SUMMARY
Cell fate decisions of pluripotent embryonic stem (ES) cells are dictated by activation and repression of lineage-specific genes. Numerous signaling and transcriptional networks progressively narrow and specify the potential of ES cells. Whether specific microRNAs help refine and limit gene expression, and thereby could be used to manipulate ES cell differentiation, has largely been unexplored. Here, we show that two serum response factor (SRF)-dependent muscle-specific microRNAs, miR-1 and miR-133 promote mesoderm formation from ES cells but have opposing functions during further differentiation into cardiac muscle progenitors. Furthermore, miR-1 and miR-133 were potent repressors of nonmuscle gene expression and cell fate during mouse and human ES cell differentiation. miR-1’s effects were in part mediated by translational repression of the Notch ligand Delta-like 1 (Dll-1). Our findings indicate that muscle-specific miRNAs reinforce the silencing of nonmuscle genes during cell lineage commitment and suggest that miRNAs may have general utility in regulating cell fate decisions from pluripotent ES cells.
INTRODUCTION
Embryonic stem (ES) cells, derived from the inner cell mass of blastocysts, are pluripotent and self-renewing cells, with the unique ability to give rise to all three germ layers—ectoderm, mesoderm, and endoderm. Precise regulation of cell fate decisions is a prerequisite for future therapeutic use of ES cells. Numerous signaling pathways, including those involving members of the Wnt, Bmp, and Notch pathways, appear to regulate cell fate during embryogenesis and can be utilized in various forms to influence lineage choices in cultured ES cells (reviewed in Loebel et al., 2003). Such pathways often culminate in transcriptional events, through either DNA-binding proteins or chromatin remodeling factors, that dictate which subset of the genome is activated or silenced in specific cell types. As a result, transcription factors that regulate pluripotency or lineage-specific gene and protein expression have been a major focus of ES cell research.
In addition to transcriptional regulation, post-transcriptional control by small noncoding RNAs such as microRNAs (miRNAs) quantitatively influences the ultimate proteome (He and Hannon, 2004; Ambros, 2004). miRNAs are naturally occurring RNAs that are transcribed in the nucleus, often under the control of specific enhancers, and are processed by the RNAses Drosha/DGCR8 and Dicer into mature ~22 nucleotide RNAs that bind to complementary target mRNAs. miRNA:mRNA interactions in RNA-induced silencing complexes can result in mRNA degradation, deadenylation, or translational repression at the level of the ribosome. Over 450 human miRNAs have been described, and each is predicted to target tens if not hundreds of different mRNAs. Because they can regulate numerous genes, often in common pathways, miRNAs are candidates for master regulators of cellular processes, much like transcription factors that regulate entire programs of cellular differentiation and organogenesis (Zhao and Srivastava, 2007).
As pluripotent cells adopt particular fates, genes are transcriptionally activated that specify lineages. For ES-derived cell types, it is equally critical to suppress the expression of genes that would otherwise drive differentiation toward alternative fates. While this occurs at the transcriptional level, it is possible that miRNAs also contribute to this process by clearing latently expressed mRNAs as cells activate expression profiles reflecting their newly adopted fates. Indeed, ES cells lacking Dicer or Drosha, and therefore most mature miRNAs, cannot differentiate into most lineages (Kanellopoulou et al., 2005; Murchison et al., 2005; Wang et al., 2007). Although ES cell–specific miRNAs have been described (Houbaviy et al., 2003;), the function or potential of specific miRNAs in ES cell differentiation has not been reported.
During differentiation of ES cells into aggregates called embryoid bodies (EBs), which to a limited extent recapitulate embryonic development, cardiomyocytes are among the first cell types to arise. They become easily visible 7 days after differentiation as small clusters of rhythmically and synchronously contracting cells. Like naturally occurring cardiac muscle cells, ES cell–derived cardiomyocytes express markers of cardiac differentiation, assemble contractile machinery, and establish cell-cell communication (Maltsev et al., 1994;).
In addition to the numerous transcription factors and signaling molecules that control development of cardiac cells (Srivastava, 2006), miRNAs have a critical role in cardiac differentiation in vivo (Zhao et al., 2005; Kwon et al., 2005; Zhao et al., 2007). In particular, miR-1 and miR-133 are cardiac and skeletal muscle–specific, bicistronic miRNAs that are transcriptionally controlled by some of the major regulators of muscle differentiation: serum response factor (SRF), MyoD and Mef2 (Zhao et al., 2005; Kwon et al., 2005, Sokol and Ambros, 2005; Rao et al., 2006). miR-1 promotes differentiation of cardiac progenitors and exit from the cell cycle in mammals and in flies (Zhao et al., 2005, 2007; Kwon et al., 2005). In contrast, miR-133 inhibits differentiation of skeletal myoblasts and maintains them in a proliferative state (Chen et al., 2006). Several direct targets of miR-1 have been described in vivo (Zhao et al., 2005; 2007), including Hand2, a transcription factor required for expansion of cardiac progenitors (Srivastava et al., 1997; Yamagishi et al., 2001), and the Notch ligand delta in Drosophila (Kwon et al., 2005).
Here, we show that miR-1 and miR-133 are enriched in ES cell-derived cardiomyocytes and are expressed at the early stages of cardiac mesoderm selection from ES cells. Expression of either miR-1 or miR-133 in ES cells resulted in enhanced mesoderm gene expression in differentiating EBs but suppressed differentiation into the ectodermal or endodermal lineages. However, miR-1 and miR-133 had opposing effects on further adoption of muscle lineages, with miR-1 promoting and miR-133 blocking differentiation into either cardiac or skeletal muscle fates. Delta-like 1 (Dll-1), a Notch ligand expressed in ES cells, was translationally repressed in miR-1-expressing ES cells and depletion of Dll-1 from ES cells resulted in a bias toward the cardiac lineage while suppressing endoderm and neuroectoderm differentiation, similar to miR-1-expressing ES cells. Our findings demonstrate that miRNAs can control cell lineage determination from pluripotent ES cells, likely by fine-tuning the transcriptome of differentiating cells during commitment to a newly adopted fate.
RESULTS
miRNA Expression in Mouse ES Cells and ES Cell–Derived Cardiomyocytes
To determine which miRNAs are enriched during differentiation of mouse ES (mES) cells into cardiomyocytes, we used a mES cell line carrying a green fluorescent protein (GFP) transgene under control of the β-myosin heavy chain promoter, which is uniquely expressed in differentiated cardiomyocytes. We isolated RNA from GFP+ and GFP− cells by fluorescence-activated cell sorting after 13 days of EB differentiation and profiled miRNA expression by microarray analysis. Seventeen miRNAs were enriched at least 3-fold in the GFP+ population (Fig. 1a). Approximately half of the miRNAs that were enriched in mES cell-derived cardiomyocytes, including the muscle-specific miRNAs miR-1 and miR-133, were undetectable in undifferentiated mES cells, indicating that they were unique to differentiating cells (Fig. 1a).
Figure 1. Identification of miRNAs expressed in ES cell–derived cardiomyocytes.
