Abstract
Therapy of chronic hepatitis B virus (HBV) infection with the polymerase inhibitor lamivudine frequently is associated with the emergence of viral resistance. Genotypic changes in the YMDD motif (reverse transcriptase [rt] mutations rtM204V/I) conferred resistance to lamivudine as well as reducing the in vitro replication efficiency of HBV. A second mutation, rtL180M, was previously reported to partially restore replication fitness as well as to augment drug resistance in vitro. Here we report the functional characterization of a third polymerase mutation (rtV173L) associated with resistance to lamivudine and famciclovir. rtV173L was observed at baseline in 9 to 22% of patients who entered clinical trials of adefovir dipivoxil for the treatment of lamivudine-resistant HBV. In these patients, rtV173L was invariably found as a third mutation in conjunction with rtL180M and rtM204V. In vitro analyses indicated that rtV173L did not alter the sensitivity of wild-type or lamivudine-resistant HBV to lamivudine, penciclovir, or adefovir but instead enhanced viral replication efficiency. A molecular model of HBV polymerase indicated that residue rtV173 is located beneath the template strand of HBV nucleic acid near the active site of the reverse transcriptase. Substitution of leucine for valine at this residue may enhance polymerization either by repositioning the template strand of nucleic acid or by affecting other residues involved in the polymerization reaction. Together, these results suggest that rtV173L is a compensatory mutation that is selected in lamivudine-resistant patients due to an enhanced replication phenotype.
Until the recent approval of adefovir dipivoxil, lamivudine (a dideoxycytidine analog in the unnatural l configuration) was the only approved oral therapy for the treatment of chronic hepatitis B. Antiviral therapy for chronic hepatitis B with famciclovir and lamivudine has been limited by the emergence of viral resistance in significant proportions of patients. Although lamivudine therapy results in potent reductions in viremia, relapse is common, as resistant viruses emerge in approximately 24% of patients after 1 year of therapy and 70% after 4 years of therapy (20). Sequencing of hepatitis B virus (HBV) isolates from patients for whom lamivudine treatment failed revealed a mutation of methionine to valine or isoleucine at position rt204 (rtM204V/I) in the YMDD motif of the C subdomain of HBV polymerase (3, 21); amino acid residues in HBV polymerase are numbered according to the consensus nomenclature developed by Stuyver et al. (34). A second mutation, of leucine 180 to methionine (rtL180M), in the upstream B subdomain of HBV polymerase frequently accompanies rtM204 mutations. The rtM204V mutation almost invariably occurs in tandem with rtL180M, while rtM204I can occur as a single mutation or in conjunction with rtL180M.
In vitro analyses have confirmed and characterized the role of the major HBV polymerase mutations in lamivudine resistance. Cell culture and enzyme assays have revealed that rtM204V/I mutations are sufficient to confer resistance to lamivudine and structurally related inhibitors (reviewed in reference 13). A molecular model of HBV polymerase (based on the crystal structure of human immunodeficiency virus [HIV] reverse transcriptase [RT]) suggested that the introduction of the β-methyl side chain of either valine or isoleucine at position rt204 creates a steric barrier to the binding of lamivudine triphosphate (12). In addition to conferring drug resistance, single rtM204V/I mutations also reduce the replication of HBV in vitro (22, 27, 28). In vitro investigation of the L180M mutation indicated that it plays a dual role in resistance by augmenting the levels of lamivudine resistance and enhancing the replication fitness of rtM204V mutant virus (1, 22, 28). Several other mutations, including rtL80V/I, rtL82M, rtF166L, rtV173L, and rtA200V, have also been reported during lamivudine therapy (reviewed in reference 15). However, in most instances, these mutations occur at relatively low frequencies and have not been characterized in vitro, and their contributions to drug resistance remain unclear.
Famciclovir, an oral prodrug of the deoxyguanosine analog penciclovir, underwent clinical trials for the treatment of chronic HBV infection but was abandoned due to limited efficacy and the frequent emergence of resistance (16, 17, 35). Nevertheless, famciclovir has received significant use, particularly in the transplant setting, where patients had few treatment options prior to the development of adefovir dipivoxil. Following famciclovir treatment failure, a variety of substitutions were found in HBV polymerase, including several within the conserved subdomains of the RT domain. Viral resistance to famciclovir, unlike that to lamivudine, does not map to a singular common locus (17, 33). The rtL180M mutation, which is also selected by lamivudine, confers clinical resistance to famciclovir and in vitro resistance to penciclovir. A distinct mutation downstream of the YMDD motif, rtV207I, also appears to be sufficient to confer resistance to penciclovir in vitro (30, 38). Mutation of valine 173 to leucine (rtV173L) has been observed during famciclovir treatment failure, as well as in lamivudine-resistant patients (2); however, the role of this mutation in drug resistance has not been established.
Adefovir is a deoxyadenosine monophosphate analog that has in vitro activity against hepadnaviruses, HIV, and herpesviruses. Preclinical analyses with enzyme, cell culture, and animal models have demonstrated that adefovir is a potent inhibitor of wild-type and lamivudine-resistant HBV. Adefovir dipivoxil, an oral prodrug of adefovir, was recently approved in the United States and European Union for the treatment of chronic hepatitis B. In contrast to the situation for lamivudine and famciclovir, the emergence of resistance to adefovir dipivoxil has been infrequent and delayed. No adefovir resistance mutations were identified in 467 chronic hepatitis B patients treated with adefovir dipivoxil for 48 weeks during phase III clinical studies or in a smaller cohort of 27 patients treated for up to 136 weeks (37, 39). Recently, a novel HBV polymerase mutation, rtN236T, was reported to have emerged in 2 of 124 patients (1.6%) who received 96 weeks of adefovir dipivoxil therapy (S. Locarnini, H. Yang, C. E. Westland, W. E. Delaney IV, D. Colledge, A. Bartholomeusz, V. Thibault, Y. Benhamou, P. Angus, G. Kitis, M. Wulfsohn, C. S. Gibbs, J. Fry, C. Brosgart, and S. Xiong, International Symposium on Viral Hepatitis and Liver Disease, Sydney, Australia, p. 117, 2003); this mutation was subsequently confirmed to have conferred decreased susceptibility to adefovir in vitro.
