Abstract
Although DNA immunization is a safe and efficient method for inducing cellular immune responses, it generates relatively weak and slow immune responses. Here, we investigated the effect of hepatitis C virus (HCV) antigen modifications on the induction of T-cell responses in DNA immunization. It is likely that the strength of T-cell responses has an inverse relationship with the length of the insert DNA. Interestingly, a mixture of several plasmids carrying each gene induced a higher level of T-cell responses than a single plasmid expressing a long polyprotein. Moreover, the presence of a transmembrane domain in HCV E2 resulted in stronger T-cell responses against E2 protein than its absence. Taken together, our results indicate that the tailored modifications of DNA-encoded antigens are capable of optimizing the induction of T-cell responses which is required for eliminating the cells chronically infected with highly variable viruses such as HCV and human immunodeficiency virus.
Since the first report of DNA immunization in 1990 (45), it has been shown to be efficacious in many disease models. (7, 36, 38). DNA vaccines mimic live attenuated vaccines in their ability to induce major histocompatibility complex (MHC) class I-restricted CD8+ T-cell responses while mitigating some of the safety concerns associated with live vaccines. In particular, DNA vaccines can eliminate the need for a cold chain (17).
However, despite early encouraging results, the level of specific immunity induced by DNA vaccines has generally been regarded as insufficient to offer protection against highly pathogenic organisms. To overcome this obstacle, many approaches have been taken to improve the efficacy of DNA vaccines, such as the incorporation of genes for cytokines and costimulatory molecules, the insertion of additional CpG motifs, in vivo electroporation, and the use of self-replicating viral replicons and prime-boost regimens (2). Several studies were performed to enhance proteasome processing of antigen and target DNA-encoded proteins to antigen-presenting cells (35). In addition, through the engineering of antigen genes, successful optimization of immune responses was obtained. For example, the poor immunogenicity of hepatitis C virus (HCV) core protein can be overcome by fusion to HBV envelope protein (15) and localization of HCV E2 protein onto the cell surface enhanced E2-specific antibody responses (14). Usage of codon-optimized DNA sequences also greatly increased antigen expression, resulting in enhancement of both humoral and cellular immune responses (1, 40, 46).
HCV is a major causative agent for liver disease, presenting a high risk of chronicity and development of cirrhosis and liver cancer. In spite of the need for a vaccine, especially in developing countries where blood screening for HCV is not well established (34) and even in developed countries where the incidence of infection among intravenous drug users is as high as 37% per year (31), none was hitherto available. The utilization of error-prone RNA-dependent RNA polymerase, together with rapid replication of HCV after infection, results in the generation of a variety of HCV quasispecies, which have been considered a major mechanism for viral escape from the host immune responses (13, 32, 37). It has been suggested that cell-mediated immune responses (CD8+ T-cell responses in particular) play an important role in protection against HCV chronic infection (9). Therefore, it is widely accepted that induction of strong multiepitope-specific cellular immunity probably overcomes the ability of HCV to escape (23).
HCV consists of a 9.5-kb genome that generates 10 proteins: a core protein and E1 and E2 as structural proteins and NS2 to NS5 nonstructural proteins. To induce multiepitope-specific cellular immunity (which is likely to be responsible for eliminating highly variable viruses such as HCV and human immunodeficiency virus [HIV]), it is undoubtedly advantageous to include as broad a range of antigens as possible. Nevertheless, little is known about the optimal way of presenting such a lengthy polyprotein for the efficient induction of multiepitope-specific T-cell responses. Two extreme approaches can be proposed; one is to construct 10 individual short plasmids with each separately encoding an antigen, and the other is to make a plasmid that encodes whole HCV polyprotein. In general, the use of a plasmid that encodes a whole polyprotein is more practical and inexpensive than the use of many plasmids with each separately carrying a gene. However, it is likely that the use of long inserts often leads to both a decrease of gene expression and instability of plasmids during amplification. Although it was reported that a DNA construct encoding a whole HCV polyprotein can induce HCV-specific T-cell responses, those T-cell responses were not directly compared to T-cell responses induced by a mixture of multiple plasmids with shorter-length inserts (16). One of the most critical concerns to be addressed is the relationship between insert length in a plasmid and the strength of induced T-cell responses.