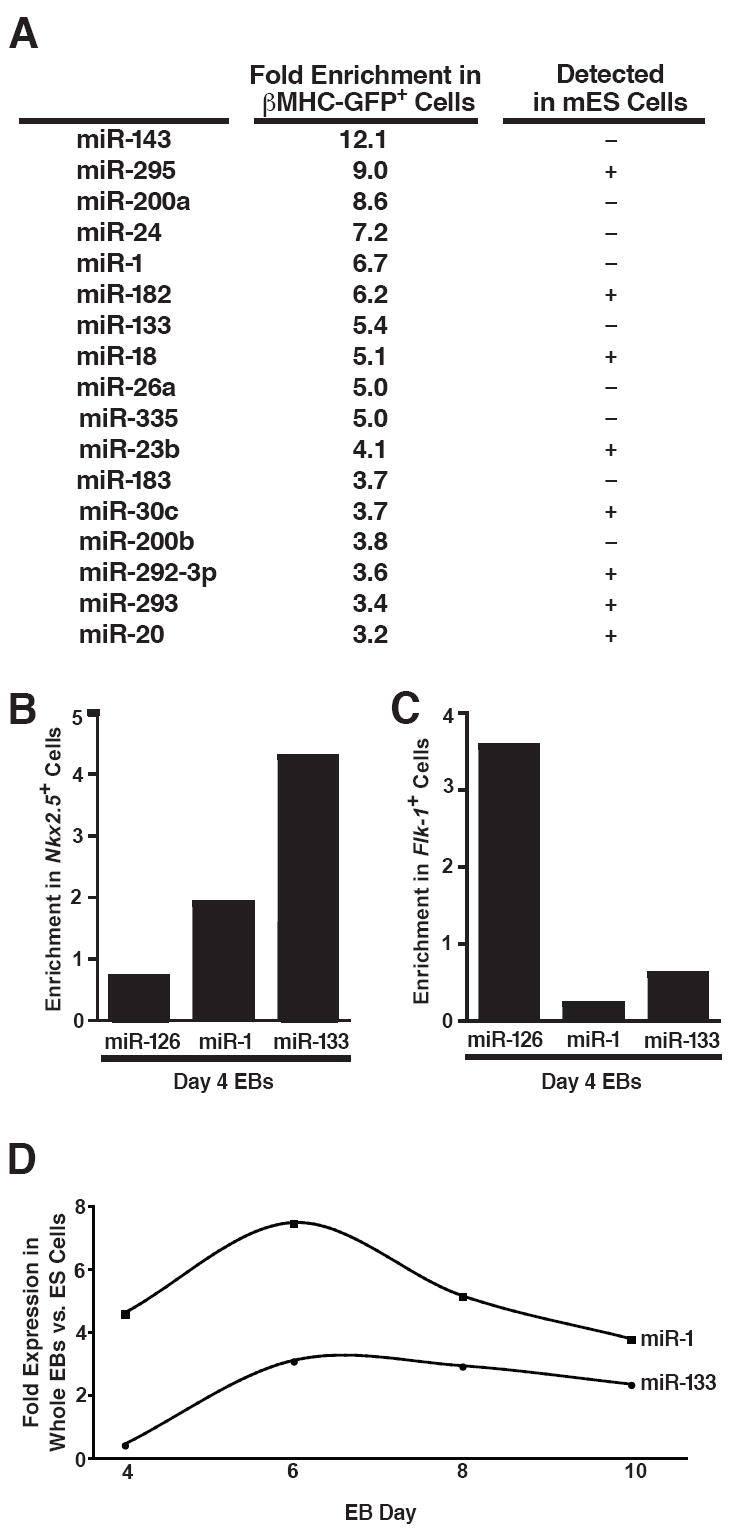
(A) mES cells carrying a GFP transgene under control of the cardiomyocyte-specific β-myosin heavy chain promoter were differentiated for 13 days as embryoid bodies (EBs), sorted by GFP expression, and analyzed by miRNA microarray. miRNAs enriched at least threefold in the GFP+ compared to GFP− cell populations are listed along with their fold enrichment and whether they were detected in ES cells.
(B, C) Quantitative RT-PCR (qRT-PCR) showing enrichment of miR-1 and miR-133 inflow-sorted Nkx2.5-GFP+ cardiac progenitors from day 4 EBs (B) but not in Flk-1+ vascular progenitors, which are enriched for the endothelial-specific miRNA, miR-126 (C).
(D) qRT-PCR showing expression kinetics of miR-1 and miR-133 during days 4–10 of EB differentiation.
To determine whether miR-1 and miR-133 were present and enriched in early cardiac progenitors, we utilized a mES cell line carrying a GFP transgene under transcriptional control of a recombinant bacterial artificial chromosome containing the Nkx2.5 enhancer (B. Conklin and E. Hsiao, unpublished results). This line effectively marks the early emergence of pre-cardiac mesoderm. Sorting of GFP-positive cells in day 4 EBs followed by quantitative RT-PCR (qRT-PCR) revealed that the muscle-specific miRNAs were expressed specifically in the early pre-cardiac mesoderm at this early stage (Fig. 1b), while the vascular endothelium-enriched miRNA, miR-126, was absent (Kuehbacher et al., 2007). Conversely, when we sorted vascular progenitors from day 4 EBs based on their cell surface expression of Flk-1, miR-1 and miR-133 were absent from the Flk-1+ mesoderm population in which miR-126 was highly expressed (Fig. 1c). We also examined the kinetics of miR-1/miR-133 expression in differentiating whole EBs (Fig. 1d). Both were detectable as early as day 4 and their expression increased until day 6 after which their relative abundance in the growing EBs diminished other cell types emerged.
miR-1 and miR-133 Can Promote Mesoderm Differentiation in mES Cells
Since miR-1 and miR-133 were not expressed in undifferentiated mES cells, but were specifically enriched in pre-cardiac mesoderm, we hypothesized that their introduction into mES cells might bias cells toward a muscle lineage. Lentiviruses were used to infect and select ES cell lines expressing miR-1 (mESmiR-1) or miR-133 (mESmiR-133) (Fig. 2a). The levels of introduced miRNAs approximated those of the endogenous miRNAs in the mouse heart (Fig. 2b). The morphology (data not shown) and doubling time of the cell lines in LIF-containing medium were unaltered (Fig. 2c), and the pluripotency markers Oct-4 and Nanog were expressed at normal levels (data not shown).
Figure 2. Effects of miR-1 and miR-133 on mesoderm differentiation.

(A) Schematic of methods used to express miRNAs in mES cells. mES cells were infected with lentiviruses expressing miR-1 or miR-133 under control of a heterologous EF-1 promoter. Stably infected cells were selected based on their resistance to blasticidin in order to generate stable miRNA-expressing mES cell lines (mESmiR-1 and mESmiR-133).
(B) qRT-PCR results confirmed the expression of miR-1 and miR-133; expression of the unintroduced miRNA was unchanged. miR-1 and miR-133 were expressed at levels comparable to those in the adult mouse heart.
(C) The population doubling times of mESmiR-1 and mESmiR-133 cells were similar to those of wild-type mES cells.
(D) qRT-PCR analyzing expression of Bry, an early mesoderm marker, in control, mESmiR-1, and mESmiR-133 EBs collected on day 4 of differentiation. Expression of miR-1 or miR-133 increased expression of Bry.
(E, F) qRT-PCR analysis of Nkx2.5 (E) and Myogenin (F) expression from day 4, 6, or 10 EBs formed from control, mESmiR-1, or mESmiR-133 cells. Control EBs displayed an induction of Nkx2.5 expression over time that was enhanced by miR-1 and suppressed by miR-133. Induction of Myogenin expression was enhanced by miR-1, but not by miR-133.
(G) Differences in Nkx2.5 expression (green fluorescent cells) were also visualized at day 10 of differentiation by expressing the miRNAs in an Nkx2.5-GFP transgenic mES cell line.
(H) Expression of miR-1 and miR-133 was undetectable in day 10 SRF−/− EBs by qRT-PCR.
(I) Overexpression of miR-1 and to a lesser extent, miR-133, in SRF−/− EBs restored the Bry and Mesp1 downregulation in day 10 EBs typical of wild-type cells.
(J) Expression of Cd53, Cxcl4, and Thbs1, which mark hematopoietic lineages, and of Mef2c, which encodes a major regulator of muscle differentiation, was partially rescued in SRF−/− EBs upon expression of miR-1 or miR-133.
To assess the lineage potential of mES cells expressing miR-1 and miR-133, we differentiated control, mESmiR-1, and mESmiR-133 cells by the hanging drop method, collected the resulting EBs on days 4, 6, and 10 of differentiation, and examined the expression of lineage markers by qRT-PCR. Since miR-1 and miR-133 were normally expressed in day 4 pre-cardiac mesoderm, we examined expression of the early mesoderm marker, Brachyury (Bry). Bry expression was detected transiently in control EBs at day 4 and then rapidly declined (Fig. 2d and data not shown). In day 4 EBs expressing miR-1 or miR-133, Bry expression was dramatically enhanced (Fig. 2d), suggesting that both can promote mesodermal gene expression in pluripotent mES cells.