During enrollment of lamivudine-resistant patients into clinical trials of adefovir dipivoxil, we observed the rtV173L mutation in baseline viral sequences from many patients. Here we report a retrospective examination of lamivudine-resistant patients to determine the frequency of the rtV173L mutation and its association with mutations at positions rt180 and rt204. To determine the role of rtV173L in viral resistance, we examined its effects on antiviral sensitivity and replication fitness in vitro. In addition, molecular modeling was used to establish the location of the mutation relative to the active site of HBV polymerase and to provide a potential explanation for the phenotype of rtV173L mutant HBV.
MATERIALS AND METHODS
Patients and baseline HBV genotyping.
Patients (n = 216) entering three trials of adefovir dipivoxil for the treatment of lamivudine-resistant HBV were studied. Study GS-98-435 included chronically infected liver transplant patients for whom lamivudine therapy failed; 122 patients were available from this study at the time of analysis. Study GS-99-460i included HIV-coinfected chronic hepatitis B patients for whom lamivudine therapy failed; 35 patients were available for analysis. Study GS-00-461 included 59 patients for investigation of the efficacy of lamivudine, adefovir dipivoxil, or the combination of lamivudine plus adefovir dipivoxil in chronic hepatitis B patients for whom lamivudine therapy failed. PCR amplification of the HBV RT domain and automated DNA sequencing were used to determine baseline viral genotypes as described previously (4, 40).
Cell culture.
The HepG2 hepatoblastoma cell line was obtained from the American Type Culture Collection (Manassas, Va.) and grown in minimal essential medium (American Type Culture Collection) supplemented with 10% heat-inactivated fetal bovine serum, 100 U of penicillin/ml, and 10 μg of streptomycin/ml (all supplements were obtained from Irvine Scientific, Santa Ana, Calif.). HepG2 cells were maintained in a humidified incubator at 37°C with 5% CO2.
Generation of HBV polymerase mutants.
The plasmid pFBHBV-wt encodes a wild-type, terminally redundant (1.3-unit-length), replication-competent HBV genome (genotype A; GenBank accession number AF305422). The rtV173L, rtL180M, and rtM204V mutations were introduced into pFBHBV-wt by site-directed mutagenesis with a QuikChange kit (Stratagene, La Jolla, Calif.) and the primers listed in Table 1. Constructs encoding the rtL180M and rtV173L mutations were generated by a single round of mutagenesis with the appropriate primers. The rtL180M-rtM204V double mutant was created by a second round of mutagenesis starting with the rtL180M single mutant. A third round of mutagenesis was used to create the triple mutant by introducing rtV173L into the rtL180M-rtM204V double mutant. The RT domains of all constructs were sequenced to confirm that no additional mutations had been introduced.
TABLE 1.
Primer sequences for introduction of the rtV173L, rtL180M, and rtM204V mutations
| Mutation | Primer | Sequence (5′→3′)a |
|---|---|---|
| rtV173L | HBV77a | AAA ATA CCT ATG GGA TTG GGC CTC AGT CCG TTT |
| HBV78 | AAA CGG ACT GAG GCC CAA TCC CAT AGG TAT TTT | |
| rtL180M | HYHBV095 | AGT CCG TTT CTC ATG GCT CAG TTT ACT |
| HYHBV096 | AGT AAA CTG AGC CAT GAG AAA CGG ACT | |
| rtM204V | HYHBV093 | GCT TTC AGC TAT GTG GAT GAT GTG GTA |
| HYHBV094 | TAC CAC ATC ATC CAC ATA GCT GAA AGC |
Bold sequences indicate the mutated polymerase codon.
Antiviral compounds.
Adefovir {9-[2-(phosphonomethoxy)ethyl]-adenine} and penciclovir [9-(4-hydroxy-3-hydroxymethylbutyl)guanine] were synthesized by Gilead Sciences Inc. (Foster City, Calif.). Lamivudine [(−)-β-l-2′,3′-dideoxy-3′-thiacytidine] was purchased from Moravek Biochemicals (Brea, Calif.).
Drug sensitivity assays.
For antiviral assays, six-well culture dishes were seeded with 7.5 × 105 HepG2 cells/well. At 16 h postseeding, cells were transfected with 5 μg of plasmid by using the Fugene 6 transfection reagent (Roche, Indianapolis, Ind.). On the following day, cells were fed fresh medium containing lamivudine, penciclovir, or adefovir. Cells were treated with fresh drug every other day for 1 week. During antiviral assays, all mutants were tested in parallel by using the same stocks of drug-containing media.
Intracellular HBV replicative intermediates were isolated by lysis of cells in phosphate-buffered saline containing 0.33% Igepal CA-630. Cell lysates were transferred to microcentrifuge tubes and spun for 5 min at 10,000 × g to pellet nuclei. Supernatants were transferred to clean tubes, adjusted to 10 mM MgCl2, and incubated with 30 U of DNase I (Roche) at 37°C for 1 h. Viral replicative intermediates were isolated from lysates by using a Masterpure DNA extraction kit (Epicentre, Madison, Wis.). Viral DNA was resuspended in 10 mM Tris-1 mM EDTA buffer (Sigma, St. Louis, Mo.) and digested with 1.5 μg of RNase (Roche) for 1 h prior to fractionation on 1% agarose gels. Each gel contained the results of a single antiviral assay wherein all four mutants were treated in parallel with one of the tested drugs. Following electrophoresis, viral DNA was transferred to nylon membranes (Roche) by standard Southern blotting procedures (32). Membranes were hybridized to a 33P-labeled HBV probe, and viral DNA was quantified with a Storm 860 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). Regression analyses of antiviral data (based on the measurement of double-stranded HBV replicative intermediates) were performed with TableCurve2D software as previously described (14). Dose-response equations were used to calculate the 50% inhibitory concentrations (IC50s) of drugs for each HBV mutant.
Replication yield assay.
For analyses of intracellular HBV replication, six-well plates were seeded and transfected with wild-type or mutant HBV as described above. On the day of harvest, media were collected from wells and spun for 5 min at 6,000 × g to pellet cellular debris. Hepatitis B e antigen (HbeAg) in the supernatants was quantified by an enzyme-linked immunosorbent assay with an ETI-EBK diagnostic kit (DiaSorin, Stillwater, Minn.); media were diluted appropriately so that HBeAg signals from transfected cells were within the linear range of the assay. Intracellular replicative intermediates were extracted and quantified as described above. To correct for differences in transfection efficiency, replication signals were normalized based on HBeAg levels.
