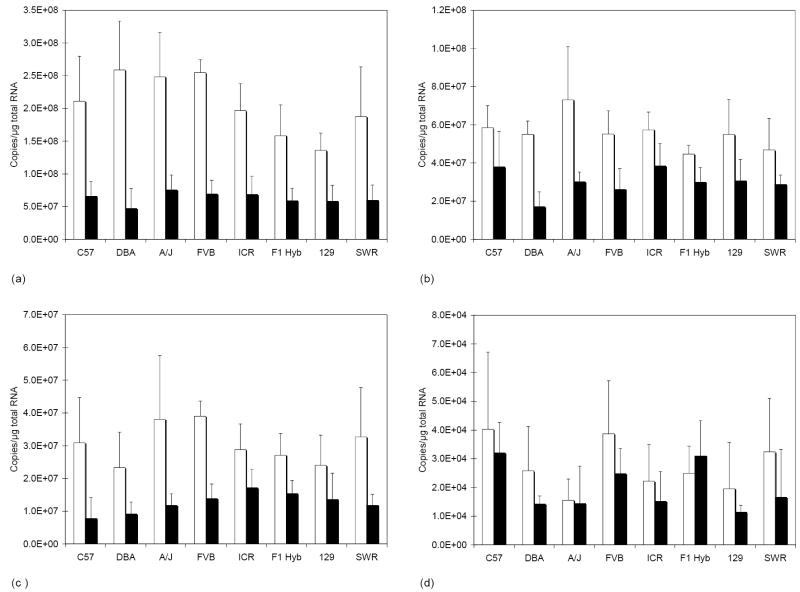
We wanted to select a housekeeping gene for normalization that was expressed at relatively constant levels from strain-to-strain and from liver to lung. We selected Hprt1, Rpl13a, and Tbp as candidates because these genes appeared to be expressed at relatively constant levels in liver and lung from humans [31]. Actb was included as it is commonly used for normalization. Expression levels were determined for all of the mouse samples. All of the housekeeping genes were expressed at higher mean levels in lung tissue than in liver (Fig. 1), although differences were not always significant. There were no statistical differences between strains with any of the four housekeeping genes. The largest mean fold difference (5.0-fold) in liver vs. lung expression was observed with Rpl13a, followed by Actb (3.8-fold), Tbp (2.8) and Hprt1 (2.2-fold). We utilized Hprt1 for normalization since the magnitude of expressed differences between liver and lung tissue was less for this gene. Hprt1 showed the least amount of variability among individual mice in both lung and liver, and on average the level of expression was closer to the message level of Fmo than either Actb or Tbp.
Fig.1.
Message level of housekeeping genes in mouse lung and liver. The mean ± S.D. strain (n = four per strain) message level in lung (white bars) and liver (black bars) is plotted for Actb (A), Hprt1 (B), Rpl13a (C) and Tbp (D).
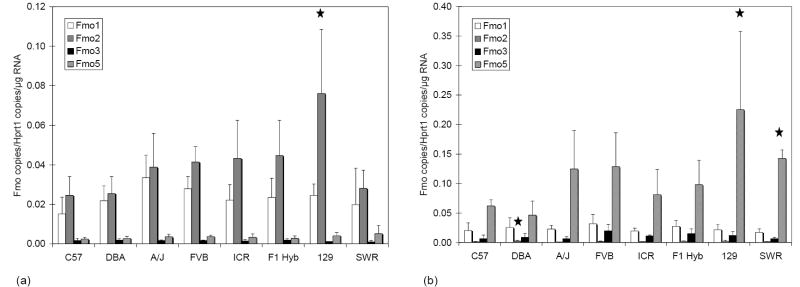
Expression levels of Fmo1, Fmo2, Fmo3, and Fmo5 mRNA were quantified in lung and liver from eight strains of mice. Levels of FMO expressed in each mouse were normalized by the amount of Hprt1 in the same mouse RNA sample (Fig. 2). In the lung (Fig. 2a), there were no significant differences in transcript levels among strains for Fmo1, Fmo3 or Fmo5. The mean Fmo2 lung transcript level among 129 mice was significantly higher than that observed among C57 (p < 0.01), DBA (p < 0.01) and SWR (p < 0.05) mice. In the liver (Fig. 2b) no significant differences in transcript levels among strains were detected for Fmo1 and Fmo3. Fmo2 transcript levels, while low in liver, were significantly higher in DBA mice than in A/J (p < 0.05) mice. Fmo5 was significantly higher in livers of 129 mice than in either C57 (p < 0.05) or DBA (P<0.01) mice, while levels detected in SWR mice were only higher than those observed in DBA (p<0.05) mice. Variance was greatest among 129 mice for lung Fmo2 and liver Fmo5 owing to a single mouse that had much lower levels of both of these isoforms than the remaining three mice; however, this variability was not observed for the other FMO isoforms. It is possible that the strain-specific statistical differences we detected are an artifact of our limited sample size. Fmo2 was the most abundant form expressed in lungs from all strains averaging 59% of total FMO message. Total FMO transcript abundance among the four isoforms in lung tissue showed Fmo2 (59%) >Fmo1 (34%) >Fmo5 (5%) >Fmo3 (2%) in the eight strains. Individual results from the eight strains from lung tissue were pooled and averages from each isoform were compared to Fmo2. The expression of Fmo1, Fmo3, and Fmo5 was 58, 4, and 8% of Fmo2 expression in lungs, respectively. Fmo5 was more abundant in liver tissue from all strains than the other three Fmo isoforms. Transcript abundance in liver tissues showed Fmo5 (76%) >Fmo3 (7%) >Fmo1 (16%) >Fmo2 (1%). When expressed as percentages of Fmo5, Fmo1, Fmo2, and Fmo3 were 21, 1, and 9% respectively of this most abundant form in liver.