To determine the effects of miR-1 and miR-133 on further differentiation, we examined expression of Nkx2.5, a transcription factor that is one of the earliest cardiac markers (Fig. 2e). In control EBs, Nkx2.5 expression was detected by day 6 and was maintained at day 10. Expression of miR-1 increased Nkx2.5 expression at day 6; by day 10, it was ~7-fold greater than in control EBs. Strikingly, expression of miR-133 blocked induction of Nkx2.5 at both time points. We performed a similar expression analysis of Myogenin, an early skeletal muscle marker, to determine the effects of miR-1 and miR-133 on skeletal muscle differentiation. qRT-PCR analysis of Myogenin expression in day 4, 6, or 10 EBs revealed that miR-1, but not miR-133, markedly enhanced Myogenin expression (Fig. 2f).
The increase in Nkx2.5 expression, as assessed by qRT-PCR, may represent either an increase in the amount of Nkx2.5 expressed per cell or in the number of cells expressing Nkx2.5. To distinguish between these two possibilities, we infected the Nkx2.5-GFP mES line with control, miR-1-, or miR-133-expressing lentivirus, selected with antibiotic, and differentiated these cells for 10 days. GFP was expressed in more miR-1-expressing EBs, and at higher levels per cell, than in wild-type EBs, and was almost undetectable in miR-133 expressing cells (Fig. 2g). Thus, miR-1 appears to promote the emergence of both cardiac and skeletal progenitors in mES cells, while miR-133 does not enhance further differentiation of mesoderm precursors into either lineage.
miR-1 or miR-133 Can Rescue Mesoderm Gene Expression in SRF−/− EBs
Efficient methods for stable miRNA knockdown studies in differentiating EBs are not yet available due to the rapid doubling time of ES cells. However, we previously showed that expression of the miR-1/miR-133 locus in embryonic mouse hearts is directly dependent on SRF (Zhao et al., 2005) and we therefore sought to use SRF-null ES cells as a model for complementation experiments that might reveal the specific contribution of these miRNAs within SRF-null cells (Zhao et al., 2005). We found that SRF-null EBs failed to activate miR-1 or miR-133 (Fig. 2h), confirming the SRF-dependency in the ES cell system, consistent with in vivo observations. Differentiation of mesodermal progenitors in EBs lacking SRF is weak and delayed (Weinhold et al., 2000). To our surprise, however, we found that Bry expression persisted in SRF-null EBs, even after 10 days of differentiation, reflecting delayed or arrested differentiation of mesodermal progenitors that normally downregulate Bry by day 5 (Fig. 2i). Despite the many genes dysregulated in SRF-null EBs, re-introduction of miR-1 in SRF-null ES cells rescued the abnormal accumulation of Bry+ progenitors at day 10 of differentiation, with Bry levels returning close to wild-type levels. Introduction of miR-133 had an intermediate effect on the level of Bry expression at day 10, but Bry levels were still significantly elevated. SRF-/- ES cells also displayed elevated expression of Mesp1, a marker of nascent cardiac mesoderm that is usually downregulated as differentiation progresses (Saga et al., 1996) and this was similarly corrected by reintroduction of miR-1 or miR-133 (Fig. 2i). These data suggest miR-1, and to a lesser degree, miR-133, can promote the progression of mesodermal progenitors and that the arrest of mesodermal progenitors in the absence of SRF may be largely due to the absence of this family of miRNAs.
Consistent with the changes in Bry expression, expression of miR-1 or miR-133 restored the expression of a number of mesodermal genes in day 10 SRF-null EBs (Fig. 2j). Blood cell–specific genes, such as Cd53, CxCl4, and Thbs1, were dramatically downregulated in SRF−/− EBs, reflecting the loss of hematopoeitic lineages in the absence of SRF. However, their expression was reinitiated upon reintroduction of miR-1 or miR-133, likely representing relief of the block to mesodermal differentiation. Even expression of Mef2c, a major regulator of muscle lineages (Li et al., 1997), was restored by miR-1 and, to a lesser extent, by miR-133.
miR-1 and miR-133 Suppress Endoderm Differentiation in mES Cells
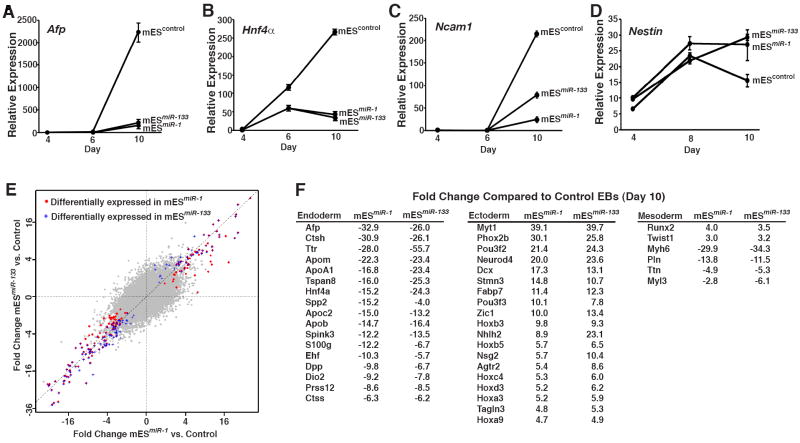
It has been proposed that in some contexts miRNAs function in a “fail-safe” mechanism to clear latent gene expression by targeting pathways that should not be activated in a particular cell type (Hornstein et al., 2005). We therefore investigated whether miR-1 and miR-133 might not only promote muscle lineage decisions, but also reinforce them by repressing nonmuscle gene expression. First, we differentiated control, mESmiR-1, and mESmiR-133 ES cells in the presence of recombinant nodal, a potent inducer of endoderm differentiation in mES cells (Vallier et al., 2004; Pfendler et al., 2005). As expected, nodal stimulated expression of the endoderm markers α-Fetoprotein (Afp) and Hnf4α in control EBs (Fig. 3a,b). These markers were expressed at dramatically lower levels in mESmiR-1 and mESmiR-133 EBs than in control EBs, indicating that miR-1 or miR-133 can each function as potent repressors of endoderm gene expression during differentiation of pluripotent mES cells (Fig. 3a,b).
Figure 3. Both miR-1 and miR-133 suppress endoderm and neuroectoderm differentiation in mES cells.
(A, B) qRT-PCR analysis of the endoderm markers Afp (A) or Hnf4α (B) from day 4, 6, or 10 nodal-treated EBs formed from control, mESmiR-1 or mESmiR-133 cells. Induction of Afp and Hnf4α expression normally observed during differentiation in the presence of nodal was suppressed by expression of miR-1 or miR-133.
(C) qRT-PCR analysis of the neural marker Ncam1 from day 4, 6, or 10 RA-treated EBs formed from control, mESmiR-1 or mESmiR-133 cells. Expression of miR-1 or miR-133 suppressed the induction of Ncam normally observed during differentiation in the presence of RA.
(D) qRT-PCR analysis of the neural progenitor marker Nestin in day 4, 8, or 10 RA-treated EBs formed from control, mESmiR-1 or mESmiR-133 cells. Nestin expression declined in wild-type EBs by day 10 as neurons differentiated, but was maintained in mESmiR-1 and mESmiR-133 EBs.
(E) Plot comparing results from mRNA expression microarray analyses of day 10 control, mESmiR-1, and mESmiR-133 EBs. Plot shows that most genes were coordinately regulated.