For analyses of extracellular HBV DNA, 100-mm plates were seeded with 5 × 106 cells/plate and transfected with wild-type or mutant HBV DNA as described above (transfection reagent and DNA quantities were scaled up accordingly). Media exposed to transfected cultures for 72 h (days 3 to 5 and days 5 to 8) were pooled for virion analysis. Viral particles were precipitated by bringing conditioned media to a final concentration of 12.5% polyethylene glycol 800 (Sigma), incubation at 4° C for 1 h, and centrifugation at 10,000 × g for 20 min. The resulting pellets were resuspended in phosphate-buffered saline and treated with 30 U of DNase I for 1 h to remove non-virion-associated DNA. Viral DNA was extracted and quantified as described above, with the exception that 5 μg of tRNA was added as a carrier prior to isopropanol precipitation.
Molecular modeling and statistical analyses.
A previously described model of HBV polymerase, generated based on the crystal structures of HIV type 1 RT and Moloney murine leukemia virus RT, was used for molecular modeling and structural interpretation of the experimental data (12). Statistical analyses were performed with Instat v3.05 (Graphpad Software, San Diego, Calif.) and two-tailed paired or unpaired t tests, as appropriate. P values of less than 0.05 were considered significant.
RESULTS
Clinical occurrence of the rtV173L mutation.
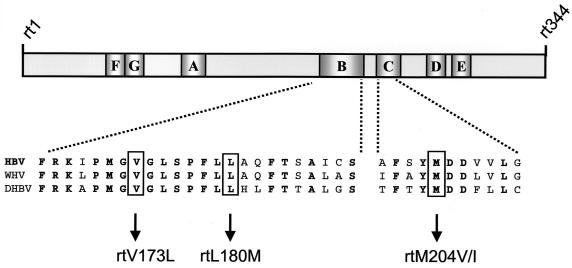
Several clinical trials have been conducted to determine the efficacy of adefovir dipivoxil in patients for whom lamivudine therapy has failed. Baseline genotypic analyses were performed on all patients (n = 216) entering these trials to identify lamivudine resistance mutations. During these analyses, mutations at the YMDD locus (rtM204V/I) as well as the rtL180M B-subdomain mutation were frequently observed, consistent with other clinical reports. The rtV173L mutation, also in the B subdomain of HBV polymerase, was observed in significant numbers of patients in the three trials that we examined (Fig. 1 and Table 2). The rtV173L mutation occurred in 9% of immunocompetent lamivudine-resistant chronic hepatitis B patients and approximately 20% of lamivudine-resistant liver transplant or HIV-coinfected patients. Interestingly, the rtV173L mutation was associated only with the rtL180M-rtM204V mutational pattern in all three trials. The rtV173L mutation was observed in patients infected with HBV genotypes A, C, and D (the predominant HBV genotypes in these trials), indicating that the mutation was not restricted by viral genotype. Due to overlap of the polymerase and surface antigen reading frames, nucleotide mutations causing the rtV173L change also caused a surface antigen mutation (sE164D) in all instances.
FIG. 1.
RT domain of HBV polymerase and location of the rtV173L mutation. The RT domain of HBV polymerase is comprised of 344 amino acids and seven subdomains (A to G) that share significant homology with other polynucleotide polymerases. The locations of the YMDD motif rtM204I/V mutation in subdomain C and the rtL180M and rtV173L mutations in subdomain B are shown. The indicated mutations occur at residues that are conserved in HBV, WHV, and DHBV; bold type indicates amino acids that are conserved in all three viruses. Amino acid numbering is based on the nomenclature of Stuyver et al. (34).
TABLE 2.
Clinical occurrence of the rtV173L mutation in 70 GenBank wild-type HBV sequences and at baseline in adefovir dipivoxil trials
| Trial | Patient population | No. of patients tested | No. (%) of patients with rtV173L at baseline |
|---|---|---|---|
| GenBank 70a | Wild-type HBV clone infected | 70 | 0 (0) |
| GS-98-435 | Lamivudine-resistant, chronic hepatitis B transplant patients | 122 | 23 (19) |
| GS-99-460ib | Lamivudine-resistant, chronic hepatitis B, HIV-coinfected patients | 35 | 8 (23) |
| GS-00-461 | Lamivudine-resistant, chronic hepatitis B patients | 59 | 5 (9) |
Seventy wild-type HBV sequences from the GenBank database; isolates of all HBV genotypes (A to G) are included. Individual accession numbers are listed in reference 40.
The occurrence of rtV173L was previously reported to be higher in this study; however, the study investigators have confirmed that the data provided in this table are accurate (4; V. Thibault and Y. Benhamou, unpublished data).
Antiviral drug sensitivity of HBV carrying the rtV173L mutation.
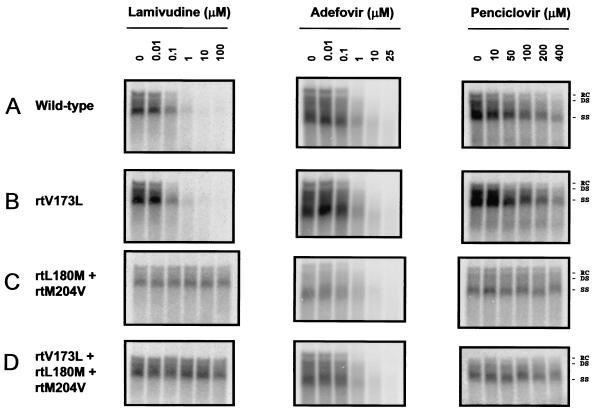
To determine the role of the rtV173L mutation in lamivudine and famciclovir resistance, we engineered this mutation into wild-type and lamivudine-resistant (rtL180M-rtM204V) HBV backgrounds and assessed the sensitivities of the mutants to lamivudine, adefovir, and penciclovir in a cell-based antiviral assay. The sensitivity of wild-type HBV to lamivudine was not significantly altered by the addition of the rtV173L mutation (Fig. 2 and Table 3). Both the rtL180M-rtM204V and the rtV173L-rtL180M-rtM204V mutants were highly resistant to lamivudine (>2,500-fold); therefore, it was not possible to determine whether the rtV173L mutation conferred additional resistance. Adefovir was active against both wild-type HBV and rtL180M-rtM204V mutant HBV, and the addition of the rtV173L mutation to either background did not result in a significant change in the IC50 (Fig. 2 and Table 3). Penciclovir was a relatively weak inhibitor of wild-type HBV (mean IC50, 63 μM), and the introduction of the rtV173L mutation did not alter sensitivity to this drug (Fig. 2 and Table 3). Viruses carrying the rtL180M-rtM204V or rtV173L-rtL180M-rtM204V mutations were highly resistant to penciclovir and, as with lamivudine, it was not possible to determine whether the rtV173L mutation conferred additional resistance.
