Abstract
We present an approach for monitoring protein–protein interactions within intact eukaryotic cells, which should increase our understanding of the regulatory circuitry that controls the proliferation and differentiation of cells and how these processes go awry in disease states such as cancer. Chimeric proteins composed of proteins of interest fused to complementing β-galactosidase (β-gal) deletion mutants permit a novel analysis of protein complexes within cells. In this approach, the β-gal activity resulting from the forced interaction of nonfunctional weakly complementing β-gal peptides (Δα and Δω) serves as a measure of the extent of interaction of the non-β-gal portions of the chimeras. To test this application of lacZ intracistronic complementation, proteins that form a complex in the presence of rapamycin were used. These proteins, FRAP and FKBP12, were synthesized as fusion proteins with Δα and Δω, respectively. Enzymatic β-gal activity served to monitor the formation of the rapamycin-induced chimeric FRAP/FKBP12 protein complex in a time- and dose-dependent manner, as assessed by histochemical, biochemical, and fluorescence-activated cell sorting assays. This approach may prove to be a valuable adjunct to in vitro immunoprecipitation and crosslinking methods and in vivo yeast two-hybrid and fluorescence energy transfer systems. It may also allow a direct assessment of specific protein dimerization interactions in a biologically relevant context, localized in the cell compartments in which they occur, and in the milieu of competing proteins.
Specific interactions between proteins in mammalian cells are the basis of many essential biological processes. For example, protein–protein interactions are involved in the assembly of enzymes and other protein homodimers and heterodimers that play important roles in the regulation of intracellular transport pathways, gene expression, receptor–ligand interactions, and in the therapeutic or toxic effects of administered drugs. To increase our understanding of these biological processes, several techniques have been developed for examining the interactions between proteins within cells. Coimmunoprecipitation experiments with antibodies are suggestive of such interactions in that they allow a determination of the affinity of a given protein for another protein, albeit in vitro following cell lysis under conditions that cannot determine whether the two proteins are present within the same compartment or at the concentrations tested (1–3). Methods for crosslinking proteins within the cell and then cofractionating them by chromatography have also proven useful, although purification, sequencing, and identification of the crosslinked proteins can be difficult when they are present in small quantities.
In addition to such biochemical techniques, the yeast two-hybrid system has been extremely useful for detecting and identifying protein–protein interactions in vivo (4–6). This system takes advantage of the properties of the GAL4 protein of the yeast Saccharomyces cerevisiae, a transcriptional activator required for the expression of genes encoding enzymes involved in galactose utilization. The GAL4 protein consists of two separate and identifiable domains, an N-terminal DNA binding domain and a C-terminal transcription activation domain. Separate fusion proteins, each comprising only one of the two GAL4 domains fused to one of two different test polypeptides, interact through affinity of the different test polypeptides, bringing two GAL4 domains into close physical proximity and reconstituting GAL4 function. A distinct advantage of this approach over biochemical approaches is that it allows identification of novel protein partners at a molecular level. However, the system requires that protein–protein interactions occur in the nucleus of a cell leading to transcriptional activation of a reporter gene and the detection of a diffusible product. Thus, the assay is indirect and is dependent on other cellular functions. Nonetheless, numerous previously unknown protein interactions have been identified using the yeast two-hybrid system.
Fluorescence ratio imaging has also been used to study protein interactions in live cells (7). This innovative system has yielded several important new findings. However, it is limited by the requirement that the fluorescent labels on the interacting proteins be sufficiently close to permit efficient energy transfer. Also, the labeled proteins need to be introduced into the cells at relatively high concentrations. Clearly, a method that would allow a direct examination of molecular interactions with fewer size constraints, at the site where they occur within a eukaryotic cell, would be advantageous.
Here we describe a novel application of the bacterial lacZ gene that may allow the direct detection of protein–protein interactions in situ in a range of cell types and species. The product of the lacZ gene, β-galactosidase (β-gal), has been used for many years as a reporter gene to measure transcriptional activity by histochemical or biochemical assays or by live cell sorting (8–11). A property of the lacZ gene, intracistronic complementation, has been known and studied for many years in prokaryotes (12–14), but has only recently been adapted for use in eukaryotes (15). Pairs of inactive β-gal deletion mutants are capable of complementing one another in trans and assembling to form an active enzyme. Pairs of deletion mutants that complement with either high or low efficiency were identified (15, 16). In previous studies of myoblast fusion, the most efficient complementing pair was employed to analyze the putative role of cell adhesion and signal transduction molecules in this poorly understood process (17).