Fig. 2.
FMO message levels in mouse lung and liver. Graphs show mean ± S.D. strain (n = four per strain) message levels of Fmo1, Fmo2, Fmo3 and Fmo5 plotted for lung (A), and liver (B). Stars indicate transcript levels for the indicated strain were significantly different than other strains as follows: (A) lung Fmo2 transcript levels for 129 were higher than those observed for C57 and DBA (p < 0.01), as well as SWR (p < 0.05); (B) liver Fmo2 transcript levels for DBA were higher than levels for A/J (p < 0.05); (B) liver Fmo5 transcript levels for 129 were higher than levels in C57 (p < 0.05) and DBA (p < 0.01), while levels in SWR were higher than in DBA (p < 0.05).
3.2 Enzyme activity
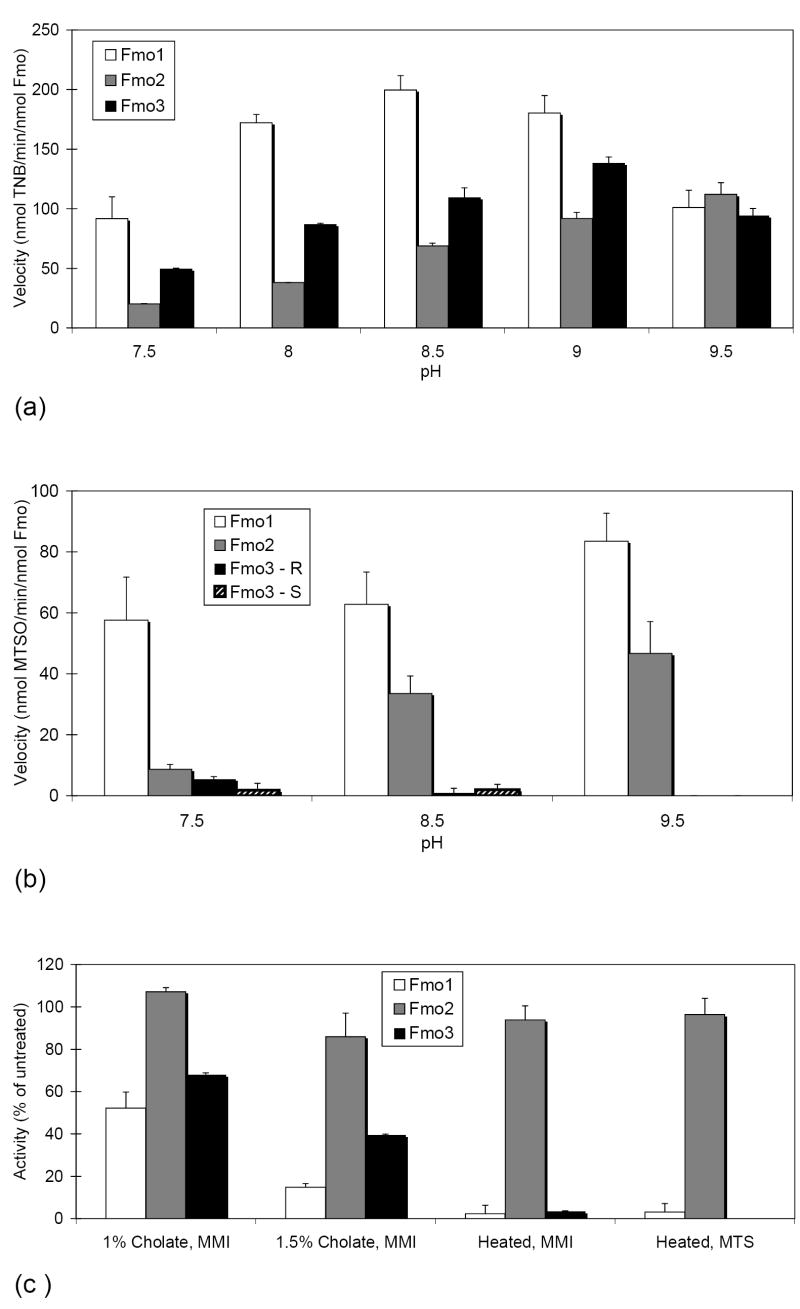
The activity of expressed Fmo1, Fmo2 and Fmo3 was assessed from pH 7.5 to 9.5 using both MMI and MTS as substrate, which are exclusively and primarily metabolized by FMOs, respectively. Fmo1, Fmo2 and Fmo3 displayed activity throughout the entire pH range when MMI was the substrate (Fig. 3a), but each isoform had a unique pattern of pH-dependent activity. Fmo1 and Fmo3 achieved higher velocities than Fmo2 at all pHs tested, except pH 9.5. Fmo2 was the only isoform that may not have achieved maximal velocity with the pH range that was tested. A different isoform-specific, pH-dependent pattern of activity was observed when MTS was the substrate (Fig. 3b). The observed velocity increased with pH for Fmo1 and Fmo2, and both of these isoforms produced only R-(+)-MTSO. Higher velocities were observed with Fmo1 than Fmo2 throughout the pH range, but the difference was the least at pH 9.5. Fmo3 achieved only low velocities when MTS was the substrate, produced mixtures of the R-(+)- and S-(-)-MTSO metabolite and generated no more than a trace amount of metabolites at pH 9.5.
Fig. 3.
Enzyme activity of mouse Fmo1, Fmo2 and Fmo3. Mean FMO activity is shown as a function of pH with 2 mM MMI (A), or 200 μM MTS (B) as substrate. Mean residual FMO activity is shown on addition of sodium cholate (pH 8.5 with 2 mM MMI) or after heating for 10 min at 50 °C (pH 8.5 with 2 mM MMI, or pH 9.5 with 200 μM MTS) (C). Error bars indicate S.D. (n = two to six assays per condition).