(F) Examples of genes that were coordinately regulated in mESmiR-1 and mESmiR-133 EBs compared to controls.
miR-1 and miR-133 Suppress Neural Differentiation From mES Cells
Next, we asked whether miR-1 or miR-133 could also suppress neuroectoderm gene expression from pluripotent mES cells. Control, mESmiR-1, and mESmiR-133 ES cells were differentiated in the presence of retinoic acid (RA), a potent inducer of neural differentiation (Bain et al., 1995; Bain et al., 1996). RA-treated, control EBs expressed high levels of neural cell adhesion molecule 1 (Ncam1), a marker of mature neurons, by day 10 of differentiation, but Ncam1 induction was suppressed in both mESmiR-1 and mESmiR-133 EBs (Fig. 3c). We also examined expression of Nestin, which is restricted largely to neural progenitor cells and is downregulated upon further neural differentiation (Hockfield and McKay, 1985). Nestin expression persisted beyond day 10 in mESmiR-1 and mESmiR-133 EBs, well after its decline in control EBs, suggesting an accumulation of neural progenitors (Fig. 3d). Suppression of endoderm or neuroectoderm differentiation was not observed when an endothelial-enriched microRNA, miR-126, was similarly introduced into mES cells (Supp. Fig. 1), indicating specificity of miR-1 and miR-133 effects. These data indicate that both miR-1 and miR-133 can curtail the differentiation of pluripotent cells into mature neurons, even as cells are pushed toward that lineage by timed administration of RA.
Coordinate Dysregulation of Gene Expression in mESmiR-1 and mESmiR-133 EBs
To more broadly assess the influence of miR-1 or miR-133 on lineage specification and gene expression, we performed mRNA expression microarray analyses on day 10 control, mESmiR-1, and mESmiR-133 EBs. Consistent with the similar effects of miR-1 and miR-133 on repression of nonmuscle gene expression, the vast majority of genes were coordinately regulated between mESmiR-1 and mESmiR-133 EBs (Fig. 3e). Among the most highly downregulated genes in both the mESmiR-1 and mESmiR-133 EBs were the early endoderm markers, Afp and Hnf4α, consistent with our qRT-PCR results from EBs treated with nodal (Fig. 3f). Expression of other genes normally enriched in endodermal structures, such as those encoding apolipoproteins, was also downregulated in both mESmiR-1 and mESmiR-133 EBs (Fig. 3f). These results support the idea that miR-1 and miR-133 can suppress endoderm specification and differentiation.
Among the most highly upregulated genes in both mESmiR-1 and mESmiR-133 EBs were those associated with neuroectoderm specification and early neural differentiation These included the early neurogenic transcription factors, Neurod4, Phox2b, and Myt1 and a number of Hox genes involved in neural specification (Fig. 3f). This is consistent with our observation of persistent Nestin expression in mESmiR-1- and mESmiR-133-derived EBs and the apparent disruption of late-stage neuronal differentiation by these miRNAs.
A number of mesodermal genes were also commonly dysregulated in both mESmiR-1 and mESmiR-133 EBs (Fig. 3f). Runx2 and Twist1, which are highly expressed in developing bone (Ducy et al., 1997; Bialek et al., 2004), were both upregulated, further supporting our conclusion that mesoderm specification is increased in miR-1- or miR-133-expressing EBs. However, a number of genes encoding sarcomeric proteins found in differentiated muscle cells were decreased in both mESmiR-1 and mESmiR-133 EBs. The mechanism for diminished sarcomeric gene expression in EBs may differ in the two cells lines: mesodermal progenitors in the mESmiR-133 EBs likely fail to differentiate into muscle, remaining in the progenitor state, while differentiating muscle cells in mESmiR-1 EBs may prematurely exit the cell cycle resulting in fewer cardiac cells, as was observed upon overexpression of miR-1 in the mouse heart (Zhao et al., 2005). Both would result in underrepresented muscle gene expression and each is consistent with our current understanding of miR-1 and miR-133 function.
miR-1 and miR-133 Suppress Neural Differentiation during Teratoma Formation
To examine the ability of miR-1 and miR-133 to suppress nonmesodermal lineages in a more in vivo setting, we injected wild-type or miRNA-expressing mES cells subcutaneously into SCID mice and monitored their differentiation in vivo. Transplanted cells of each line formed teratomas in the recipients and were analyzed 6 weeks after inoculation. Teratomas from control, mESmiR-1, or mESmiR-133 cells included derivatives of all three embryonic germ layers, but the control teratomas were much more homogeneous (Fig. 4). As shown by immunostaining with βIII-tubulin antibodies, teratomas from control mES cells were composed mostly of differentiated neurons (Fig. 4a,d). In contrast, teratomas formed from mESmiR-1 or mESmiR-133 cells had far fewer differentiated neuronal cells (Fig. 4b,c,e,f).
Figure 4. Differentiation of neural cells is suppressed by miR-1 or miR-133 in teratomas.
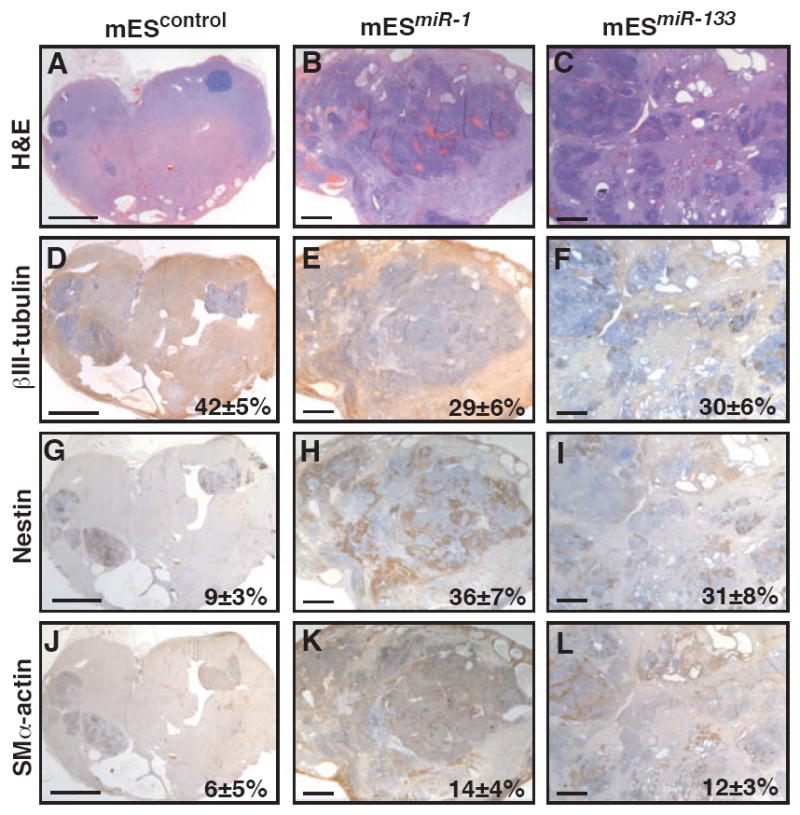
Teratomas were generated by injecting control, mESmiR-1, or mESmiR-133 cells into the rear flank of SCID mice. After 6 weeks, hematoxylin/eosin-stained teratomas derived from control ES cells were strikingly homogeneous (A) and composed mostly of βIII-tubulin-immunoreactive neural cells (D). Teratomas from mESmiR-1 or mESmiR-133 cells were more heterogeneous (B,C) and contained fewer βIII-tubulin-positive cells (E,F). An accumulation of nestin-positive neural precursors was observed in miR-1 or miR-133 expressing teratomas compared to control (G-I). Expression of miR-1 or miR-133 enhanced muscle specification, as shown by immunostaining with smooth muscle α-actin antibody (J-L). Quantification of areas immunostained with each antibody is indicated as percentages with standard deviation. Scale bars represent ~2mm.
Based on our analyses of neural differentiation in EBs, we also immunostained teratomas using an antibody to nestin. Control teratomas were fully differentiated and contained only rare pockets of nestin-positive neural progenitors, as expected (Fig. 4g). However, mESmiR-1 and mESmiR-133 teratomas contained abundant nestin-positive cells even after 6 weeks of development, suggesting an arrest of neural differentiation at the progenitor stage (Fig. 4h,i). The accumulation of nestin-positive progenitors in these teratomas further supports the idea that miR-1 and miR-133 permit specification of the ectodermal lineage from pluripotent mES cells, but inhibit complete differentiation of neural progenitor cells into neurons.