As reported here, pairs of inactive β-gal deletion mutants were used to produce chimeric proteins to test whether the complemented enzyme could serve as a marker of other protein–protein interactions (see Fig. 1). Several features were critical in the application of this approach. Most important was the necessity to select β-gal mutants with sufficiently low affinity for each other so that they monitored rather than drove the association of the test proteins. Our biochemical, histochemical, and fluorescence-activated cell sorting (FACS) experiments show that appropriate β-gal mutants can be incorporated into fusion proteins that serve as tracers of protein–protein interactions in intact eukaryotic cells.
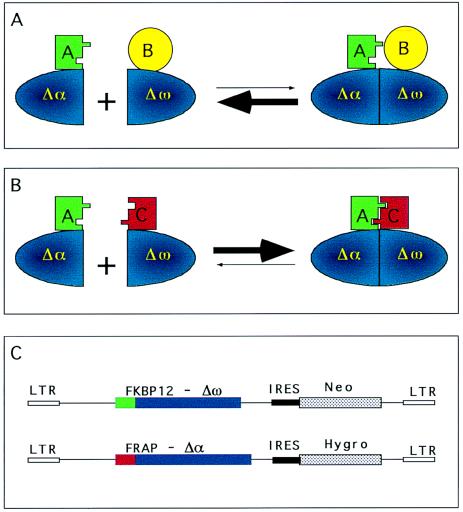
Figure 1.
Experimental design. (A) When the Δα and Δω β-gal mutants are fused to proteins that do not dimerize, their association is not favored and β-gal activity is not detected. (B) When the Δα and Δω β-gal mutants are fused to proteins that can dimerize, the formation of active β-gal is favored. (C) Schematic representation of the FKBP12-Δω-Neo and the FRAP-Δα-Hygro constructs. IRES, internal ribosome entry sequence; LTR, long terminal repeat.
MATERIALS AND METHODS
Construction of pWZL FRAP-Δα and pWZL FKBP12-Δω Viruses.
To be able to fuse the sequences coding for FKBP12 and the FKBP12–rapamycin binding domain in-frame with β-gal, an adaptor oligonucleotide (CATGGAGCTCGAGAG) containing an XhoI site was inserted in the NcoI site at the ATG of the previously described Δα and Δω β-gal mutants (15). Two XhoI–SalI DNA fragments corresponding to amino acids 2025–2114 of human FRAP and to the complete coding sequence of human FKBP12 (kind gifts of Gerald Crabtree, Stanford University School of Medicine) were cloned into the XhoI site of the modified Δα and Δω mutants, generating FRAP-Δα and FKBP12-Δω. For both constructs, conservation of the appropriate reading frame was confirmed by sequencing.
To insert the FRAP-Δα and FKBP12-Δω coding sequences in pWZL-neo and pWZL-hygro retroviruses (P. J. Morgenstern, unpublished work), an adaptor oligonucleotide containing NcoI and BamHI sites (GATCACCATGGACGCGTGGATCCC) was inserted in the BamHI and XhoI sites of the pWZL vectors. Both of the original sites were destroyed by this insertion. The FRAP-Δα and FKBP12-Δω coding sequences were then inserted in the modified pWZL vectors as NcoI–BamHI fragments.
Virus Production and Infection Protocol.
Proviral constructs were introduced into the Phoenix ecotropic packaging cells (P. L. Achacoso and G. P. Nolan, unpublished work) by calcium phosphate transfection. The media-containing retrovirus from the packaging cells was harvested 24–72 hr after transfection and used to infect C2C12 myoblasts (18) in the presence of 8 μg/ml polybrene (Sigma). Singly and doubly infected cells were selected with the appropriate drugs. Both G418 and hygromycin were used at a final concentration of 1 mg/ml. The selected cells were expanded as populations for subsequent experiments.
Quantitation of β-Gal Activity.
Unless otherwise stated, rapamycin (Calbiochem) was used at a concentration of 10 ng/ml.
Biochemical quantitation of β-gal activity.
β-gal activity was measured by chemiluminescence as described (15). Briefly, cells cultured in microtiter plates were lysed in situ in 50 μl of a 1:1 mixture of lysis and assay buffers containing Galacton Plus substrate from the Galacto-Light Plus assay kit (Tropix, Bedford, MA). Reactions were terminated after 1 hr at room temperature. After addition of Light Emission Accelerator solution, luminescence was measured using a MicroBeta 1450 scintillation counter (Wallac, Gaithersburg, MD).
Histochemical detection of β-gal using fluorescence histochemistry (Fluor-X-Gal).