We selected pH 8.5 for further studies with MMI as we observed more reliability in the Fmo1 spectrophotometric output (the oxidation rate detected displayed little deviation during the observed time interval) at this pH (data not shown), and pH 9.5 for additional studies with MTS because Fmo2 was our primary isoform of interest and we could eliminate virtually all Fmo3 contributions under these conditions. Addition of 1% sodium cholate to the reaction mixture for MMI metabolism marginally increased the activity of Fmo2 to 107% of untreated, but decreased the activity of Fmo1 and Fmo3 to 52 and 68%, respectively (Fig. 3c). Increasing the detergent concentration to 1.5% decreased the activity of all of the isoforms. Fmo2 retained 86% of its activity under these conditions, while Fmo1 and Fmo3 retained 15 and 39% activity respectively. Heat-treating the microsomal samples prior to assay was a more effective means of eliminating Fmo1 and Fmo3 activity while retaining most of the Fmo2 activity (Fig. 3c). The activity retained after heating with either MMI or MTS as the substrate was very similar, 94 to 96% for Fmo2, and 2-3% retention for Fmo1.
Calculated kinetic parameters demonstrate that Fmo1 is more efficient at MTS oxidation than Fmo2 (Table 4) under our standard assay conditions. MTS was a good substrate for both isoforms but the Km for Fmo2, 10.2 μM, was more than three times higher than that observed for Fmo1, 3.2 μM. Coupling the low Km of Fmo1 with the higher kcat for this enzyme yielded a mean specificity constant estimate of 56.1 min-1 μM-1, compared with the significantly lower (P < 0.05) but very respectable specificity constant of 7.5 min-1 μM-1 for Fmo2. To our knowledge the specificity constant that we have estimated for MTS oxidation by Fmo1 is about six-fold higher than what has been published for any FMO with any substrate, and in our own laboratory have only estimated a higher specificity constant for the oxidation of tetrahydrothiophene oxidation by human FMO2.1 (unpublished results).
Table 4.
Kinetic constants for MTS metabolism at pH 9.5 by Fmo1 and Fmo2.
The Km is in micromolar units.