FIG. 2.
Activities of lamivudine, adefovir, and penciclovir against wild-type and lamivudine-resistant HBV in cell cultures. HepG2 cells were transfected with plasmids encoding wild-type, rtV173L, rtL180M and rtM204V, or rtV173L, rtL180M, and rtM204V HBV DNAs and then treated with the indicated concentrations of lamivudine, adefovir, or penciclovir. After 1 week of treatment, intracellular HBV replicative intermediates were isolated and analyzed by Southern blotting. Relaxed circular (RC), double-stranded (DS), and single-stranded (SS) forms of HBV DNA are indicated.
TABLE 3.
Sensitivities of rtV173L mutants to lamivudine, adefovir, and penciclovir
| HBV mutant(s) | Lamivudine
|
Adefovir
|
Penciclovir
|
|||
|---|---|---|---|---|---|---|
| IC50 (μM)a | Fold resistanceb | IC50 (μM) | Fold resistance | IC50 (μM) | Fold resistance | |
| None (wild type) | 0.04 ± 0.01 | 1.0 | 0.23 ± 0.21 | 1.0 | 65.34 ± 34.0 | 1.0 |
| rtV173L | 0.03 ± 0.01 | 0.8 | 0.18 ± 0.12 | 0.8 | 76.2 ± 6.6 | 1.2 |
| rtL180M + rtM204V | >100 | >2,500 | 0.20 ± 0.04 | 0.9 | >400 | >6 |
| rtV173L + rtL180M + rtM204V | >100 | >2,500 | 0.12 ± 0.05 | 0.5 | >400 | >6 |
IC50 values were calculated by nonlinear regression of antiviral data generated by Southern blotting (Fig. 2) and PhosphorImager analysis and represent the mean and standard deviation of three or more independent experiments.
Fold resistance was calculated as the ratio of mutant IC50 to wild-type IC50.
Replication fitness of HBV carrying the rtV173L mutation.
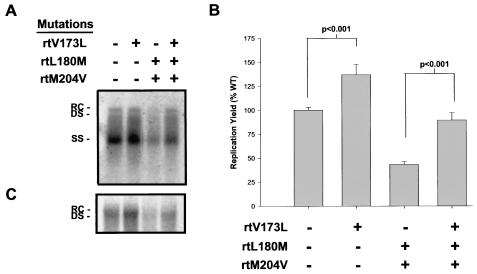
We next tested whether the rtV173 mutation had any effect on the in vitro replication of HBV. Wild-type and mutant HBVs were introduced into HepG2 cells by transient transfection and were allowed to replicate in the absence of drug for 1 week. Intracellular replicating HBV DNA was extracted from cells and quantified by Southern blotting and PhosphorImager analysis; the results of a typical experiment are presented in Fig. 3A. Experiments were performed three separate times in triplicate, and secreted HBeAg, which is transcribed from the input plasmid DNA, was used to normalize transfection efficiency. In comparison to wild-type HBV, rtL180M-rtM204V mutant HBV was found to replicate to about 40% wild-type levels (Fig. 3A and B). The addition of the rtV173L mutation resulted in average 37 and 48% increases in both wild-type and rtL180M-rtM204V mutant HBV levels, respectively (Fig. 3B); these increases were statistically significant (P < 0.001; paired t tests). rtV173L-rtL180M-rtM204V mutant HBV replicated to an average of 90% wild-type levels. To determine whether the increased intracellular replication conferred by the rtV173L mutation resulted in parallel increases in the secretion of mature virions, viral DNA was extracted from media conditioned with transected cells. In agreement with the intracellular data, levels of extracellular HBV DNA were increased by the addition of the rtV173L mutation to the wild-type and rtL180M-rtM204V backgrounds (Fig. 3C).
FIG. 3.
Replication yields of wild-type HBV and lamivudine-resistant HBV after transient transfection into HepG2 cells. (A) HepG2 cells were transfected with plasmids encoding wild-type, rtV173L, rtL180M and rtM204V, or rtV173L, rtL180M, and rtM204V HBV DNAs. HBV was allowed to replicate in the absence of drug for 1 week, after which replicative intermediates were isolated and analyzed by Southern blotting. Relaxed circular (RC), double-stranded (DS), and single-stranded (SS) forms of HBV DNA are indicated. (B) Three experiments were performed in triplicate, and the mean levels of viral replication relative to those of the wild type (WT) are plotted (error bars indicate standard errors). HBeAg, which is secreted from the input plasmid DNA, was used to normalize transfection efficiency. Paired t tests were used to compare replication levels between wild-type and rtV173L HBVs and between rtL180M-rtM204V and rtV173L-rtL180M-rtM204V HBVs; P values are indicated. (C) In a subsequent experiment, the secretion of mature HBV virions was measured by extraction of extracellular HBV DNA from media conditioned with transfected cells and Southern blot analysis.
Molecular modeling of HBV carrying the rtV173L mutation.