Cells were processed as described (15). Briefly, cells grown on glass coverslips were fixed in 4% paraformaldehyde in PBS and rinsed twice with PBS. For triple-labeling (actin, β-gal, and nuclei), fixed cells were first stained for actin by incubation in a solution of 1 μM biotin-XX-phalloidin (Molecular Probes) for 30 min at room temperature, followed by PBS rinses and incubation in Cy5-labeled streptavidin (Amersham) (1:250 in PBS, 30 min, room temperature). β-gal was detected by incubation of fixed cells in a solution of 25 μg/ml 5-bromo-6-chloro-3-indolyl β-d galactopyranoside (Fluka) plus 100 μg/ml fast red violet LB (Sigma) in PBS for 45 min at 37°C. Coverslips were rinsed 4 times with PBS, nuclei labeled with Hoechst 33258 (Calbiochem; 1:10,000 in PBS), rinsed again, mounted in PBS, and sealed to glass slides. Triple-labeled deconvolved images were collected using a DeltaVision deconvolution microscope (Applied Precision, Mercer Island, WA). Images were represented in false color using Adobe Photoshop software. Fluor-X-Gal staining can be detected with either fluorescein or rhodamine filters; in this case, detection was with the rhodamine filter, and is depicted in green (see Fig. 3 C and D). Cy5 was detected with a Cy5 filter and is depicted in red; Hoechst was detected with a DAPI filter and is depicted in blue (see Fig. 3 C and D). Double-labeled cells were photographed using a Zeiss Axiophot fluorescence microscope. Fluor-X-Gal staining was detected with a rhodamine filter and appears red; Hoechst was detected with a UV filter (see Fig. 3 A and B).
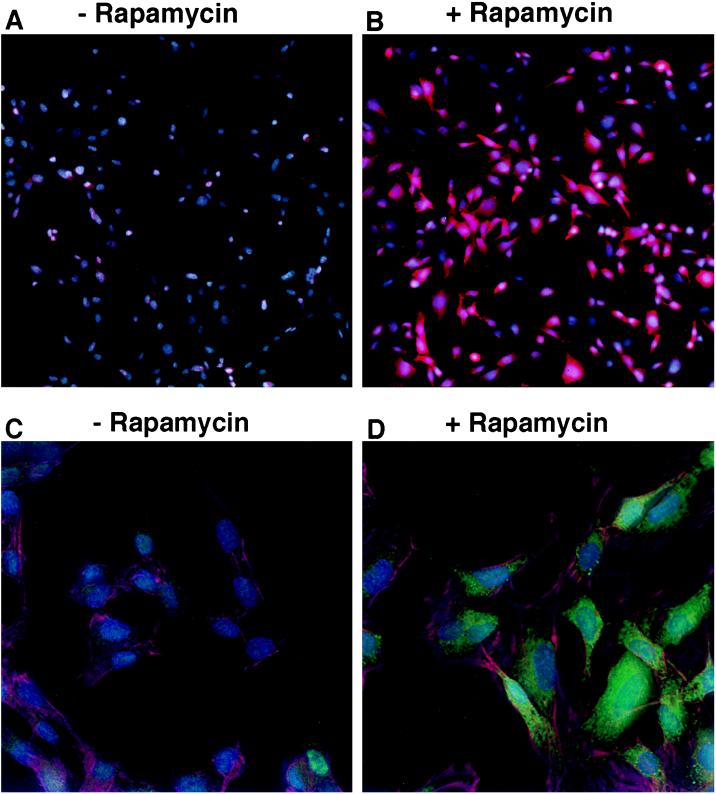
Figure 3.
Histochemical assay of induction of β-gal activity upon chimera complementation. C2C12 cells expressing both FKBP12-Δω and FRAP-Δα were maintained overnight either in the absence (A and C) or the presence (B and D) of 10 ng/ml rapamycin. β-gal activity was visualized by fluorescence microscopy using Fluor-X-Gal as substrate. (A and B) Double-labeled samples showing Hoechst stained nuclei (blue) and β-gal activity using Fluor-X-Gal as substrate viewed with a rhodamine filter set (red). (C and D) Triple-labeled samples obtained by imaging with a DeltaVision microscope showing β-gal activity using Fluor-X-Gal as substrate (green), Hoechst stained nuclei (blue), and Cy5-labeled actin filaments (red) to visualize the contour of each cell at higher magnification.
FACS.
β-gal expression in live cells was determined on a FACS as described (10), except that the fluorescein di-β-d-galactopyranoside substrate was used at a concentration of 1 mM.
RESULTS
Design of Fusion Protein Test System for Monitoring Interactions Using Complementing β-Gal Mutants.