The reported kcat was calculated as nmoles MTSO formed min-1 nmol-1 FMO.
Mean and (S.D.) from analysis of two independently expressed batches of protein.
We used our un-normalized measurement of FMO transcript levels together with our estimates of activity (unheated vs. heated), with MMI as substrate, to predict the level of retained activity that we should observe with heated lung and liver microsomes (Table 5). We predicted that we would observe 35.6% retained activity with lung microsomes and 4.2% activity with liver microsomes. Unheated activity for lung and liver was 21.2 (S.D. 7.1) and 22.0 (S.D. 1.6) nmol TNB oxidized min-1 mg-1 protein, respectively. We observed 41.4% (S.D. 6.4%) retained activity with lung microsomes and 5.8% (S.D. 2.8%) retained activity with liver microsomes.
Table 5.
Predicted pre- and post-heat activity for the major drug-metabolizing FMOs in an average mouse lung and liver.
| Isoform contribution | |||||
|---|---|---|---|---|---|
| Parameter | Units | Fmo1 | Fmo2 | Fmo3 | Total activity |
| Unheated activitya | nmol/min/nmol Fmo | 199.8 | 68.9 | 109.5 | |
| Heated activityb | nmol/min/nmol Fmo | 4.6 | 64.6 | 3.6 | |
| Lung | |||||
| Observed transcript copy numberc | 1.4 × 106 | 2.4 × 106 | 9.2 × 104 | ||
| Relative abundance of Fmod | 0.58 | 1.0 | 0.04 | ||
| Contribution to unheated activitye | nmol/min/nmol Fmo | 115.9 | 68.9 | 4.4 | 189.2 |
| Percent of total | 61.3 | 36.4 | 2.3 | 100.0 | |
| Contribution to heated activityf | nmol/min/nmol Fmo | 2.7 | 64.6 | 0.1 | 67.4 |
| Percent of total | 4.0 | 95.8 | 0.1 | 99.9 | |
| Percent retained activity | 35.6 | ||||
| Liver | |||||
| Observed transcript copy numberc | 7.1 ×105 | 4.8 × 104 | 3.3 × 105 | ||
| Relative abundance of Fmod | 1.0 | 0.07 | 0.47 | ||
| Contribution to unheated activitye | nmol/min/nmol Fmo | 199.8 | 4.8 | 51.5 | 256.1 |
| Percent of total | 78.0 | 1.9 | 20.1 | 100.0 | |
| Contribution to heated activityf | nmol/min/nmol Fmo | 4.6 | 4.5 | 1.7 | 10.8 |
| Percent of total | 42.6 | 41.7 | 15.7 | 100.0 | |
| Percent retained activity | 4.2 | ||||
Mean velocity at pH 8.5 for metabolism of MMI by a single isoform; from Fig. 3a.
Mean velocity after heating; metabolism of MMI by a single isoform at pH 8.5.
Mean un-normalized FMO transcript copy number/μg RNA from eight mouse strains (n = 32).
Proportion of total transcript copy number of Fmo1 + Fmo2 + Fmo3.
Relative abundance × unheated activity.
Relative abundance × heated activity.
4. Discussion
This paper explores the co-expression of Fmo genes in mouse lung and liver and provides an initial systematic characterization of enzymatic properties of Fmo1, Fmo2 and Fmo3. Fmo5 was included for transcriptional studies for methods verification, but was not included for enzymatic studies since it has not so far been demonstrated to metabolize any drugs or xenobiotics to a significant degree. The goal of this work was to determine if a single mouse Fmo2 knockout would model human Fmo2 expression, or if a double knockout would be required. Enzyme characterization was undertaken to determine the best conditions for studying contributions from individual FMOs in the event that multiple isoforms were co-expressed in target organs.
If correct, recent findings [26] indicating that Fmo2 transcript was nearly absent from the lungs of 129 and C57 mice, from 0 to 8 weeks of age, would render meaningless any efforts to develop an Fmo2 knockout mouse. We hypothesized that strain differences might exist that would account for the lack of Fmo2 transcript they measured and would explain the presence of both Fmo2 transcript and Fmo2 enzyme activity from the lungs of ICR (CD-1) mice [25]. We included mice from these three strains in our study and added five additional strains. We worked with eight-week old mice and used the same forward and reverse primer sequences for Fmo1, Fmo3 and Fmo5 as were used to isolate cDNA clones for RNAse protection studies [26]. When we initiated the study, we designed a different set of primers for Fmo2, as the published forward primer sequence did not match (28.5% overall mismatch; 11.1% 3’ end mismatch) NCBI’s Fmo2 cDNA reference sequence (NM_018881). Interestingly, new real-time PCR studies performed by the Shephard and Phillips laboratory, in response to our findings, with Fmo2 primers designed to a different upstream region of the gene, and RNA isolated from a new set of male C57 mice reconfirmed the pattern of Fmo1-4 transcript abundance that they originally reported (personal communication, E. Shephard). Thus the source of inter-laboratory differences in relative Fmo2 transcript levels is, at present, unknown.