Immunological research in the HCV field has been difficult, because there is no reliable small-animal model. A chimpanzee is the only relevant animal model for HCV infection; however, chimpanzees are costly and rare and have to be used judiciously. The limitations of the chimpanzee model have hampered testing of a variety of strategies designed to enhance immunity to HCV antigens. Although the use of mice does not permit HCV replication, studies using a mouse model have contributed to our understanding of the generic aspects of immunization, such as the nature of antigens and the route, time course, and order of delivery of immunogens for the generation of optimal immune responses (including the use of cytokines or other possible immunomodulators). In this study, we investigated the effect of HCV antigen modifications on the optimization of the induction of strong cellular immune responses. HCV E2 was selected as a reporter antigen, because the analysis of immune responses to E2 has been previously established (18). Using different lengths of HCV antigen-expressing DNA constructs, we demonstrated that the T-cell response was inversely correlated with the length of the insert. In addition, immunization with a mixture of separate plasmids which each expressed a protein induced stronger T-cell responses than immunization with a plasmid expressing a long polyprotein. Finally, deletion of the transmembrane domain (TMD) of E2 resulted in a reduction of T-cell responses.
MATERIALS AND METHODS
Animals and immunization.
Female BALB/c mice (H-2d) 5 to 6 weeks of age were purchased from Japan SLC (Shizuoka, Japan) and maintained under specific pathogen-free conditions. Mice (five to six per group) were immunized with 100 μg of an indicated plasmid DNA injected bilaterally into the anterior tibialis muscle. A control group of mice were injected with vector DNA alone. At the indicated time after immunization, mice were sacrificed and splenocytes were removed for the analysis of cell-mediated immune responses. Animals are maintained in accordance with applicable portions of the Animal Welfare Act and with the guidelines of the United States Department of Health and Human Services, Public Health Services, the National Institutes of Health, and the Guide for the Care and Use of Laboratory Animals.
Construction of expression plasmids.
pTZ-HCV (encoding a whole genomic region of HCV type 1b, which consists of a structural region from a Korean isolate [accession number AY308072] and a nonstructural region from a JS strain) (8, 41) was generated using restriction enzyme sites within HCV sequences on the basis of a standard molecular cloning technique (39) and used as a template for PCR amplifications in this study. pTV2-sE2t, which localized E2 protein into the endoplasmic reticulum by inserting the signal sequence of herpes simplex virus glycoprotein D into the N terminus of E2, was described previously (27). To construct pTV2-ST, pTV2-SN2, and pTV2-ΔSN2, PCR amplifications using Core PstIS (5′-aaa ctg cag acc atg agc aca aat cct aaa cct-3′)/E2 XbaIA (5′-ccc tct aga tgc gtc cgc cag gag aag gaa-3′), Core PstIS/NS2 XbaIA (5′-aaa tct aga tca gtc tcg cag gcc cgc gtg ggc-3′), and ΔCore PstIS (5′-aaa ctg cag acc atg ggc ccc agg ttg ggt-3′)/NS2 XbaIA, respectively, were performed and the product was digested with PstI/XbaI for insertion into a pTV2 vector. ΔCore XbaIS (5′-aaa tct aga acc atg ggc ccc agg ttg ggt-3′) and E2 XbaIA primers were used for PCR and were inserted into pTV2 after digestion with XbaI to construct pTV2-ΔST.
pTV2-SN5 was constructed through digestion of pTV2-ST and pTZ-HCV with NotI/XbaI (6.25 and 7.50 kb, respectively) and ligation to each other. To construct pTV2-s, a PCR product from pTV2-sE2t prepared using cytomegalovirus AscI+ (5′-aag gcg cgc ccg atg tac ggg cca gat ata-3′)/gDs AscIA (5′-aag gcg cgc cag aga ggc atc cgc caa ggc-3′) was digested with SpeI (1.28 kb) and ligated to the digestion product of pTV2-sE2t with SpeI/EcoRV (3.62 kb). For the construction of pTV2-sE2, pTV2-sΔST, and pTV2-sΔSTt, PCR amplifications using gE2 AscIS (5′-aag gcg cgc cgc acc cgc gtg aca gga gga-3′)/E2 XbaIA, ΔCore fusion AscIS (5′-aag gcg cgc cgc ccc agg ttg ggt gtg cgc-3′)/E2 XbaIA, and ΔCore fusion AscIS/E2t XbaIA (5′-aaa aat cta gat taa tac tgg gac ttg atc act at-3′), respectively, were performed and the products were digested with AscI/XbaI for insertion into pTV2-s vector. Fragments of ACP vector (from J. H. Byun) and pTV2 digested with NcoI/EcoRI (4.84 and 0.8 kb, respectively) were ligated to produce ACP30.