We have recently adapted lacZ intracistronic complementation for use in eukaryotic cells (15). For the purpose of testing the potential application of β-gal complementation as a method to monitor protein–protein interactions, a pair of mutants was selected that contained the domains postulated to be necessary for trans-complementation, but which were impaired in their ability to restore enzymatic activity upon coexpression in mammalian cells. These mutants were the classical ω donor M15 (hereafter referred to as Δα), which lacks amino acids 11–41 of the wild-type molecule, and an unusually long α donor containing the first 788 amino acids of β-gal (Δω) (15). Since both polypeptides are capable of efficiently complementing a third deletion mutant (Δμ, lacking 553 amino acids in the central portion of the molecule), it seemed unlikely that their poor ability to complement each other was due to instability or to low expression. Inefficient α donors have long been known to exist in prokaryotes, and differences in their folded structure that render the α domain unavailable for complementation have been proposed as a basis for their biochemical properties (16). One model separates the β-gal complementation mechanism into two steps: rapid formation of a complex between α and ω donor peptides, followed by a slow conformational change from an inactive to an active conformation (19, 20). Which of these two steps was impaired in inefficient α donors was unclear. We reasoned that if complex formation but not catalytic activity were impaired, it should be possible to obtain efficient complementation by promoting the heterodimerization of weakly complementing Δα and Δω β-gal mutants.
To test this hypothesis, we used a well-characterized protein complex (21–24). The intracellular rapamycin binding protein, FK506-binding protein-12 (FKBP12), interacts with the intracellular FKBP–rapamycin associated protein (FRAP) only when rapamycin is present in the culture medium. This interaction is well documented to increase over time and with the dose of rapamycin. Rapamycin is a small, cell-permeable molecule that binds to the two intracellular proteins via independent determinants. Since rapamycin is unable to bind two FKBP12 molecules at the same time and FRAP only binds rapamycin within the FKBP12-rapamycin complex, heterodimers do not form unless rapamycin is present (25).
Specifically, the test system involved combining the weakly complementing β-gal mutants, Δα or Δω, with the FKBP12/FRAP/rapamycin system by producing fusion proteins. Two different retroviral constructs were designed that encoded fusion proteins of either the FKBP12–rapamycin binding domain of FRAP or the entire FKBP12 peptide together with the β-gal Δα or Δω mutants, respectively (FRAP-Δα and FKBP12-Δω). A prediction of the hypothesis is that introduction of these fusion proteins into cells would result in negligible β-gal activity in the absence of rapamycin (Fig. 1A). Thus, β-gal activity would first become detectable upon addition of drug. As a result, β-gal activity would monitor the association of FRAP and FKBP12 proteins (green and red proteins and cDNAs in Fig. 1 B and C, respectively).
A critical feature of the design of the system presented here was to reduce protein expression levels as much as possible to avoid perturbing the intracellular protein milieu. Accordingly, the fusion proteins should serve as “tracers” of naturally occurring protein–protein interactions, overcoming the problems previously experienced upon overexpression of introduced proteins either by transient or stable transfection, or transduction with retroviruses. To reduce expression levels, in the experiments reported here the cDNAs encoding FRAP-Δα and FKBP12-Δω were not inserted into MFG, but into the pWZL retroviral vector (P. J. Morgenstern, unpublished work), upstream of the gene encoding hygromycin or neomycin, respectively. The levels of protein expression are reduced in the pWZL vector due to the presence of mutations that delete the splice donor/acceptor sequences upstream of the ATG of the fusion proteins. These mutations result in a lower translation efficiency of the first coding sequence of a bicistronic message, but do not affect the translation of the second sequence, in this case the selectable marker, which is solely dependent on an encephalomyocarditis virus internal ribosomal entry sequence. Using a pWZL vector, 50% less of the upstream protein is expressed compared with vectors containing wild-type splice donor/acceptor sequences (data not shown). As a result of the reduced levels of expression the frequency of spontaneous interactions of β-gal mutants, which is concentration dependent, should be significantly reduced.
Induction of β-Gal Activity on Coexpression of FRAP-Δα and FKBP12-Δω Fusion Proteins in the Presence of Rapamycin.
The FRAP-Δα-Neo and FKBP12-Δω-Hygro vectors were tested in an established line of myoblasts, C2C12 (18). Infectious viral particles were produced by transient transfection of each construct into the Phoenix packaging cell line (P. L. Achacoso and G. P. Nolan, unpublished work). C2C12 myoblasts were infected either singly with each retrovirus alone or simultaneously with both. All experiments were performed after selection with hygromycin and G418 to ensure that 100% of the cells contained the constructs.