Our findings verify that there is significant production of Fmo1 transcript in mouse lung, which is in keeping with both earlier studies [25,26]. However, in contrast to the 2004 study [26], we found Fmo2 transcript levels to be about 1.7-fold higher than Fmo1 transcript levels in mouse lung. We did not detect strain differences in the pattern of FMO isoforms expressed. Absolute transcript levels that we measured were consistently lower (except for Fmo2) than those estimated by RNase protection [26]. However when we expressed FMO levels in female mice as ratios the methods provided quite consistent results. We estimated that the ratio of Fmo1:Fmo5 was 7.0 in lung and 0.2 in liver, compared with 4.5 and 0.2, respectively, by RNase protection. We estimated that the ratio of Fmo1:Fmo3 was 15.2 in lung and 2.2 in liver, compared with approximately 14.0 and 0.5, respectively, by RNase protection. Thus our data for Fmo1, Fmo3 and Fmo5 is consistent with published work, except that our mice, for unknown reasons, had relatively less Fmo3 in liver.
MTS sulfoxidation has been used to simultaneously discriminate FMO1, FMO3 and FMO5 from rabbit [32] based on kinetic parameters and prochiral selectivity. MTS was primarily metabolized by FMOs, although cytochrome P450 can contribute to the sulfoxidation of MTS. MTS was not a significant substrate for rabbit FMO5, but was a substrate for FMO1, FMO2 and FMO3. FMO1 and FMO2 both had an apparent Km < 10 μM, while FMO3 had a low affinity Km of 270 μM. FMO3 was additionally distinguishable from FMO1 and FMO2 because it produced a mixture of the (R)- and (S)-sulfoxide, while FMO1 and FMO2 essentially produced only the (R)-sulfoxide product but could be distinguished because they are not co-expressed in rabbit tissues.
Commercially available human FMO5 generated insignificant levels of MTS sulfoxidation and no oxidative activity at all toward the environmentally important FMO substrates alpha-naphthylthiourea and phorate (data not shown). Thus, while we found abundant Fmo5 transcript in liver, in agreement with earlier work [26], we postulate a negligible enzymatic contribution from this isoform.
Our results for MTS metabolism with mouse Fmo1, Fmo2 and Fmo3 follow the same pattern reported for rabbit isoforms [32]. Fmo3 produced only low levels of sulfoxide, but depending on pH, produced varying proportions of the (R)- and (S)-sulfoxides. By restricting analysis to pH 9.5 we could virtually eliminate any contribution of Fmo3 to activity. This strategy should be particularly useful for the study of Fmo1 in mouse liver that expresses almost no Fmo2. Because both Fmo1 and Fmo2 had high affinity for MTS and produced only detectable (R)-sulfoxide, we will have to couple this assay with an additional parameter to separate contributions by Fmo1 and Fmo2 in tissues such as lung that co-express these FMOs. In our study, heating the expressed baculosome samples for 10 min at 50 °C virtually eliminated the Fmo1 contribution (and Fmo3 as well); thus in lung microsomes 96% of the measured activity after heat treatment was predicted to be due to Fmo2.
By combining our estimates of transcript level with data from characterization of expressed isoform activity, we were able to predict the relative activity we should observe in unheated and heated microsomes prepared from mouse lung and liver tissue. The high degree of concordance in predicted and observed activity from lung and liver microsomes is independent confirmation of FMO transcript levels we measured in these tissues. By carefully controlling protein concentration and buffer conditions during microsomal pre-heating, previously demonstrated to be critical for generating reproducible data [16,33], we have identified conditions where at least 95% of the activity in mouse lung microsomes should be due to Fmo2. This multiple assay procedure to assess Fmo2 contributions will be necessary until an Fmo2-specific substrate can be identified.
In studies designed to determine which member of a gene family, e.g. CYPs, are responsible for metabolism of a given xenobiotic, antibody inhibition has proven to be a valuable approach. The relative contribution of a specific CYP can be determined by adding increasing amounts of polyclonal antibody, specific for that CYP, to the reaction mixture. If that CYP is the major or sole form active toward that substrate, 90% or more of the activity can be eliminated. Unfortunately, without exception, polyclonal antibodies raised to FMOs have weak or no effect on catalytic activity. Additionally, most antibodies to FMO have at least some cross-reactivity to multiple isoforms.