pVax1 was digested with BamHI/BspHI (1.4 kb; Kanr) and was filled in for insertion into the PvuI site of ACP or ACP30 to produce pGX101 or pGX103, respectively. Fragments of pTV2-sΔST digested with PstI/XbaI were inserted into pGX10 (19), pGX101, or pGX103 to produce pGX10-sΔST, pGX101-sΔST, or pGX103-sΔST, respectively. The NS34 or NS5 region was PCR amplified using NS3 PstIS (5′-ccc ctg cag acc atg ccc atc acg gcc tac tcc caa-3′)/NS4 XbaIA (5′-aaa tct aga tta gca tgg cgt gga gca gtc ctc-3′) or NS5 KpnIS (5′-aaa ggt acc atg tcc ggc tcg tgg cta agg gat-3′)/NS5 XbaIA2 (5′-aat cta gaa gcg gtt ggg gag cag gta gac-3′) and digested with PstI/XbaI or Asp718/XbaI for insertion into pGX10 to generate pGX10-NS34 or pGX10-NS5, respectively. The joining of the parts of all these constructs was confirmed by nucleotide sequencing, and the expression of their proteins were confirmed by Western blot analysis.
In vitro transient expression.
COS-7 (monkey kidney) or C2C12 (mouse myoblast) cells were electroporated with 16 μg of the indicated expression plasmid together with 4 μg of luciferase reporter plasmid as an internal control. After incubation for 48 h, cells were harvested and analyzed for their expression of HCV E2 protein. Luciferase activities were determined, and an amount of cell lysate corresponding to each luciferase activity was loaded on sodium dodecyl sulfate-polyacrylamide electrophoresis gels for comparison of the expression levels among different plasmid constructs. Anti-HCV E2 monoclonal antibodies (25) were used for the detection of E2-specific bands by Western blot analysis.
IFN-γ ELISPOT assay.
At the indicated time after immunization, splenocytes from two to three mice were pooled and used in this study. A gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assay was performed as described before (33). Briefly, splenocytes were serially diluted threefold in triplicate, starting from 106 cells per well on an IFN-γ capture antibody (BD PharMingen; 5 μg/ml)-coated 96-well ELISPOT plate (Millipore, Bedford, Mass.) and incubated at 37°C in a CO2 incubator. The splenocytes were stimulated either with 2 × 104 CT26-hghE2t cells (H-2d; MHC I+/MHC II−) expressing HCV E2 protein or with an E2 peptide pool (20-mer with a 10-amino-acid [aa] overlap) at 1 μg/ml per peptide. After 20 h, the plates were washed four times with phosphate-buffered saline containing 0.1% Tween 20 (washing buffer). A total of 50 μl of 2 μg of biotinylated anti-mouse IFN-γ antibody (BD PharMingen)/ml was added to each well and incubated for 2 h at room temperature. After washing buffer was used for four washings, 50 μl of 1:1,000-diluted alkaline phosphatase-coupled streptavidin (BD PharMingen) was added and the mixture was incubated for 1 h. After washing was performed six times, spots were visualized by adding 5-bromo-4-chloro-3-indolyl phosphate-tetranitroblue tetrazolium substrate solution (Calbiochem, San Diego, Calif.). The reaction was stopped by washing with tap water. The plates were dried, and the spots were enumerated under a dissecting microscope. The average number of IFN-γ-secreting cells was shown per 106 splenocytes.
Cytotoxic T lymphocyte (CTL) assay.
A total of 1.5 × 107 splenocytes (pooled from two or three immunized mice) were expanded with 106 CT26-hghE2t cells for 5 days in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 50 μM β-mercaptoethanol, and 10 U of recombinant murine interleukin-2/ml. Cytotoxicity was examined for 104 CT26-hghE2t (H-2d) target cells/well that were labeled with 51Cr. The cells were incubated for 5 h in triplicate at 37°C at different effector-to-target cell (E:T) ratios. The maximum or spontaneous release of 51Cr was determined from the cells treated with either 2% Triton X-100 or medium alone, respectively. Percentages of specific lysis were calculated as [(experimental release − spontaneous release)/(maximum release − spontaneous release)] × 100.
RESULTS
Construction of E2-containing plasmids and their expression.