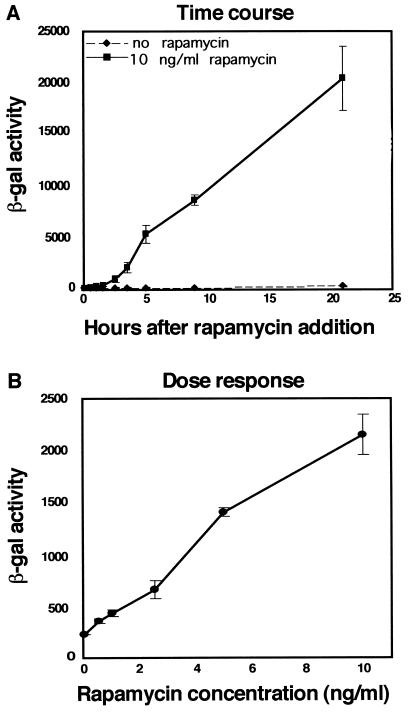
β-gal activity was measured in biochemical assays and found to be dependent on the presence of rapamycin in the medium. C2C12 cells expressing both fusion proteins were plated in replicate in 96-well plates. Rapamycin was added to the culture medium, and the β-gal activity was measured at different time points. For each time point, six replicate samples were assayed with a sensitive chemiluminescence assay, as described (15). In untreated control samples, no β-gal activity was detected above background. Rapamycin at a concentration of 10 ng/ml induced a 30-fold increase in β-gal activity within 5 hr. After 5 hr, β-gal activity continued to increase, reaching a level 2 orders of magnitude above background within 20 hr (Fig. 2A). In control populations of cells expressing only one of the two constructs, β-gal activity did not increase above background when rapamycin was added (data not shown). The linear increase in β-gal activity observed between 5 and 20 hr after rapamycin addition may be due to an increase in protein concentration resulting from cell proliferation, since the C2C12 cells have a doubling time of 12 hr. It is also possible that α donor peptides are stabilized when they are incorporated into a multimeric complex. In support of the latter possibility, Western blotting of cellular extracts with antibodies to β-gal revealed an increase in steady-state levels of β-gal mutant peptides upon rapamycin treatment (data not shown); in addition, stabilization of complementing β-gal peptides upon assembly of active multimeric enzymes has been reported by others (26).
Figure 2.
Biochemical assay of induction of β-gal activity upon chimera complementation. (A) Kinetics of induction of β-gal activity upon treatment with rapamycin. Pure populations of C2C12 cells stably expressing both FKBP12-Δω and FRAP-Δα were plated in 96-well plates and 10 ng/ml rapamycin was added at time zero. Cells were then lysed at different time intervals thereafter, and the β-gal activity in the lysates was quantitated by chemiluminescence. (B) Dose response of β-gal activity upon rapamycin treatment. C2C12 cells expressing both FKBP12-Δω and FRAP-Δα were plated in 96-well plates and treated with different concentrations of rapamycin for 3.5 hr. β-gal activity is expressed as luminescence counts per second. Each point represents the average of six replicate samples. Error bars indicate standard deviations from the mean.
In Fig. 2B, the dose response curve is shown. β-gal activity increased linearly with the dose of rapamycin in the 0–10 ng/ml range. This linearity suggests that β-gal enzymatic activity can serve as a reporter to quantitate protein–protein interactions. The results of these experiments demonstrate that the interaction of the FKBP12- and FRAP-β-gal fusion proteins in the presence of rapamycin is specific and exhibits a comparable dose-response curve to results previously obtained by others for the FKBP12/FRAP/rapamycin complex alone (25). Thus, fusion to β-gal peptides does not interfere with the interaction of the FKBP12 and FRAP proteins. Moreover, endogenous FKBP12 and FRAP proteins are ubiquitously expressed and will interact in the presence of rapamycin, thereby competing with the introduced FRAP-Δα and FKBP12-Δω fusion proteins, yet not generating β-gal activity. Our results indicate that productive FRAP-Δα and FKBP12-Δω dimers will also form, generating β-gal activity that will reflect the interaction of FRAP with FKBP12-rapamycin in that cellular environment even in the presence of the competing endogenous proteins. This finding suggests that it will be possible to use β-gal complementation as a tool to analyze protein–protein interactions generally.