Another tool used in such studies are enzyme-specific inhibitors, including mechanism-based (or suicide substrate) inhibitors. Most CYPs have at least one inhibitor identified with high specificity. However, other than competitive substrates, there are no inhibitors of FMO and certainly no isoform-specific inhibitors. The use of substrate specificity has provided only limited value in identification of the activity of a particular FMO.
We have not attempted to directly compare the total content of FMO in lung and liver. Because housekeeping gene levels were not equivalent in these tissues, normalized data could not be used for this purpose. Although transcript levels of Fmo1, Fmo2 and Fmo3 were higher in lung tissue, the RNA content per cell is higher in mouse liver than lung [34]. Nonetheless, as unheated activity toward MMI was virtually identical in unheated lung and liver microsomes and Fmo1 has much higher activity toward MMI than does Fmo2, the combined Fmo1-3 content of lung microsomes must have been higher than in liver microsomes.
Our evidence that Fmo2 transcript is the major lung Fmo isoform and evidence that activity in lung microsomes verifies results of mouse [25] and rat [24] studies that identify both Fmo1 and Fmo2 in the lung of these species by transcript detection and enzyme activity. We found that our predictions of the relative amount of activity that would be retained in heated lung and liver microsomes, based on determinations of FMO transcript levels and activity retention by individual isoforms, closely mirrored the observed activity in liver and lung microsomes. The somewhat higher level of observed retained activity than we had predicted indicates a possibility that we may have underestimated Fmo2 transcript levels, that there has been some compromise of Fmo1 activity in the tissues we purchased, or that transcript abundance and enzyme assays need to be determined from the same animals for absolute concordance of methods.
Because Fmo1 message levels were significant in the lung of all mouse strains evaluated, and given that substrates of Fmo2 are usually substrates of Fmo1, it will be necessary to develop an Fmo1/Fmo2 double knockout to confidently model the phenotypes associated with the human FMO2 genetic polymorphism. An Fmo1 knockout mouse will model the Hispanic and African American individuals that express active FMO2, while the Fmo1/Fmo2 double knockout will model the majority of the human population that does not express active FMO2 protein. Given that all mouse strains tested had similar FMO profiles, any genetic background should work in future studies.
Acknowledgments
Part of this work was presented at the Fourteenth North American meeting of the International Society for the Study of Xenobiotics (October 22-26, 2006, Rio Grande, Puerto Rico). This study was supported by PHS Grant HL038650. We also acknowledge support from the Cell Culture Facility Core of the Oregon State University Environmental Health Sciences Center (ES 00210).
Abbreviations
- Actb
β-actin
- Hprt1
hypoxanthine guanine phosphoribosyl transferase 1
- FMO
flavin-containing monooxygenase
- MMI
methimazole
- MTS
methyl p-tolyl sulfide
- MTSO
methyl p-tolyl sulfoxide
- Rpl13a
large ribosomal protein 13a
- Tbp
TATA box binding protein
- and the following mouse strains:
- A/J
A/J
- C57BL/6J
C57
- DBA/2J
DBA
- FVB/NJ
FVB
- SWR/J
SWR
- 129S1/SvImJ
129
- Hsd:ICR (CD-1)
ICR
- B6129SF1/J
F1 Hyb
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Krueger SK, Williams DE. Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther. 2005;106:357–87. doi: 10.1016/j.pharmthera.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo WX, Poulsen LL, Ziegler DM. Use of thiocarbamides as selective substrate probes for isoforms of flavin-containing monooxygenases. Biochem Pharmacol. 1992;44:2029–37. doi: 10.1016/0006-2952(92)90106-s. [DOI] [PubMed] [Google Scholar]
- 3.Kim YM, Ziegler DM. Size limits of thiocarbamides accepted as substrates by human flavin-containing monooxygenase 1. Drug Metab Dispos. 2000;28:1003–6. [PubMed] [Google Scholar]