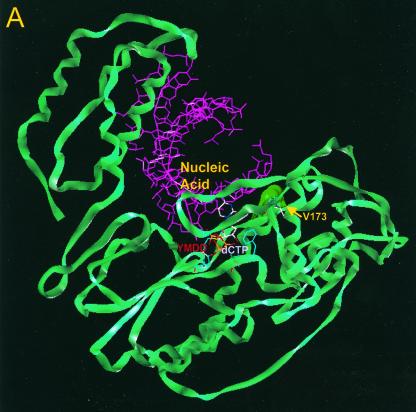
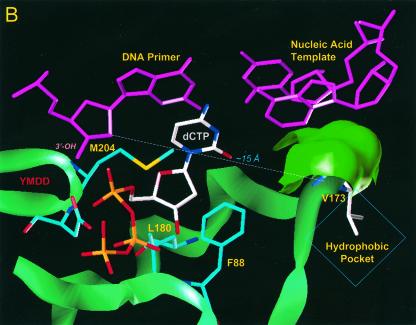
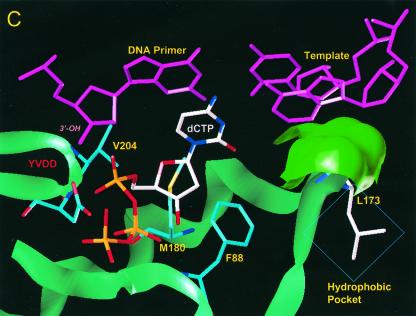
Using a previously described molecular model of HBV polymerase (12), we investigated the location of residue rtV173 and the potential effects of the leucine mutation. In our proposed model, residue rtV173 is located near the active site of HBV polymerase and, along with two flanking glycine residues (rtG172 and rtG174), is positioned directly under the template strand of HBV nucleic acid (Fig. 4A). This region is well conserved in HBV isolates as well as in woodchuck hepatitis B virus (WHV) and duck hepatitis B virus (DHBV). Based on the location, it seems likely that the amino acid backbone of rtV173, along with that of rtG172 and rtG174, is involved in positioning of the template strand during the polymerization reaction. rtV173 and neighboring amino acids are also in close proximity to rtF88, a residue that (by analogy to residue Y115 of HIV RT) interacts with the sugar ring of the deoxynucleoside triphosphate substrate and is expected to undergo significant conformational changes during the polymerization reaction (Fig. 4B). It is therefore possible that the rtV173L mutation affects replication capacity either by repositioning the template nucleic acid strand or by altering the environment around rtF88 in such a way that polymerization efficiency is enhanced (Fig. 4C).
FIG. 4.
Location of residue rtV173 within HBV polymerase and modeling of the leucine substitution. (A) A molecular model of HBV polymerase based on the crystal structures of HIV type 1 RT and Moloney murine leukemia virus RT has been described elsewhere (12). The location of residue rtV173 and its molecular surface (indicated in bright green) are shown in the context of the entire enzyme. Nucleic acids representing the primer and template strands of DNA are indicated in magenta. The location of the YMDD motif is indicated. dCTP is docked in the nucleotide binding pocket of the polymerase. (B) Close-up view of the wild-type polymerase active site, showing the position of rtV173 beneath the template DNA strand. (C) Similar close-up view, modeling the rtV173L-rtL180M-rtM204V mutant active site. Note the position shift of residue F88.
DISCUSSION
Several investigators have reported the emergence of the rtV173L mutation in lamivudine- or famciclovir-treated patients. Two early reports of HBV drug resistance first described the clinical appearance of rtV173L. Bartholomew et al. observed the rtV173L mutation in one of three transplant patients for whom lamivudine therapy failed (3). In this patient, rtV173L was found in association with the hallmark lamivudine resistance mutations rtL180M and rtM204V. A case report by Aye et al. described a similar patient who had virologic breakthrough during famciclovir therapy after liver transplantation (2). Genotyping of the virus obtained from this patient revealed the rtV173L-rtL180M mutational pattern, and phenotypic analysis with an in vitro polymerase assay indicated that the virus was not sensitive to penciclovir triphosphate; these results suggested a role of either or both mutations in resistance. The rtV173L mutation was subsequently described in other clinical reports of lamivudine or famciclovir treatment failure (1, 8, 10, 23, 35). In the large majority of these cases, rtV173L was identified along with rtL180M (in famciclovir-resistant patients) or rtL180M and rtM204V (in lamivudine-resistant patients). During our studies, rtV173L was associated only with the rtL180M-rtM204V double mutation, strongly suggesting that rtV173L specifically emerges in viruses with this pattern of lamivudine resistance mutations.
Although the rtV173L mutation emerged during antiviral therapy in several previous studies, little functional characterization was performed. Thus, it was unclear whether this mutation was directly involved in altering the binding or incorporation of nucleoside analogs by HBV polymerase or whether it was a compensatory mutation that affected the replication fitness of drug-resistant HBV. After observing rtV173L at baseline in many patients enrolled in clinical trials of adefovir dipivoxil for the treatment of lamivudine-resistant HBV, we sought to clarify the role of this mutation in drug resistance. Our analyses indicated that this mutation by itself did not affect the in vitro sensitivity of wild-type HBV to lamivudine, penciclovir, or adefovir. We were unable to determine whether the rtV173L mutation affected the resistance of the rtL180M-rtM204V mutant, since viruses carrying the double or triple mutations were already highly resistant to both penciclovir and lamivudine.
Since the rtL180M-rtM204V double mutation is sufficient to confer clinical resistance to both lamivudine and famciclovir and the rtV173L mutation by itself did not cause in vitro resistance to nucleoside analogs, it is reasonable to hypothesize that the clinical emergence of rtV173L occurs for a reason other than drug resistance. Hence, we investigated the effect of this mutation on in vitro viral replication. During these experiments, HBV carrying the L180M and M204V mutations replicated to about 40% wild-type levels, consistent with previous in vitro reports (9, 26). However, the addition of the rtV173L mutation conferred an increase in replication efficiency to both wild-type HBV and rtL180M-rtM204V-carrying HBV. With respect to the rtL180M-rtM204V mutant, the addition of rtV173L restored viral replication to nearly wild-type levels (90%). Thus, these data suggest that rtV173L functions as a compensatory mutation for lamivudine-resistant HBV and provide a potential explanation for its emergence in lamivudine-treated patients infected with the rtL180M-rtM204V mutant.
Results from the molecular modeling analysis (Fig. 4B and C) indicated that residue rtV173 is located near the active site of HBV polymerase, consistent with the effect of the leucine mutation on viral replication. Based on the modeling data, at least two hypotheses could explain the observed enhancement of replication. First, the rtV173L mutation might result in a repositioning of the template strand of HBV nucleic acid; this repositioning could subsequently alter the interaction between the primer-template and the nucleotide substrate in a way that is more favorable for either catalysis or translocation following polymerization. Second, the proximity of rtV173 to rtF88 might allow the rtV173L mutation to affect the positioning or flexibility of the phenylalanine side chain. Since rtF88 is predicted to undergo a conformational change during polymerization, an alteration induced by rtV173L might allow this change to occur in a more efficient manner. Further experimental and molecular modeling studies will be required to understand the exact mechanism by which rtV173L acts to enhance viral replication.