We also immunostained teratomas using an antibody to smooth muscle α-actin, a marker of smooth muscle and immature striated muscle cells (cardiac and skeletal). Consistent with the promesodermal effects of miR-1 and miR-133 in EBs, teratomas derived from mESmiR-1- and mESmiR-133-derived teratomas had more cells on average expressing smooth muscle α-actin (Fig. 4k,l) than control (Fig. 4j). High magnification views of immunostained sections demonstrate the specificity of each antibody (Supp.Fig. 2).
The Notch Ligand, Delta-like 1, is Translationally Repressed by miR-1
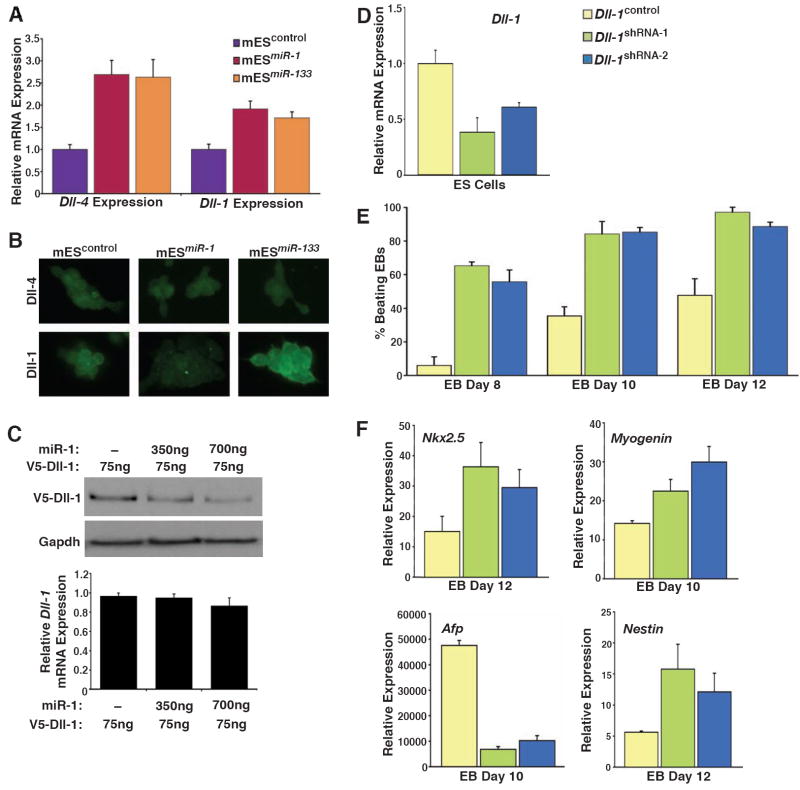
miRNAs likely function by regulating numerous pathways, but in some cases a subset serve as the “major” effectors. Since Notch signaling can promote neural differentiation and inhibit muscle differentiation in ES cells (Nemir et al., 2006; Lowell at al., 2006), which is opposite of miR-1’s effects, we hypothesized that miR-1-mediated repression of Notch signaling may contribute to the observed effects of miR-1 in mES cells. Indeed, we had previously shown that miR-1 directly targets the Notch ligand delta in Drosophila for repression (Kwon et al., 2005). Three orthologs of Drosophila delta have been identified in mice—Dll-1, Dll-3, and Dll-4. Dll-1 and Dll-4, but not Dll-3, contained putative miR-1 or miR-133 binding sites in their 3’ UTR. As shown by qRT-PCR analysis, mRNA expression of Dll-1 and Dll-4 was similar in mESmiR-1 and mESmiR-133 cells and somewhat higher than in control mES cells (Fig. 5a).
Figure 5. Dll-1 protein levels are negatively regulated by miR-1 in mES cells, and knockdown of Dll-1 expression promotes cardiac mesoderm and suppresses non-mesodermal gene expression.
(A) Dll-1 and Dll-4 mRNA levels, assessed by qRT-PCR, were somewhat higher in mESmiR-1 and mESmiR-133 cells than in controls.
(B) Immunostaining with Dll-4 or Dll-1 antibody showed equivalent Dll-4 protein levels in mESmiR-1, mESmiR-133 cells, and control mES cells; Dll-1 protein levels were lower in mESmiR-1 cells and higher in mESmiR-133 cells than wild–type mES cells.
(C) miR-1 expression caused a dose-dependent decrease in epitope (V5)-tagged Dll-1 protein levels by Western blot without affecting RNA expression of Dll-1 assessed by qRT-PCR (graph). Gapdh protein levels reflect equal loading of protein.
(D) Dll-1 mRNA levels, assessed by qRT-PCR, were 62% and 40% lower in response to two distinct short hairpin RNAs targeting Dll-1 mRNA (Dll-1shRNA-1 and Dll-1shRNA-2), compared to control cell line.
(E) EBs formed from Dll-1control, Dll-1shRNA-1 and Dll-1shRNA-2 ES cells were scored for beating cardiomyocytes on days 8, 10, and 12 of differentiation. Beating cardiomyocytes appeared earlier and were more numerous in EBs from Dll-1shRNA cell lines than in EBs from the control line.
(F) qRT-PCR analyses of Nkx2.5, Myogenin, Afp, and Nestin expression in EBs generated from Dll-1control, Dll-1shRNA-1 and Dll-1shRNA-2 ES cells. Knocking down Dll-1 increased Myogenin expression, decreased Afp expression and sustained Nestin expression compared to controls.
Since miRNAs can block the translation of target mRNAs, we examined Dll-1 and Dll-4 protein levels in all three mES cell lines (Fig. 5b). mESmiR-1, mESmiR-133, and control cells had similar levels of Dll-4 by immunocytochemistry (Fig. 5b) and Western analysis (data not shown). Quantitative analysis of endogenous Dll-1 protein was not possible due to the lack of published Dll-1 antibodies that function in Western blots. However, mESmiR-1 cells had consistently decreased Dll-1 protein levels by immunocytochemistry despite having normal levels of Dll-1 mRNA, consistent with translational inhibition of Dll-1 by miR-1. Although a potential miR-1 binding site in the Dll-1 3’-UTR has extensive, conserved sequence matching (Supp. Fig. 3a) and is present in an accessible region with little secondary structure (data not shown), repression through this site was not transferable to the luciferase 3’-UTR in the surrogate assay commonly employed to test specific binding sites (Supp. Fig. 3b). However, miR-1 potently repressed protein, but not mRNA expression of an epitope-tagged Dll-1 containing the full 3’UTR in a dose-dependent manner indicating translational inhibition of Dll-1 in mammalian cells (Fig 5c).
Dll-1 Knockdown in mES Cells Promotes Cardiac Mesoderm and Suppresses Non-mesoderm Gene Expression
To determine whether downregulation of Dll-1 protein by miR-1 could account for a subset of the effects of miR-1 on cell lineage decisions, we used short hairpin RNA (shRNA) constructs directed against distinct regions of Dll-1 to generate two different Dll-1shRNA cell lines (Dll-1shRNA-1 and Dll-1shRNA-2). The Dll-1 mRNA level was about 62% lower in Dll-1shRNA-1 cells and 40% lower in Dll-1shRNA-2 cells than in a control line expressing a scrambled shRNA construct (Fig. 5d). Oct3/4 levels and cell morphology were unaltered (data not shown). EBs formed from Dll-1shRNA cells had a much greater propensity toward cardiomyocyte differentiation and formed beating cardiomyocytes earlier than control EBs (Fig. 5e). By day 12 of differentiation, 89% of EBs formed from Dll-1shRNA-1 cells and 97% of EBs from Dll-1shRNA-2 cells contained beating cardiomyocytes compared to 48% of Dll-1control EBs. Nkx2.5 expression, marking cardiac progenitors, was also more highly induced in Dll-1shRNA than in control EBs (Fig. 5f), as were Nkx2.5-GFP-positive cells (data not shown). In addition, Myogenin expression was higher in Dll-1shRNA EBs compared to controls (Fig. 5f). Although the effect of Dll-1 knockdown on Nkx2.5 and myogenin expression was not as robust as miR-1 expression, the trends were similar. These results indicate that depletion of Dll-1 increases muscle differentiation from mES cells and suggest that miR-1 may promote cardiac differentiation, in part, by downregulating Dll-1 protein.