β-gal activity was assayed using the sensitive Fluor-X-Gal histochemical stain, which allows simultaneous tricolor fluorescence analysis, as described (15). Because of its broad emission spectrum, the fluorescence of this substrate can be visualized at wavelengths that yield either a red (Fig. 3 A and B) or a green signal (Fig. 3 C and D). In the absence of rapamycin, only a very weak β-gal fluorescence was observed. For example, in Fig. 3A, in which two fluorochromes were used at low magnification, the blue nuclei of the myoblasts was primarily evident with little evidence of red β-gal activity. Similarly, in Fig. 3C, in which three fluorochromes were used at higher magnification, blue nuclei and red phalloidin-staining of actin were evident, but green β-gal activity was barely detectable. In both histochemical assays, in the presence of 10 ng/ml rapamycin, the activity of β-gal increased significantly to produce an intense stain that was primarily localized in the cytoplasm, as expected (Fig. 3 B and D). Detection of the complemented β-gal enzyme was enhanced by imaging with a deconvolution microscope (Fig. 3D). These images show that protein complexes forming outside the nucleus are readily detectable using this assay.
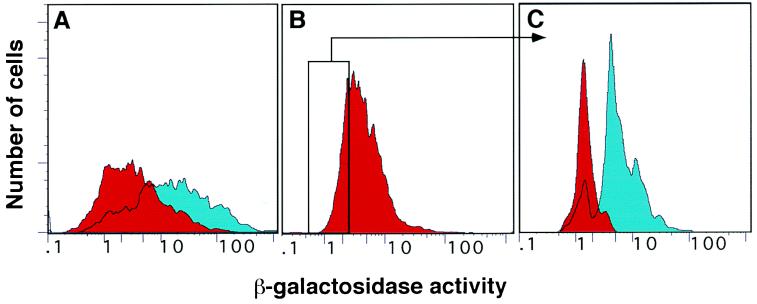
The β-gal activity of a population of cells was assayed in the presence and absence of 10 ng/ml rapamycin by FACS. Using this sensitive assay, we were able to detect increased β-gal activity in most of the cells after only 90 min of rapamycin treatment (Fig. 4A). A range of expression levels was seen, as evidenced by the breadth of the peak of emission in the presence and absence of the drug (Fig. 4A, blue and red profiles). This breadth is presumably due to variable efficiency of expression of each of the retroviral vectors following integration in the target cell. This inference is supported by the finding that when the 25% of the cells expressing the lowest β-gal activity in the absence of rapamycin were collected (Fig. 4B) and then reassayed in the presence and absence of rapamycin, a clear distinction between the two populations was seen (Fig. 4C). Thus, nonoverlapping populations of cells that do or do not express complementing fusion proteins can be identified and isolated by FACS.
Figure 4.
FACS analysis of induced β-gal activity upon chimera complementation. The red peaks represent the untreated samples and the blue peaks represent samples treated with 10 ng/ml rapamycin. (A) Induction of β-gal activity in a population of C2C12 cells expressing both FKBP12-Δω and FRAP-Δα after 90 min of rapamycin treatment. The majority of the cells respond to rapamycin treatment with an increase in β-gal activity. (B) Subpopulation of cells selected on the basis of low β-gal activity in uninduced conditions. (C) The same population was maintained overnight in the absence (red peak) or in the presence (blue peak) of rapamycin. The induced and uninduced populations yield essentially nonoverlapping peaks. The vertical axis represents relative cell number and the horizontal axis represents intensity of β-gal fluorescence on a logarithmic scale.
Rapamycin-Dependent Induction of β-Gal Activity in Cell Lysates.
To test whether the heterodimerization of FRAP-Δα and FKBP12-Δω induced by rapamycin required cotranslational assembly or could occur with fully synthesized and folded proteins, β-gal activity was assayed in a cell free system. Cells expressing both constructs were grown in the absence of rapamycin and lysed in situ in 50 μl of a 1:1 mixture of lysis and assay buffers containing Galacton Plus substrate from the Galacto-Light Plus assay kit (Tropix). Rapamycin was then added to the lysates, and β-gal activity was quantitated immediately, 1 and 3 hr later. As a control, β-gal activity was measured in a parallel set of lysates that were not exposed to rapamycin at all. No statistically significant increase in β-gal activity was detected in the samples that did not receive rapamycin. By contrast, a more than 2-fold increase in β-gal activity was observed in the rapamycin-treated lysates 1 hr after drug administration. The increase in activity detected in lysates is only a fraction of the increase observed upon rapamycin treatment of intact cells, a finding that very likely reflects the lower concentration of FRAP-Δα and FKBP12-Δω in the lysates. This is probably due to the combined effects of the instability of β-gal mutants within lysates and of their dilution during lysis. Nevertheless, these experiments demonstrate that de novo synthesis is not required for complementation and that folded proteins can be induced to form complexes that can be monitored by β-gal activity.
DISCUSSION
Prerequisites for Monitoring Protein–Protein Interactions by β-Gal Complementation.