- 4.Smith PB, Crespi C. Thiourea toxicity in mouse C3H/10T12 cells expressing human flavin- dependent monooxygenase 3. Biochem Pharmacol. 2002;63:1941–8. doi: 10.1016/s0006-2952(02)00978-4. [DOI] [PubMed] [Google Scholar]
- 5.Henderson MC, Krueger SK, Stevens JF, Williams DE. Human flavin-containing monooxygenase form 2 S-oxygenation: sulfenic acid formation from thioureas and oxidation of glutathione. Chem Res Toxicol. 2004;17:633–40. doi: 10.1021/tx034253s. [DOI] [PubMed] [Google Scholar]
- 6.Lawton MP, Cashman JR, Cresteil T, Dolphin CT, Elfarra AA, Hines RN, et al. A nomenclature for the mammalian flavin-containing monooxygenase gene family based on amino acid sequence identities. Arch Biochem Biophys. 1994;308:254–7. doi: 10.1006/abbi.1994.1035. [DOI] [PubMed] [Google Scholar]
- 7.Hernandez D, Janmohamed A, Chandan P, Phillips IR, Shephard EA. Organization and evolution of the flavin-containing monooxygenase genes of human and mouse: identification of novel gene and pseudogene clusters. Pharmacogenetics. 2004;14:117–30. doi: 10.1097/00008571-200402000-00006. [DOI] [PubMed] [Google Scholar]
- 8.Hines RN, Hopp KA, Franco J, Saeian K, Begun FP. Alternative processing of the human FMO6 gene renders transcripts incapable of encoding a functional flavin-containing monooxygenase. Mol Pharmacol. 2002;62:320–5. doi: 10.1124/mol.62.2.320. [DOI] [PubMed] [Google Scholar]
- 9.Williams DE, Ziegler DM, Nordin DJ, Hale SE, Masters BS. Rabbit lung flavin-containing monooxygenase is immunochemically and catalytically distinct from the liver enzyme. Biochem Biophys Res Commun. 1984;125:116–22. doi: 10.1016/s0006-291x(84)80342-3. [DOI] [PubMed] [Google Scholar]
- 10.Tynes RE, Sabourin PJ, Hodgson E. Identification of distinct hepatic and pulmonary forms of microsomal flavin-containing monooxygenase in the mouse and rabbit. Biochem Biophys Res Commun. 1985;126:1069–75. doi: 10.1016/0006-291x(85)90294-3. [DOI] [PubMed] [Google Scholar]
- 11.Lawton MP, Gasser R, Tynes RE, Hodgson E, Philpot RM. The flavin-containing monooxygenase enzymes expressed in rabbit liver and lung are products of related but distinctly different genes. J Biol Chem. 1990;265:5855–61. [PubMed] [Google Scholar]
- 12.Nikbakht KN, Lawton MP, Philpot RM. Guinea pig or rabbit lung flavin-containing monooxygenases with distinct mobilities in SDS-PAGE are allelic variants that differ at only two positions. Pharmacogenetics. 1992;2:207–16. doi: 10.1097/00008571-199210000-00003. [DOI] [PubMed] [Google Scholar]
- 13.Yueh M-F, Krueger SK, Williams DE. Pulmonary flavin-containing monooxygenase (FMO) in rhesus macaque: expression of FMO2 protein, mRNA and analysis of the cDNA. Biochim Biophys Acta. 1997;1350:267–71. doi: 10.1016/s0167-4781(97)00004-3. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Cashman JR. Quantitative analysis of FMO gene mRNA levels in human tissues. Drug Metab Dispos. 2006;34:19–26. doi: 10.1124/dmd.105.006171. [DOI] [PubMed] [Google Scholar]
- 15.Dolphin CT, Beckett DJ, Janmohamed A, Cullingford TE, Smith RL, Shephard EA, et al. The flavin-containing monooxygenase 2 gene (FMO2) of humans, but not of other primates, encodes a truncated, nonfunctional protein. J Biol Chem. 1998;273:30599–607. doi: 10.1074/jbc.273.46.30599. [DOI] [PubMed] [Google Scholar]
- 16.Krueger SK, Martin SR, Yueh MF, Pereira CB, Williams DE. Identification of active flavin-containing monooxygenase isoform 2 in human lung and characterization of expressed protein. Drug Metab Dispos. 2002;30:34–41. doi: 10.1124/dmd.30.1.34. [DOI] [PubMed] [Google Scholar]
- 17.Whetstine JR, Yueh M-F, McCarver DG, Williams DE, Park C-S, Kang JH, et al. Ethnic differences in human flavin-containing monooxygenase 2 (FMO2) polymorphisms: detection of expressed protein in African-Americans. Toxicol Appl Pharmacol. 2000;168:216–24. doi: 10.1006/taap.2000.9050. [DOI] [PubMed] [Google Scholar]