Compensatory mutations that enhance the viral replication fitness of drug-resistant HIV have been reported for both the protease and the RT enzymes (19, 24, 25, 29, 31). With respect to HIV RT, a virus bearing the Q151M multinucleoside resistance mutation subsequently can develop V75I, a mutation that greatly enhances the in vitro replication of this virus (18). The V75I mutation as well as the V74L/I mutations was also reported to enhance the replication of drug-resistant HIV carrying the quinoxaline (nonnucleoside RT inhibitor) resistance mutation G190E (19). Interestingly, positions 74 and 75 in HIV RT are located adjacent to the template strand of HIV nucleic acid, and mutation of these residues has been postulated to enhance replication fitness by repositioning the template nucleic acid (5, 19). Recent in vitro studies indicated that the Y115W mutation in RT (located at the substrate binding site) impaired HIV replication but that a second mutation, M230I, that emerged during in vitro passaging restored replication efficiency. The M230I mutation, located in the “primer grip” of HIV RT, is predicted to influence the positioning of the primer strand of HIV nucleic acid. Thus, data from the HIV field as well as those presented in this report suggest that repositioning of the template-primer complex may be a common mechanism by which viral polymerases can overcome catalytic deficiencies caused by drug resistance mutations.
Although we found that the rtV173L single mutation enhanced in vitro viral replication by 37%, this mutation was not observed as a single change in chronic hepatitis B patients. Indeed, valine is highly conserved at position rt173 in wild-type HBV isolates (Table 2) as well as in published DHBV and WHV polymerase sequences. Although our modeling study identified two potential indirect effects of rtV173L, it did not provide an obvious reason why an rtV173L single mutant would be selected against in vivo. However, it is possible that rtV173 is strongly preferred in wild-type hepadnavirus polymerases for a functional reason. For example, rtV173L may have a negative effect on replication fidelity, which would be counterselective in patients with an otherwise wild-type HBV polymerase. However, the lamivudine resistance mutations rtL180M and rtM204V alter the geometry of the nucleotide binding pocket and are known to affect the substrate specificity of HBV polymerase (12). Thus, in the context of the rtL180M-rtM204V double mutation, rtV173L may not have an adverse effect on fidelity (or other polymerase functions), a suggestion which may not hold true for an rtV173L single mutation.
It should also be noted that the nucleotide change that causes rtV173L produces a simultaneous sE164D mutation in the overlapping surface antigen reading frame. Residue sE164 is located just downstream of the second antigenic loop of the “a” determinant in the hepatits B surface antigen (HbsAg). Mutations in or near the “a” determinant are known to cause escape from vaccine-induced immunization or passive prophylaxis with HBsAg-specific immunoglobins (6, 7, 11). Indeed, Torresi and colleagues recently reported that sE164D alters the antigenicity of HBsAg and changes its in vitro binding affinity for anti-HBsAg antibodies (36). It is possible that viruses carrying the sE164D mutation are slightly less fit than wild-type virus in an immunocompetent host but that the replication advantage bestowed upon the rtL180M-rtM204V mutant polymerase by rtV173L changes this balance in lamivudine-resistant patients. In an immunocompromised host, this process might be accelerated due to better tolerance of the sE164D mutation; this notion would explain why rtV173L was observed more frequently in the HIV coinfection and liver transplant settings (Table 2).
In conclusion, the studies described here have characterized the clinical prevalence and in vitro phenotype of HBV carrying the rtV173L mutation. This mutation emerges in immunocompetent and, more frequently, immunocompromised lamivudine-resistant patients and was observed only in patients infected with L180M-M204V mutant HBV. Collectively, our in vitro analyses suggested that rtV173L is a compensatory mutation that restores replication fitness to an rtL180M-rtM204V mutant while maintaining high-level drug resistance to lamivudine and penciclovir; importantly, the rtV173L-rtL180M-rtM204V mutant remains sensitive to adefovir in vitro. Based on the position of the rtV173L mutation within HBV polymerase, it is plausible that this mutation affects the positioning of either the template strand of HBV nucleic acid or other residues that are critical for the polymerization reaction. Further studies are warranted to determine the exact mechanism by which rtV173L enhances viral replication.
Acknowledgments
We gratefully acknowledge NIH grants AI27690 (MERIT Award to E.A.) and PO1GM066671-01 for support for HIV RT structural studies.
We thank Mick Hitchcock for critical review of the manuscript.