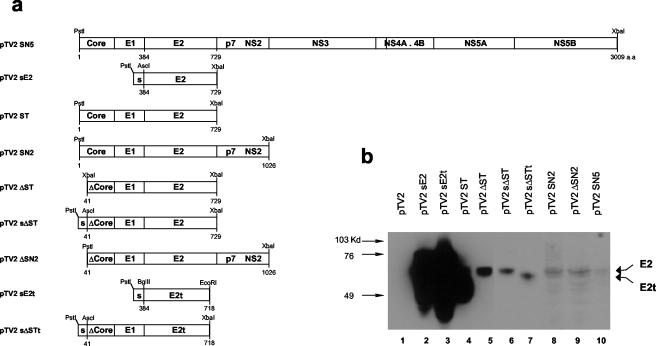
We constructed several plasmids, starting from a plasmid expressing E2 alone and proceeding to a plasmid expressing the whole HCV polyprotein (Fig. 1a). Since it was previously reported that the HCV core has immunosuppressive function in vivo and in vitro (22, 24) and since the truncation of 40 aa from the N terminus of the core appeared to abolish its suppressive function in terms of interleukin-12 and tumor necrosis factor alpha secretion in the stimulated macrophage cells (C. H. Lee, unpublished data), we constructed plasmids that expressed the core from which 40 aa at the N terminus were deleted, namely, pTV2-ΔST, pTV2-ΔSN2, and pTV2-sΔST. To investigate the effect of the presence of the E2 TMD on the strength of the cellular immune response, the TMD of E2 was truncated from the pTV2-sE2 and pTV2-sΔST plasmids to construct pTV2-sE2t and pTV2-sΔSTt, respectively.
FIG. 1.
Schematic diagram of the HCV expression plasmids and their identification in vitro. (a) pTV2 vector was previously described (27). Corresponding amino acid positions in HCV polypeptides are indicated under each diagram. s, glycoprotein D signal sequence from herpes simplex virus type 1. (b) COS7 cells were transfected with the indicated plasmids and a luciferase-expressing plasmid as an internal control. Corresponding amounts of cell lysates were processed for sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis followed by Western blot analysis with anti-E2 monoclonal antibodies. The positions of molecular mass markers are indicated at the left of the panel; those of E2 and E2t are indicated on the right.
To identify the expression of the above constructs, COS7 cells were transiently transfected with the indicated plasmids followed by Western blot analysis using anti-E2 monoclonal antibodies (25). A luciferase-expressing DNA was also cotransfected with the constructed plasmids, and transfection efficiency was normalized according to the luciferase activity. Expected E2 glycoproteins were detected at around 65 to 70 kDa. The highest and lowest levels of expression were detected in pTV2-sE2t and pTV2-SN5, respectively (Fig. 1b, lanes 3 and 10). pTV2-ST-related constructs appeared to express intermediate levels of E2 protein (lanes 4 to 7). Expression levels of pTV2-SN2 and pTV2-ΔSN2 were between those of pTV2-ST-related constructs and pTV2-SN5 (lane 8 and 9). These results suggest that the expression levels have an inverse relationship with the insert length of the plasmid. Truncation of E2 TMD in pTV2-sE2t and pTV2-sΔSTt plasmids was identified by the slight difference in their migration characteristics compared to those of pTV2-sE2 and pTV2-sΔST (Fig. 1b, lanes 2 and 6 versus lanes 3 and 7) and resulted in the secretion of E2 protein (data not shown). Although pTV2-ST appeared to express a higher level of E2 than any other pTV2-ST derivatives, it was due to the spreading over of the pTV2-sE2t band, as confirmed by independent experiments. The pattern of relative expression levels of the above constructs were also confirmed when a C2C12 myoblast cell line was used (data not shown).
The effect of insert length on the strength of E2-specific T-cell responses induced by DNA immunization.
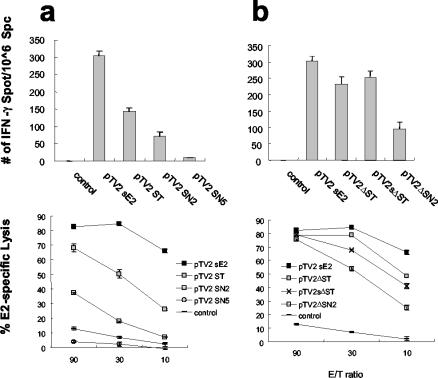
To investigate the effect of insert length on the induction of cellular immune responses, different plasmids with inserts of increasing length were used for immunization. T-cell responses were analyzed using an E2-expressing CT26-hghE2t cell line at 5 weeks after immunization. pTV2-sE2 appeared to induce the highest IFN-γ ELISPOT response and 84% of cytolytic activity at an E:T ratio of 30. In contrast, pTV2-ST, pTV2-SN2, and pTV2-SN5 induced 47, 23, and 3% of the IFN-γ ELISPOT number induced by pTV2-sE2 and 50, 18, and 2% of specific lysis at an E:T ratio of 30, respectively (Fig. 2a). These results suggest that there is an inverse relationship between insert length and the strength of T-cell responses.