- 18.Furnes B, Feng J, Sommer SS, Schlenk D. Identification of novel variants of the flavin-containing monooxygenase gene family in African Americans. Drug Metab Dispos. 2003;31:187–93. doi: 10.1124/dmd.31.2.187. [DOI] [PubMed] [Google Scholar]
- 19.Krueger SK, Williams DE, Yueh MF, Martin SR, Hines RN, Raucy JL, et al. Genetic polymorphisms of flavin-containing monooxygenase (FMO) Drug Metab Rev. 2002;34:523–32. doi: 10.1081/dmr-120005653. [DOI] [PubMed] [Google Scholar]
- 20.Krueger SK, Siddens LK, Martin SR, Yu Z, Pereira CB, Cabacungan ET, et al. Differences in Fmo2*1 allelic frequency between hispanics of Puerto Rican and Mexican descent. Drug Metab Dispos. 2004;32:1337–40. doi: 10.1124/dmd.104.001099. [DOI] [PubMed] [Google Scholar]
- 21.Krueger SK, Siddens LK, Henderson MC, Andreasen EA, Tanguay RL, Pereira CB, et al. Haplotype and functional analysis of four flavin-containing monooxygenase isoform 2 (FMO2) polymorphisms in Hispanics. Pharmacogenet Genomics. 2005;15:245–56. doi: 10.1097/01213011-200504000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson MC, Krueger SK, Siddens LK, Stevens JF, Williams DE. S-oxygenation of the thioether organophosphate insecticides phorate and disulfoton by human lung flavin-containing monooxygenase 2. Biochem Pharmacol. 2004;68:959–67. doi: 10.1016/j.bcp.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 23.Lattard V, Longin-Sauvageon C, Krueger SK, Williams DE, Benoit E. The FMO2 gene of laboratory rats, as in most humans, encodes a truncated protein. Biochem Biophys Res Commun. 2002;292:558–63. doi: 10.1006/bbrc.2002.6656. [DOI] [PubMed] [Google Scholar]
- 24.Hugonnard M, Benoit E, Longin-Sauvageon C, Lattard V. Identification and characterization of the FMO2 gene in Rattus norvegicus: a good model to study metabolic and toxicological consequences of the FMO2 polymorphism. Pharmacogenetics. 2004;14:647–55. doi: 10.1097/00008571-200410000-00002. [DOI] [PubMed] [Google Scholar]
- 25.Venkatesh K, Blake B, Levi PE, Hodgson E. The flavin-containing monooxygenase in mouse lung: evidence for expression of multiple forms. J Biochem Toxicol. 1992;7:163–9. doi: 10.1002/jbt.2570070305. [DOI] [PubMed] [Google Scholar]
- 26.Janmohamed A, Hernandez D, Phillips IR, Shephard EA. Cell-, tissue-, sex-and developmental stage-specific expression of mouse flavin-containing monooxygenases (Fmos) Biochem Pharmacol. 2004;68:73–83. doi: 10.1016/j.bcp.2004.02.036. [DOI] [PubMed] [Google Scholar]
- 27.Guengerich FP. Analysis and characterization of enzymes. In: Hayes AW, editor. Principles and Methods of Toxicology. Raven Press; New York: 1989. pp. 777–814. [Google Scholar]
- 28.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 29.Dixit A, Roche TE. Spectrophotometric assay of the flavin-containing monooxygenase and changes in its activity in female mouse liver with nutritional and diurnal conditions. Arch Biochem Biophys. 1984;233:50–63. doi: 10.1016/0003-9861(84)90600-3. [DOI] [PubMed] [Google Scholar]
- 30.Rettie AE, Bogucki BD, Lim I, Meier GP. Stereoselective sulfoxidation of a series of alkyl p-tolyl sulfides by microsomal and purified flavin-containing monooxygenases. Mol Pharmacol. 1990;37:643–51. [PubMed] [Google Scholar]
- 31.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(RESEARCH0034):1–12. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rettie AE, Lawton MP, Sadeque AJ, Meier GP, Philpot RM. Prochiral sulfoxidation as a probe for multiple forms of the microsomal flavin-containing monooxygenase: studies with rabbit FMO1, FMO2, FMO3, and FMO5 expressed in Escherichia coli. Arch Biochem Biophys. 1994;311:369–77. doi: 10.1006/abbi.1994.1250. [DOI] [PubMed] [Google Scholar]
- 33.Krueger SK, Yueh M-F, Martin SR, Pereira CB, Williams DE. Characterization of expressed full-length and truncated FMO2 from rhesus monkey. Drug Metab Dispos. 2001;29:693–700. [PubMed] [Google Scholar]
- 34.Kanno J, Aisaki K, Igarashi K, Nakatsu N, Ono A, Kodama Y, et al. “Per cell” normalization method for mRNA measurement by quantitative PCR and microarrays. BMC Genomics. 2006;7:64. doi: 10.1186/1471-2164-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]