We have shown that β-gal activity can be used to monitor the interaction of chimeric proteins. Critical to the success of this system was the choice of two poorly complementing β-gal mutants, since strongly complementing mutants spontaneously assemble and produce functional β-gal activity detectable in the absence of any other protein constituents (15). By contrast, the weakly interacting Δα or Δω β-gal mutants expressed from the pWZL vector do not yield detectable enzymatic activity, unless their local concentration is increased by forcing them to heterodimerize. This is only achieved by synthesizing them as fusion proteins in which the non-β-gal portions of the chimeras have sufficient affinity to drive the interaction.
The molecular basis of the impaired ability of the Δα and Δω mutants to associate spontaneously and recreate active β-gal is unclear. Potential insights derive from prokaryotic studies and the recently published crystal structure of intact β-gal (27). It is well known that efficient β-gal complementation in prokaryotes requires intermolecular interactions resulting in the sharing of domains between α acceptor and ω donor mutants (20). Moreover, the recently published structure of wild-type β-gal suggests that both the α and ω domains are involved in a dimerization step critical to the assembly of functional enzymes. These domains are also involved in essential contacts with the relatively large central region of the molecule (27). Δα β-gal can be efficiently and spontaneously complemented in mammalian cells by an α donor lacking the central μ portion of the molecule (15). Similarly, Δω β-gal can complement spontaneously with an ω donor lacking the central domain. In contrast, the weakly complementing mutant pairs used here each contain a large intact central μ domain. Thus, one possible explanation for the observed lack of spontaneous complementation between the Δω and the Δα peptides is that the presence of the μ domain in both mutants may diminish their affinity for each other by steric hindrance or by sequestering either the α or the ω domain in an intramolecular interaction. Accordingly, we postulate that the lack of spontaneous assembly and generation of β-gal activity can be overcome by increasing the local concentration of Δα and Δω by forced association of these two mutants in chimeras, thereby counteracting the potential inhibitory effect imposed by the presence of two central domains. Testing of this model will require further mutagenesis, activity assays, and x-ray crystallographic analysis of the resulting structures.
Irrespective of the precise molecular nature of the interactions, the data presented here provide strong evidence that by engineering constructs in which domains or proteins of interest drive the dimerization of Δα or Δω β-gal mutants, it will be possible to monitor such interactions by measuring the level of β-gal activity following coexpression of these fusion proteins in intact cells. Moreover, the present system should theoretically allow detection of complexes in subcellular compartments, including the nucleus, the cytoplasm, or the membrane. Finally, as shown here, protein dimerization can be monitored in the context of the cell in the presence of endogenous competing protein partners.
Potential for the Development of a “Mammalian Two-Hybrid” System.
The experiments described in this report show that two distinct β-gal mutants that do not readily assemble can be forced to interact and yield significantly increased levels of active enzyme. This is achieved by coupling each mutant with one of a pair of highly interactive proteins, as chimeras or fusion proteins. In this case, we have used FRAP and FKBP12, proteins that only interact in the presence of a small molecule, rapamycin. Using this tripartite complex, we were able to show that the affinity of the non-β-gal proteins drove the interaction and that the β-gal components generated enzymatic activity as a result, serving to monitor the extent of that interaction. The levels of β-gal activity in the presence and absence of forced dimerization were clearly separable by both biochemical and FACS assays, suggesting that this system could be used to screen for as yet unidentified protein partners. The target protein fused to a complementing β-gal mutant (bait) could be stably expressed in a well-characterized cell line. Expression libraries containing cDNAs fused to a β-gal deletion mutant could be introduced into these cells with high efficiency using retroviral vectors (28). Finally, gene products that interact with the bait could be isolated by identifying β-gal positive clones. A potential advantage of this system over systems described previously is that the screen could be carried out in any cell type with its own particular milieu of competing resident proteins. An attractive possibility is that the bait could be targeted to a given cellular compartment, with the aim of identifying proteins involved in interactions restricted to that specific location. This mammalian “two-hybrid” screen could also be carried out in the presence of extracellular signaling molecules, growth factors, or differentiation factors, that might alter the potential for dimerization of two given proteins in particular cell types. Taken together, this approach to the study of protein–protein interactions should greatly enhance our understanding of the development of diverse cell types and organisms and how that development goes awry.
Acknowledgments
We are grateful to Bruce Blakely for constructive comments on the manuscript, to Oivin Guicherit and Norris Turner for technical advice, and to Gerald Crabtree for useful discussions. F.R. was supported by a long-term postdoctoral fellowship from the Human Frontiers in Science Program. H.M.B. is the recipient of an National Institutes of Health MERIT award.