We also performed qRT-PCR analyses on EBs formed from Dll-1shRNA cell lines to determine if suppression of ectodermal and endodermal lineages by miR-1 might also involve Dll-1 downregulation. Expression of the endoderm markers Afp (Fig. 5f) and Hnf4α (data not shown) was lower in Dll-1shRNA EBs than in Dll-1control EBs. Moreover, expression of Nestin, which decreased between days 10 and 12 as neurons differentiated in Dll-1control EBs, was increased during this period in both lines of Dll-1shRNA EBs (Fig. 5f). Thus, loss of Dll-1 also repressesendoderm differentiation and and results in persistence of neural progenitor gene expression.
Effects of miR-1 or miR-133 in Human ES Cells
Human ES (hES) cells often behave differently than mES cells. To investigate whether miR-1 or miR-133 function similarly in the two cell types, we infected the H9 hES cell line with the same lentiviruses encoding either miR-1 or miR-133. Expression was verified by qRT-PCR (Fig. 6a). The resulting hESmiR-1 and hESmiR-133 cell lines were differentiated as EBs in suspension and collected on days 4, 6, and 8. NKX2.5 expression was detectable by qRT-PCR in control human EBs by day 6 and decreased overall by day 8 (Fig. 6b). As in the mouse EBs, hESmiR-1 EBs had higher levels of NKX2.5 expression than controls, while hESmiR-133 EBs failed to induce NKX2.5 expression to the levels observed in controls (Fig. 6b). Consistent with this, we also found that the percentage of hESmiR-1 EBs with beating cardiac cells on day 18 of differentiation was more than 3-fold higher than in wild-type EBs, while hESmiR-133 EBs did not display enhanced cardiomyocyte formation (Fig. 6c). Thus, regulation of cardiac differentiation by miR-1 and miR-133 appears to be grossly similar in hES and mES cells.
Figure 6. Effects of miR-1 or miR-133 expression in hES cells.

(A) Lentivirus-mediated expression of miR-1 or miR-133 in human ES (hES) cells was verified by qRT-PCR.
(B) NKX2.5 mRNA expression assessed by qRT-PCR in hEBs collected on days 4, 6, and 8. Overexpression of miR-1 in hES cells increased NKX2.5 expression compared to wild type, while miR-133 expression led to decreased NKX2.5 induction.
(C) Human EBs were scored for beating on day 18 of differentiation. Expression of miR-1 increased the number of beating human EBs, while expression of miR-133 did not.
(D) Day 18 human EBs were immunostained with antibodies to nestin or βIII-tubulin.
To examine the effects of miR-1 or miR-133 expression on neuroectoderm differentiation in hES cells, we also immunostained day 18 control, hESmiR-1, and hESmiR-133 EBs with antibodies recognizing nestin or βIII-tubulin (Fig. 6d). Like miRNA-expressing mouse EBs, hESmiR-1 and hESmiR-133 EBs accumulated more nestin-positive progenitors than control human EBs. As in our mouse ES cells studies, there were fewer βIII-tubulin positive neural cells in hESmiR-133 EBs compared to controls, although this effect was not consistent for hESmiR-1 cells. These results demonstrate that the muscle-specific miRNAs miR-1 and miR-133 have similar, but somewhat unique effects on the differentiation of hES and mES cells, and suggest that miRNAs may be useful for coaxing and repressing differentiation of human or mouse ES cells into particular lineages.
DISCUSSION
This study shows that miR-1 can promote differentiation of both mouse and human ES cells into the cardiac lineage, while miR-133, which is normally co-expressed with miR-1 in developing muscle, blocks differentiation of myogenic precursors. Both miRNAs enhanced mesoderm specification and suppressed the differentiation of ES cells into neuroectoderm or endoderm within EBs or teratomas. miR-1 expression resulted in translational repression of Dll-1, a mammalian ortholog of delta, and reducing the level of Dll-1 expression in mES cells using shRNAs caused similar cell fate trends as miR-1 expression.
The onset of miR-1 and miR-133 expression in mES cells occurred just as mesoderm is becoming specified at day 4, consistent with the early twist-dependent expression of miR-1 throughout Drosophila mesoderm, preceding mef2 expression (Kwon et al., 2005; Sokol and Ambros, 2005). The ability of miR-1 and miR-133 to promote early mesoderm gene expression when misexpressed suggests that this early onset in ES cells may promote mesoderm lineages. Strikingly, further differentiation of mesoderm into the muscle lineage was promoted by miR-1 but inhibited by miR-133. This is similar to in vivo observations (Zhao et al., 2005, Chen et al., 2006).
The findings in SRF-null ES cells that do not express miR-1 or miR-133 provide important data that compliment the gain-of-function studies. SRF-null cells are known to have a block of final muscle differentiation (Weinhold et al., 2000), but the persistence of Bry expression, indicative of an arrest of early mesodermal progenitors, had not been noted. This observation allowed us to determine whether loss of miR-1/miR-133 transcription in SRF-null cells might play a causative role in this interesting developmental block. The ability of miR-1 to rescue the further differentiation of mesodermal progenitors suggests that it can push arrested mesoderm in SRF-null ES cells past the stage of Bry expression, although it did not induce sarcomeric gene expression.
Unexpectedly, miR-1 and miR-133 potently repressed endoderm and neuroectoderm gene expression. This was observed during in vitro differentiation experiments despite the presence of potent inducers of each lineage, and in vivo during teratoma formation. In contrast to their roles during muscle differentiation, miR-1 and miR-133 functioned in concert to repress nonmuscle gene expression, suggesting that they may have many common targets, although competitive increases in mesoderm specification may account for some of the observed alternative lineage suppression. Gene expression analyses of ES cells expressing miR-1 and miR-133 also suggested that the two microRNAs regulate many pathways in common. To our knowledge, this is the first example of miR-1 and miR-133 functioning in a parallel rather than opposing fashion, consistent with their bicistronic derivation. Non-muscle gene expression was not detected in miR-1-2-null mouse hearts (data not shown). However, conclusively determining whether miR-1/miR-133 are required to repress non-muscle gene expression in vivo awaits the creation of compound miR-1-1 and miR-1-2 knockout mice.
The combined effects of the miRNAs in regulating mesoderm differentiation and preventing endoderm and neuroectoderm differentiation reflect a novel but elegant mechanism for controlling lineage decisions (Fig. 7). By initiating the expression of specific miRNAs, a cell might promote active clearance of transcripts that it has “outgrown” and expedite further differentiation. The repression of undesired gene expression may also be useful in efforts to differentiate and utilize ES cells for therapeutic purposes, as strict control of lineage potential is of utmost concern to avoid tumor formation and introduction of harmful cell types.
Figure 7. Model of miR-1/miR-133 effects during ES cell differentiation.
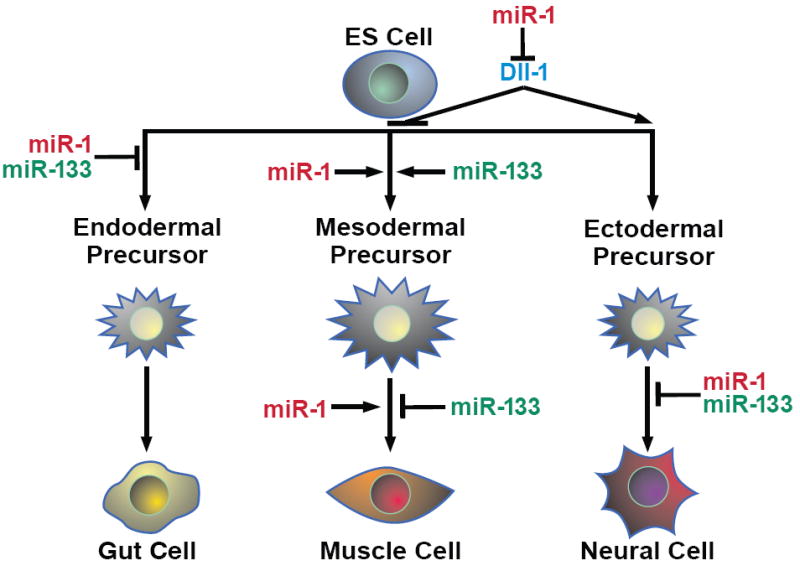
miR-1 and miR-133 promotion of mesoderm and inhibition of endoderm and ectoderm differentiation at specific stages are indicated. Opposing effects of the two miRNAs in later steps of muscle differentiation are also shown. miR-1 inhibition of Dll-1 translation, along with yet unknown targets, likely contribute to the some of the observed effects of miR-1.