REFERENCES
- 1.Allen, M. I., M. Deslauriers, C. W. Andrews, G. A. Tipples, K. A. Walters, D. L. Tyrrell, N. Brown, L. D. Condreay, et al. 1998. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Hepatology 27:1670-1677. [DOI] [PubMed] [Google Scholar]
- 2.Aye, T. T., A. Bartholomeusz, T. Shaw, S. Bowden, A. Breschkin, J. McMillan, P. Angus, and S. Locarnini. 1997. Hepatitis B virus polymerase mutations during antiviral therapy in a patient following liver transplantation. J. Hepatol. 26:1148-1153. [DOI] [PubMed] [Google Scholar]
- 3.Bartholomew, M. M., R. W. Jansen, L. J. Jeffers, K. R. Reddy, L. C. Johnson, H. Bunzendahl, L. D. Condreay, A. G. Tzakis, E. R. Schiff, and N. A. Brown. 1997. Hepatitis-B-virus resistance to lamivudine given for recurrent infection after orthotopic liver transplantation. Lancet 349:20-22. [DOI] [PubMed] [Google Scholar]
- 4.Benhamou, Y., M. Bochet, V. Thibault, V. Calvez, M. H. Fievet, P. Vig, C. S. Gibbs, C. Brosgart, J. Fry, H. Namini, C. Katlama, and T. Poynard. 2001. Safety and efficacy of adefovir dipivoxil in patients co-infected with HIV-1 and lamivudine-resistant hepatitis B virus: an open-label pilot study. Lancet 358:718-723. [DOI] [PubMed] [Google Scholar]
- 5.Boyer, P. L., H. Q. Gao, and S. H. Hughes. 1998. A mutation at position 190 of human immunodeficiency virus type 1 reverse transcriptase interacts with mutations at positions 74 and 75 via the template primer. Antimicrob. Agents Chemother. 42:447-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carman, W. F., C. Trautwein, F. J. van Deursen, K. Colman, E. Dornan, G. McIntyre, J. Waters, V. Kliem, R. Muller, H. C. Thomas, and M. P. Manns. 1996. Hepatitis B virus envelope variation after transplantation with and without hepatitis B immune globulin prophylaxis. Hepatology 24:489-493. [DOI] [PubMed] [Google Scholar]
- 7.Carman, W. F., A. R. Zanetti, P. Karayiannis, J. Waters, G. Manzillo, E. Tanzi, A. J. Zuckerman, and H. C. Thomas. 1990. Vaccine-induced escape mutant of hepatitis B virus. Lancet 336:325-329. [DOI] [PubMed] [Google Scholar]
- 8.Chayama, K., Y. Suzuki, M. Kobayashi, M. Kobayashi, A. Tsubota, M. Hashimoto, Y. Miyano, H. Koike, M. Kobayashi, I. Koida, Y. Arase, S. Saitoh, N. Murashima, K. Ikeda, and H. Kumada. 1998. Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild type after cessation of therapy. Hepatology 27:1711-1716. [DOI] [PubMed] [Google Scholar]
- 9.Chin, R., T. Shaw, J. Torresi, V. Sozzi, C. Trautwein, T. Bock, M. Manns, H. Isom, P. Furman, and S. Locarnini. 2001. In vitro susceptibilities of wild-type or drug-resistant hepatitis B virus to (−)-β-d-2,6-diaminopurine dioxolane and 2′-fluoro-5-methyl-β-l-arabinofuranosyluracil. Antimicrob. Agents Chemother. 45:2495-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooley, L., A. Ayres, A. Bartholomeusz, S. Lewin, S. Crowe, A. Mijch, S. Locarnini, and J. Sasadeusz. 2003. Prevalence and characterization of lamivudine-resistant hepatitis B virus mutations in HIV-HBV co-infected individuals. AIDS 17:1649-1657. [DOI] [PubMed]
- 11.Cooreman, M. P., G. Leroux-Roels, and W. P. Paulij. 2001. Vaccine- and hepatitis B immune globulin-induced escape mutations of hepatitis B virus surface antigen. J. Biomed. Sci. 8:237-247. [DOI] [PubMed] [Google Scholar]
- 12.Das, K., X. Xiong, H. Yang, C. E. Westland, C. S. Gibbs, S. G. Sarafianos, and E. Arnold. 2001. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC). J. Virol. 75:4771-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delaney, W. E., IV, A. Bartholomeusz, and S. A. Locarnini. 2002. Evolving therapies for the treatment of chronic hepatitis B virus infection. Expert Opin. Investig. Drugs 11:169-187. [DOI] [PubMed] [Google Scholar]
- 14.Delaney, W. E., IV, R. Edwards, D. Colledge, T. Shaw, J. Torresi, T. G. Miller, H. C. Isom, C. T. Bock, M. P. Manns, C. Trautwein, and S. Locarnini. 2001. Cross-resistance testing of antihepadnaviral compounds using novel recombinant baculoviruses which encode drug-resistant strains of hepatitis B virus. Antimicrob. Agents Chemother. 45:1705-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delaney, W. E., IV, S. Locarnini, and T. Shaw. 2001. Resistance of hepatitis B virus to antiviral drugs: current aspects and directions for future investigation. Antivir. Chem. Chemother. 12:1-35. [DOI] [PubMed] [Google Scholar]
- 16.de Man, R. A., P. Marcellin, F. Habal, P. Desmond, T. Wright, T. Rose, R. Jurewicz, and C. Young. 2000. A randomized, placebo-controlled study to evaluate the efficacy of 12-month famciclovir treatment in patients with chronic hepatitis B e antigen-positive hepatitis B. Hepatology 32:413-417. [DOI] [PubMed] [Google Scholar]
- 17.Gunther, S., F. von Breunig, T. Santantonio, M. C. Jung, G. B. Gaeta, L. Fischer, M. Sterneck, and H. Will. 1999. Absence of mutations in the YMDD motif/B region of the hepatitis B virus polymerase in famciclovir therapy failure. J. Hepatol. 30:749-754. [DOI] [PubMed] [Google Scholar]
- 18.Iversen, A. K., R. W. Shafer, K. Wehrly, M. A. Winters, J. I. Mullins, B. Chesebro, and T. C. Merigan. 1996. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J. Virol. 70:1086-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleim, J. P., M. Rosner, I. Winkler, A. Paessens, R. Kirsch, Y. Hsiou, E. Arnold, and G. Riess. 1996. Selective pressure of a quinoxaline nonnucleoside inhibitor of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) on HIV-1 replication results in the emergence of nucleoside RT-inhibitor-specific (RT Leu-74→Val or Ile and Val-75→Leu or Ile) HIV-1 mutants. Proc. Natl. Acad. Sci. USA 93:34-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai, C. L., J. Dienstag, E. Schiff, N. W. Leung, M. Atkins, C. Hunt, N. Brown, M. Woessner, R. Boehme, and L. Condreay. 2003. Prevalence and clinical correlates of YMDD variants during lamivudine therapy for patients with chronic hepatitis B. Clin. Infect. Dis. 36:687-696. [DOI] [PubMed] [Google Scholar]