ABBREVIATIONS
- β-gal
β-galactosidase
- FACS
fluorescence-activated cell sorting
- FKBP12
FK506-binding protein-12
- FRAP
FKBP–rapamycin associated protein
References
- 1.Gu W, Schneider J W, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 2.Halevy O, Novitch B G, Spicer D B, Skapek S X, Rhee J, Hannon G J, Beach D, Lassar A B. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 3.Schneider J W, Gu W, Zhu L, Mahdavi V, Nadal-Ginard B. Science. 1994;264:1467–1471. doi: 10.1126/science.8197461. [DOI] [PubMed] [Google Scholar]
- 4.Bai C, Elledge S J. Methods Enzymol. 1996;273:331–347. doi: 10.1016/s0076-6879(96)73029-x. [DOI] [PubMed] [Google Scholar]
- 5.Fields S, Song O. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 6.Chien C T, Bartel P L, Sternglanz R, Fields S. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams S R, Harootunian A T, Buechler Y J, Taylor S S, Tsien R Y. Nature (London) 1991;349:694–697. doi: 10.1038/349694a0. [DOI] [PubMed] [Google Scholar]
- 8.Lis J T, Simon J A, Sutton C A. Cell. 1983;35:403–410. doi: 10.1016/0092-8674(83)90173-3. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen D A, Chou J, MacKrell A J, Casadaban M J, Steiner D F. Proc Natl Acad Sci USA. 1983;80:5198–5202. doi: 10.1073/pnas.80.17.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nolan G P, Fiering S, Nicolas J F, Herzenberg L A. Proc Natl Acad Sci USA. 1988;85:2603–2607. doi: 10.1073/pnas.85.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price J, Turner D, Cepko C. Proc Natl Acad Sci USA. 1987;84:156–160. doi: 10.1073/pnas.84.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ullmann A, Jacob F, Monod J. J Mol Biol. 1967;24:339–343. doi: 10.1016/0022-2836(67)90341-5. [DOI] [PubMed] [Google Scholar]
- 13.Ullmann A, Jacob F, Monod J. J Mol Biol. 1968;32:1–13. doi: 10.1016/0022-2836(68)90140-x. [DOI] [PubMed] [Google Scholar]
- 14.Ullmann A, Perrin D, Jacob F, Monod J. J Mol Biol. 1965;12:918–923. doi: 10.1016/s0022-2836(65)80338-2. [DOI] [PubMed] [Google Scholar]
- 15.Mohler W A, Blau H M. Proc Natl Acad Sci USA. 1996;93:12423–12427. doi: 10.1073/pnas.93.22.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villarejo M, Zamenhof P J, Zabin I. J Biol Chem. 1972;247:2212–2216. [PubMed] [Google Scholar]
- 17.Charlton, C. A., Mohler, W. A., Radice, G. L., Hynes, R. O. & Blau, H. M. (1997) J. Cell Biol., in press. [DOI] [PMC free article] [PubMed]
- 18.Blau H M, Chiu C P, Webster C. Cell. 1983;32:1171–1180. doi: 10.1016/0092-8674(83)90300-8. [DOI] [PubMed] [Google Scholar]
- 19.Langley K E, Zabin I. Biochemistry. 1976;15:4866–4875. doi: 10.1021/bi00667a018. [DOI] [PubMed] [Google Scholar]
- 20.Zabin I. Mol Cell Biochem. 1982;49:87–96. doi: 10.1007/BF00242487. [DOI] [PubMed] [Google Scholar]
- 21.Brown E J, Albers M W, Shin T B, Ichikawa K, Keith C T, Lane W S, Schreiber S L. Nature (London) 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Zheng X F, Brown E J, Schreiber S L. Proc Natl Acad Sci USA. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belshaw P J, Ho S N, Crabtree G R, Schreiber S L. Proc Natl Acad Sci USA. 1996;93:4604–4607. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi J, Chen J, Schreiber S L, Clardy J. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 25.Ho S N, Biggar S R, Spencer D M, Schreiber S L, Crabtree G R. Nature (London) 1996;382:822–826. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- 26.Lin S, Zabin I. J Biol Chem. 1972;247:2205–2211. [PubMed] [Google Scholar]
- 27.Jacobson R H, Zhang X J, DuBose R F, Matthews B W. Nature (London) 1994;369:761–766. doi: 10.1038/369761a0. [DOI] [PubMed] [Google Scholar]
- 28.Kitamura T, Onishi M, Kinoshita S, Shibuya A, Miyajima A, Nolan G P. Proc Natl Acad Sci USA. 1995;92:9146–9150. doi: 10.1073/pnas.92.20.9146. [DOI] [PMC free article] [PubMed] [Google Scholar]