Several targets for miR-1 and some for miR-133 have been described, and scores more have been predicted but not validated. Therefore, it is likely that these miRNAs control cell fate decisions by regulating numerous genes and pathways. miR-1 regulated the translation of Dll-1 protein, thereby negatively influencing Notch signaling, consistent with the observation that miR-1 negatively regulates Notch signaling by targeting delta in Drosophila. Specific knockdown of Dll-1 caused similar cell fate trends as miR-1 expression although combinatorial targeting of multiple mRNAs likely results in the full effect of miR-1. Consistent with the effects of miR-1 expression and Dll-1 knockdown, recent reports indicate that Notch1 inhibition promotes cardiac differentiation and that stimulation of the Notch pathway positively regulates neuronal differentiation (Nemir et al., 2006; Lowell et al., 2006). Thus, despite the many pathways likely repressed by miR-1, our findings suggest that negative regulation of Notch signaling may be one of the major mechanisms by which miR-1 influences cell fate decisions (Fig. 7). Whether miR-133 also functions by regulating other components of Notch signaling or if it targets independent pathways remains to be determined.
In summary, our results indicate that the muscle-specific miRNAs miR-1 and miR-133 act comparably during mES and hES differentiation to promote mesoderm differentiation while suppressing gene expression of alternative lineages. Our results also suggest that miRNAs may offer a means to direct the differentiation of ES cells into desired fates and inhibit the formation of undesired lineages.
EXPERIMENTAL PROCEDURES
Mouse ES Cell Culture and Flow Cytometry
The mouse E14 ES cell line was maintained as a monolayer in medium supplemented with 10% fetal bovine serum, LIF-conditioned medium, pyruvate, glutamine, and β-mercaptoethanol in gelatin-coated tissue-culture plates and passaged with trypsin. Cells were differentiated by the hanging drop method. Briefly, cells were trypsinized and resuspended at 25,000 cells/ml in differentiation medium (20% fetal bovine serum, pyruvate, glutamine, and β-mercaptoethanol). Droplets (20 μl) were transferred to each well of a 96-well v-bottom tissue culture plate, which was then inverted. After 2 days of incubation at 37°C, the plates were turned upright, and 200 μl of differentiation medium was added to each well. For neuroectodermal or endodermal induction, 0.5 μM retinoic acid (Sigma) or 50 ng/ml recombinant nodal (R&D Systems), respectively, was added to the wells 96 h after formation of the hanging drops. The medium was changed every 2 days. The β-myosin heavy chain (β-MHC)-GFP E14 cells were a gift of W. Tingley and R. Shaw. For flow cytometry studies, EBs were dissociated via trypsin and passed through a nylon cell strainer. Flk-1+ cells were labeled with a PE-conjugated Flk-1 antibody (BD Pharmingen) and a Becton Dickinson (Franklin Lakes, NJ) FACS Diva flow cytometer and cell sorter was used for detecting and sorting Flk-1+, Nkx2.5-GFP+, or βMHC-GFP+ cells.
miRNA and mRNA Expression Microarray Analyses
ES cells or EBs were harvested in Trizol (Invitrogen) for total RNA isolation. For mRNA expression microarray analysis, 1 μg total RNA was labeled and hybridized to a mouse mRNA expression microarray (Affymetrix). Gene expression values were obtained from Affymetrix CEL files using the GC-RMA package from Bioconductor (Dudoit et al. 2003; Wu et al. 2004). To identify transcripts differing in mean expression across the three experimental groups (mESwt, mESmiR-1, and mESmiR-133 EBs), p values were calculated by permutation test with the F-statistic function from the multtest package of Bioconductor (Dudoit et al. 2003) and a t test comparing each miRNA-expressing group to wild-type EBs. Fold changes in transcript levels were calculated from the mean log2 expression values versus the mean of control EBs.
For miRNA expression microarray, 100 ng of total RNA from each sample was labeled with Cy3 or Cy5 using miRCURY™ LNA microRNA Power labeling kit (Exiqon) and then hybridized to miRCURY™ LNA arrays (Exiqon). Hybridization quality was assessed with Bioconductor marray package and log2 ratios of Cy5 to Cy3 signals were calculated with limma package.
Quantitative RT-PCR
ES cells or EBs were harvested in Trizol (Invitrogen) for total RNA isolation. For mRNA qRT-PCR, 2 μg of total RNA from each sample was reversed transcribed with Superscript III (Invitrogen). 1/16 of the reverse transcription reaction was used for subsequent PCRs, which were performed in duplicate on an ABI 7900HT instrument (Applied Biosystems) using Taqman primer probe sets (Applied Biosystems) for each gene of interest and a GAPDH endogenous control primer probe set for normalization. Each qRT-PCR was performed on at least 3 different experimental samples; representative results are shown as fold expression relative to undifferentiated ES cells, unless otherwise stated. Error bars reflect one standard deviation from the mean of technical replicates.
miRNA qRT-PCR was performed with miRNA Taqman Expression Assays (Applied Biosystems) and the miRNA Reverse Transcription kit (Applied Biosystems). For each miRNA analyzed, 10 ng of total RNA was reverse transcribed with a miRNA-specific primer. A ubiquitous miRNA, miR-16, was used as the endogenous control. Each qRT-PCR was performed on at least three different experimental samples; representative results are shown as fold expression relative to undifferentiated ES cells, unless otherwise noted. Error bars indicate one standard deviation from the mean of technical replicates.
Lentiviral Production and ES Cell Infection
Lentiviruses for miRNA expression were generated with the ViraPower Promoterless Lentiviral Gateway Expression System with MultiSite Gateway Technology (Invitrogen). The EF-1α promoter was recombined into the pLenti vector upstream of a cassette containing either miR-1 or miR-133 pre-miRNA sequence with an additional ~100 nucleotides flanking each end, which was cloned by PCR from a bacterial artificial chromosome containing the mouse genomic miR-1-2 or miR-133a-1 sequences. Details of virus production and introduction into ES cells can be found in Supplemental Methods.
Teratoma Formation
Teratomas were formed by subcutaneous injection of approximately 1×106 control or miRNA-expressing mES cells into the rear flank of 8-week-old male SCID mice (n=10 mice per cell line). Transplanted cells of each line formed teratomas in the recipients and were analyzed 6 weeks after inoculation.
Immunostaining
For immunocytochemistry studies, ES cells were plated on gelatinized cover slips and allowed to settle, rinsed with PBS, fixed in 4% paraformaldehyde for 1 h at room temperature with shaking, and stored in PBS at 4°C. The fixed cells were rinsed in PBS, blocked in blocking solution (1% bovine serum albumin, 1% Tween-20, and PBS) for 30 min at room temperature and incubated in primary antibody in a humidified chamber for 1 h at room temperature. The antibodies were diluted in blocking buffer as follows: Dll-1, 1:100 (AbCam, ab10554); Jag-1, 1:100 (AbCam, ab7771); Dll-4, 1:50 (AbCam, ab7280). After washing in PBS, the cells were incubated for 1 h with FITC-conjugated secondary antibodies (1:200) at room temperature in a darkened chamber, rinsed with PBS, and mounted on slides with Vectashield containing DAPI (Vector Laboratories).