- 21.Ling, R., D. Mutimer, M. Ahmed, E. H. Boxall, E. Elias, G. M. Dusheiko, and T. J. Harrison. 1996. Selection of mutations in the hepatitis B virus polymerase during therapy of transplant recipients with lamivudine. Hepatology 24:711-713. [DOI] [PubMed] [Google Scholar]
- 22.Melegari, M., P. P. Scaglioni, and J. R. Wands. 1998. Hepatitis B virus mutants associated with 3TC and famciclovir administration are replication defective. Hepatology 27:628-633. [DOI] [PubMed] [Google Scholar]
- 23.Mutimer, D., D. Pillay, P. Cook, D. Ratcliffe, K. O'Donnell, D. Dowling, J. Shaw, E. Elias, and P. A. Cane. 2000. Selection of multiresistant hepatitis B virus during sequential nucleoside-analogue therapy. J. Infect. Dis. 181:713-716. [DOI] [PubMed] [Google Scholar]
- 24.Nijhuis, M., R. Schuurman, D. de Jong, J. Erickson, E. Gustchina, J. Albert, P. Schipper, S. Gulnik, and C. A. Boucher. 1999. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 13:2349-2359. [DOI] [PubMed] [Google Scholar]
- 25.Olivares, I., V. Sanchez-Merino, M. A. Martinez, E. Domingo, C. Lopez-Galindez, and L. Menendez-Arias. 1999. Second-site reversion of a human immunodeficiency virus type 1 reverse transcriptase mutant that restores enzyme function and replication capacity. J. Virol. 73:6293-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ono, S. K., N. Kato, Y. Shiratori, J. Kato, T. Goto, R. F. Schinazi, F. J. Carrilho, and M. Omata. 2001. The polymerase L528M mutation cooperates with nucleotide binding-site mutations, increasing hepatitis B virus replication and drug resistance. J. Clin. Investig. 107:449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ono-Nita, S. K., N. Kato, Y. Shiratori, T. Masaki, K. H. Lan, F. J. Carrilho, and M. Omata. 1999. YMDD motif in hepatitis B virus DNA polymerase influences on replication and lamivudine resistance: a study by in vitro full-length viral DNA transfection. Hepatology 29:939-945. [DOI] [PubMed] [Google Scholar]
- 28.Ono-Nita, S. K., N. Kato, Y. Shiratori, H. Yoshida, J. Kato, T. Goto, R. F. Schinazi, F. J. Carrilho, and M. Omata. 2000. Influence of B domain mutation (L528M) of the hepatitis B virus polymerase on replication ability and resistance to nucleoside analogues. Hepatology 32:393A. [Google Scholar]
- 29.Pelemans, H., R. Esnouf, K. L. Min, M. Parniak, E. De Clercq, and J. Balzarini. 2001. Mutations at amino acid positions 63, 189, and 396 of human immunodeficiency virus type 1 reverse transcriptase (RT) partially restore the DNA polymerase activity of a Trp229Tyr mutant RT. Virology 287:143-150. [DOI] [PubMed] [Google Scholar]
- 30.Pichoud, C., B. Seigneres, Z. Wang, C. Trepo, and F. Zoulim. 1999. Transient selection of a hepatitis B virus polymerase gene mutant associated with a decreased replication capacity and famciclovir resistance. Hepatology 29:230-237. [DOI] [PubMed] [Google Scholar]
- 31.Resch, W., R. Ziermann, N. Parkin, A. Gamarnik, and R. Swanstrom. 2002. Nelfinavir-resistant, amprenavir-hypersusceptible strains of human immunodeficiency virus type 1 carrying an N88S mutation in protease have reduced infectivity, reduced replication capacity, and reduced fitness and process the Gag polyprotein precursor aberrantly. J. Virol. 76:8659-8666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Analysis and cloning of eukaryotic genomic DNA, p. 9.1-9.62. In N. Ford, C. Nolan, and M. Ferguson (ed.), Molecular cloning, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Seigneres, B., C. Pichoud, S. S. Ahmed, O. Hantz, C. Trepo, and F. Zoulim. 2000. Evolution of hepatitis B virus polymerase gene sequence during famciclovir therapy for chronic hepatitis B. J. Infect. Dis. 181:1221-1233. [DOI] [PubMed] [Google Scholar]
- 34.Stuyver, L. J., S. A. Locarnini, A. Lok, D. D. Richman, W. F. Carman, J. L. Dienstag, and R. F. Schinazi. 2001. Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology 33:751-757. [DOI] [PubMed] [Google Scholar]
- 35.Tillmann, H. L., C. Trautwein, T. Bock, K. H. Boker, E. Jackel, M. Glowienka, K. Oldhafer, I. Bruns, J. Gauthier, L. D. Condreay, H. R. Raab, and M. P. Manns. 1999. Mutational pattern of hepatitis B virus on sequential therapy with famciclovir and lamivudine in patients with hepatitis B virus reinfection occurring under HBIg immunoglobulin after liver transplantation. Hepatology 30:244-256. [DOI] [PubMed] [Google Scholar]
- 36.Torresi, J., L. Earnest-Silveira, G. Deliyannis, K. Edgtton, H. Zhuang, S. A. Locarnini, J. Fyfe, T. Sozzi, and D. C. Jackson. 2002. Reduced antigenicity of the hepatitis B virus HBsAg protein arising as a consequence of sequence changes in the overlapping polymerase gene that are selected by lamivudine therapy. Virology 293:305-313. [DOI] [PubMed] [Google Scholar]
- 37.Westland, C., H. Yang, W. Delaney IV, C. Gibbs, M. Miller, M. Wulfsohn, J. Fry, C. Brosgart, and S. Xiong. 2003. Week 48 resistance surveillance in two phase 3 clinical studies of adefovir dipivoxil for chronic hepatitis B. Hepatology 38:96-103. [DOI] [PubMed] [Google Scholar]
- 38.Xiong, X., H. Yang, C. E. Westland, R. Zou, and C. S. Gibbs. 2000. In vitro evaluation of hepatitis B virus polymerase mutations associated with famciclovir resistance. Hepatology 31:219-224. [DOI] [PubMed] [Google Scholar]
- 39.Yang, H., C. Westland, W. E. Delaney IV, V. Ho, M. D. Miller, C. S. Gibbs, J. Fry, C. L. Brosgart, and S. Xiong. 2001. Resistance monitoring in chronic hepatitis B patients exposed to adefovir dipivoxil for 72 to 136 weeks. Hepatology 34:316A. [DOI] [PubMed] [Google Scholar]
- 40.Yang, H., C. E. Westland, W. E. Delaney IV, E. J. Heathcote, V. Ho, J. Fry, C. Brosgart, C. S. Gibbs, M. D. Miller, and S. Xiong. 2002. Resistance surveillance in chronic hepatitis B patients treated with adefovir dipivoxil for up to 60 weeks. Hepatology 36:464-473. [DOI] [PubMed] [Google Scholar]