FIG. 2.
HCV E2-specific T-cell responses inversely correlated with insert lengths of DNA constructs. BALB/c mice were immunized intramuscularly with 100 μg of the indicated expression plasmids. Splenocytes were removed at 5 weeks after immunization and used for CTL and IFN-γ ELISPOT assays with CT26-hghE2t cells for stimulation. Plasmids containing intact core (a) or Δcore (b) were compared for their levels of induction of T-cell responses. Standard deviations are indicated as error bars. Data are representative of the results of one of three independent experiments. Statistical analysis was performed using Student's t test. P values between different groups of immunization were compared, and those of less than 0.01 were considered significant.
When plasmids with core deletions were tested for the ability to induce E2-specific T-cell responses, pTV2-ΔST, pTV2-sΔST, and pTV2-ΔSN2 induced 77, 84, and 31% of the IFN-γ ELISPOT number induced by pTV2-sE2 and 79, 68, and 53% of specific lysis at an E:T ratio of 30, respectively (Fig. 2b) and thus revealed a similar inverse relationship between insert length and strength of T-cell responses. Interestingly, it is likely that there is an enhancement of IFN-γ ELISPOT response in the expression of plasmids with core deletions compared to that seen with plasmids with intact core; pTV2-ΔST and pTV2-ΔSN2 induced 163 and 134% of the number of IFN-γ secreting cells induced by pTV2-ST (P < 0.01) and pTV2-SN2 (P < 0.1), respectively (Fig. 2a and b, respectively), which might be explained by the elimination of the putative immunosuppressive function of core protein.
Effect of antigen expression on the induction level of T-cell responses.
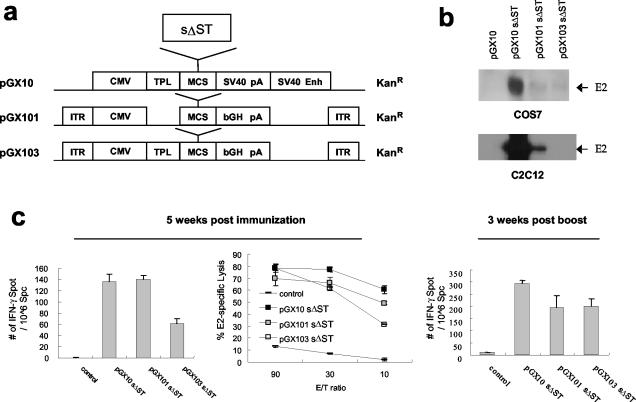
It is likely that there is an overall inverse relationship between expression level and insert size, since the expression levels were decreased overall as the insert became longer (E2 > ST > SN2 > SN5) (Fig. 1b). To investigate whether the magnitude of T-cell responses correlated with expression level, we constructed three different plasmids expressing HCV structural proteins, pGX10-sΔST, pGX101-sΔST, and pGX103-sΔST, which express different levels of E2 protein (Fig. 3a); the relative ratios of the E2 expression levels of pGX10-sΔST, pGX101-sΔST, and pGX103-sΔST were approximately 500:10:1 (Fig. 3b). pGX101-sΔST and pGX103-sΔST induced 103 and 45% of the ELISPOT numbers induced by pGX10 sΔST, respectively, at 5 weeks after the first immunization (Fig. 3c). In addition, pGX10-sΔST, pGX101-sΔST, and pGX103-sΔST showed 77, 66, and 61% of CTL activity at an E:T ratio of 30, respectively. At 3 weeks after boosting, the number of spots induced by pGX101-sΔST and pGX103-sΔST were 67 and 68% of the number induced by pGX10-sΔST. These results indicated that the expression level slightly affects the induction level of T-cell response but is not directly proportional to T-cell responses in DNA vaccination. These results are partially consistent with a recent report that antigen expression does not correlate with immunogenicity (28).
FIG. 3.