For immunohistochemical studies, teratomas were submerged in CPT (Sakuro), flash frozen in liquid nitrogen, and sectioned. Details of immunostaining and antibodies are in Supplemental Methods.
For EB immunohistochemistry, EBs were fixed in 4% paraformaldehyde, blocked in 5% goat serum, and incubated overnight in βIII-tubulin antibody (1:100; Chemicon, CBL412). The following day, EBs were rinsed, placed in rhodamine-conjugated anti-mouse IgG diluted 1:400 for 2 h, rinsed, mounted with Vectashield containing DAPI (Vector Laboratories), and visualized.
Dll-1 knockdown
mES cells were infected with lentiviral constructs encoding shRNAs against mouse Dll-1 or a control shRNA (Sigma). After transduction and 2 days of recovery, infected mES cells were selected for 7 days with 1 μg/ml puromycin. Colonies were isolated, expanded, and assayed for Dll-1 knockdown compared to control-infected mES cells by qRT-PCR. The pluripotency of the resulting cell lines was assessed by measuring the proliferation rate and Oct3/4 expression and comparing the value to those of uninfected mES cells. Only lines that maintained normal levels of Oct3/4 expression and normal proliferation rates were used for further study.
miR-1 Target analyses
12-well plates of Cos-1 cells were transfected for either luciferase assays or transient expression analyses using Lipofectamine 2000 (Invitrogen). For luciferase assays, a luciferase expression construct containing the 3’UTR of mouse Dll-1 (50ng) was co-transfected alone or with miR-1 or miR-133 expression constructs (300ng) and a LacZ expression construct. Empty expression plasmid was used to normalize the total DNA mass. After 24 hours, cells were harvested and the luciferase assays were performed using a Luciferase Assay Kit (Promega). β-galactosidase assays were also performed and the results were used to normalize for transfection efficiency. For transient expression analyses, a Dll-1 expression construct lacking Dll-1-derived 5’UTR sequence elements, but with the full mouse Dll-1 3’UTR and an n-terminal V5 epitope tag (75ng) was co-transfected with increasing amounts of miR-1 expression construct (0ng, 350ng, or 700ng). Empty expression vector was included to ensure equal DNA mass in each condition. After 24 hours, cells were harvested in modified RIPA buffer or Trizol (Invitrogen). Western analyses to detect V5-tagged Dll-1 protein were performed using an HRP-conjugated V5 antibody diluted 1:1500 (Invitrogen).
Human ES cell culture
The human ES cell line, H9 (WiCell), was maintained on mouse embryonic feeder cells in proliferation medium consisting of Knockout DMEM (GIBCO) supplemented with 20% Knockout serum replacement (GIBCO), pyruvate, glutamine, β-mercaptoethanol and human basic fibroblast growth factor. Details of hES cell differentiation and immunostaining can be found in Supplemental Methods.
Supplementary Material
Acknowledgments
We are grateful to members of the Srivastava lab and to B.G. Bruneau for critical discussion and comments on the manuscript; to B. Taylor and G. Howard for editorial assistance and manuscript preparation; to C. Barker and the Gladstone Genomics core and J. Fish in the Histopathology core for technical assistance; to W. Tingley and R. Shaw for sharing the β-MHC ES cell line; to N. Salomonis for assistance with microarray data analysis. D.S. is supported by grants from NHLBI/NIH, March of Dimes Birth Defects Foundation, and the California Institute of Regenerative Medicine and is an Established Investigator of the American Heart Association. K.N.I. is a post-doctoral scholar of the California Institute of Regenerative Medicine (T2-0003). H.S.B. was supported by a grant from NHLBI/NIH (HL085377) and F.W.K. was supported by NRSA/NIH (HL007544). This work was also supported by NIH/NCRR grant (C06RR018928) to the Gladstone Institutes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Kitchens D, Yao M, Huettner JE, Gottlieb DI. Embryonic stem cells express neuronal properties in vitro. Dev Biol. 1995;168:342–357. doi: 10.1006/dbio.1995.1085. [DOI] [PubMed] [Google Scholar]
- Bain G, Ray WJ, Yao M, Gottlieb DI. Retinoic acid promotes neural and represses mesodermal gene expression in mouse embryonic stem cells in culture. Biochem Biophys Res Commun. 1996;223:691–694. doi: 10.1006/bbrc.1996.0957. [DOI] [PubMed] [Google Scholar]
- Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, Justice MJ, Karsenty G. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Dudoit S, Gentleman RC, Quackenbush J. Open source software for the analysis of microarray data. Biotechniques. 2003;(Suppl):45–51. [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hockfield S, McKay RD. Identification of major cell classes in the developing mammalian nervous system. J Neurosci. 1985;5:3310–3328. doi: 10.1523/JNEUROSCI.05-12-03310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, Baskerville S, Bartel DP, Tabin CJ. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–35. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci U S A. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Arnold J, Hsiao EC, Taketo MM, Conklin BR, Srivastava D. Canonical Wnt signaling is a positive regulator of mammalian cardiac progenitors. Proc Natl Acad Sci U S A. 2007;104:10894–10899. doi: 10.1073/pnas.0704044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Capetanaki Y. AN E box in the desmin promoter cooperates with the E box and Mef-2 sites of a distal enhancer to direct muscle-specific transcription. EMBO J. 1997;13:3580–3589. doi: 10.1002/j.1460-2075.1994.tb06665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebel DA, Watson CM, De Young RA, Tam PP. Lineage choice and differentiation in mouse embryos and embryonic stem cells. Dev Biol. 2003;264:1–14. doi: 10.1016/s0012-1606(03)00390-7. [DOI] [PubMed] [Google Scholar]
- Lowell S, Benchoua A, Heavey B, Smith AG. Notch promotes neural lineage entry by pluripotent embryonic stem cells. PLoS Biol. 2006;4:e121. doi: 10.1371/journal.pbio.0040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltsev VA, Wobus AM, Rohwedel J, Bader M, Hescheler J. Cardiomyocytes differentiated in vitro from embryonic stem cells developmentally express cardiac-specific genes and ionic currents. Circ Res. 1994;75:233–244. doi: 10.1161/01.res.75.2.233. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemir M, Croquelois A, Pedrazzini T, Radtke F. Induction of cardiogenesis in embryonic stem cells via downregulation of Notch1 signaling. Circ Res. 2006;98:1471–1478. doi: 10.1161/01.RES.0000226497.52052.2a. [DOI] [PubMed] [Google Scholar]
- Pfendler KC, Catuar CS, Meneses JJ, Pedersen RA. Overexpression of Nodal promotes differentiation of mouse embryonic stem cells into mesoderm and endoderm at the expense of neuroectoderm formation. Stem Cells Dev. 2005;14:162–172. doi: 10.1089/scd.2005.14.162. [DOI] [PubMed] [Google Scholar]
- Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y, Hata N, Kobayashi S, Magnuson T, Seldin MF, Taketo MM. Mesp1: a novel basic helix-loop-helix protein expressed in the nascent mesodermal cells during mouse gastrulation. Development. 1996;122:2769–2778. doi: 10.1242/dev.122.9.2769. [DOI] [PubMed] [Google Scholar]
- Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Vallier L, Reynolds D, Pedersen RA. Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev Biol. 2004;275:403–421. doi: 10.1016/j.ydbio.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhold B, Schratt G, Arsenian S, Berger J, Kamino K, Schwarz H, Ruther U, Nordheim A. Srf(-/-) ES cells display non-cell-autonomous impairment in mesodermal differentiation. EMBO J. 2000;19:5835–5844. doi: 10.1093/emboj/19.21.5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA. Preprocessing of oligonucleotide array data. Nat Biotechnol. 2004;22:656–658. doi: 10.1038/nbt0604-656b. author reply 658. [DOI] [PubMed] [Google Scholar]
- Yamagishi H, Yamagishi C, Nakagawa O, Harvey RP, Olson EN, Srivastava D. The combinatorial activities of Nkx2.5 and dHAND are essential for cardiac ventricle formation. Dev Biol. 2001;239:190–203. doi: 10.1006/dbio.2001.0417. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.