Effect of E2 expression level on the strength of T-cell responses against E2. Three different expression vectors were used to carry the same HCV structural gene, sΔST (a), and their E2 expression levels in COS7 as well as in C2C12 cells were compared by Western blot analysis (b). BALB/c mice were immunized intramuscularly at 0 and 4 weeks with 100 μg of the indicated expression plasmids, and IFN-γ ELISPOT and CTL responses were examined (c). Standard deviations are indicated as error bars. CMV, cytomegalovirus early promoter/enhancer; TPL, adenovirus tripartite leader; MCS, multicloning site; SV40 pA, simian virus 40 poly(A) signal; SV40 Enh, simian virus 40 enhancer; ITR, inverted terminal repeat of adeno-associated virus; bGH pA, bovine growth hormone poly(A) signal.
The effect of nonstructural gene coexpression on the induction of E2-specific CD8+ T-cell responses.
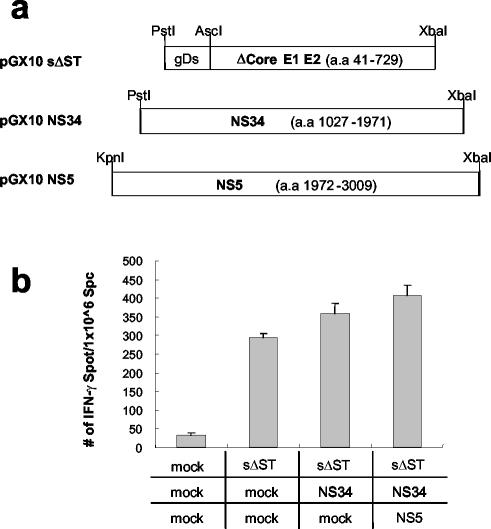
T-cell responses induced by pTV2-SN5 were significantly lower than those induced by pTV2-sE2 or pTV2-sΔST (Fig. 2). It is possible that nonstructural proteins simultaneously expressed with E2 protein in pTV2-SN5 might cause antigenic competition with E2 protein and thus hinder efficient induction of E2-specific CD8+ T-cell responses. To investigate this possibility, we coimmunized pGX10-sΔST with or without pGX10-NS34 and pGX10-NS5 plasmids to mimic the simultaneous expression of nonstructural proteins in pTV2-SN5 and then compared the induction of E2-specific IFN-γ ELISPOT responses (Fig. 4a). Interestingly, codelivery of pGX10-NS34 with pGX10-sΔST rather slightly (albeit statistically insignificantly) enhanced E2-specific IFN-γ ELISPOT numbers (122%) compared to injection of pGX10-sΔST alone (Fig. 4b). In addition, coinjection of pGX10-NS34 plus pGX10-NS5 appeared to further enhance IFN-γ ELISPOT response (139%). Successful induction of T-cell responses toward nonstructural proteins was confirmed by IFN-γ ELISPOT assays using peptide pools encompassing NS3 or NS5A regions (data not shown). These results suggest that coexpression of nonstructural proteins does not inhibit the induction of T-cell response against E2 in DNA immunization and that antigenic competition by nonstructural proteins is unlikely to be the reason for the low immunogenicity of pTV2-SN5.
FIG. 4.
E2-specific T-cell response was not affected by the coadministration of separate plasmids expressing HCV nonstructural genes. (a) Plasmids pGX10-NS34 and pGX10-NS5, carrying nonstructural regions of HCV, were used together with pGX10-sΔST (100 μg of plasmids total, consisting of 33 μg of each plasmid with or without mock DNA) for immunization at 0 and 4 weeks. (b) At 3 weeks after booster injections were administered, an IFN-γ ELISPOT assay was performed using CT26-hghE2t cells for stimulation. Standard deviations are indicated as error bars. Mock, mock DNA.
The effect of TMD on the strength of cellular immune responses induced by DNA immunization.
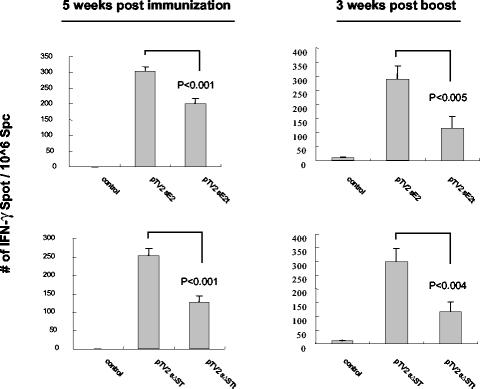
To investigate the effect of the TMD on the induction of T-cell responses, E2-expressing plasmids (pTV2-sE2 and pTV2-sΔST) and TMD-truncated E2-expressing plasmids (pTV2-sE2t and pTV2-sΔSTt) were used for DNA immunization. To exclude the effect of the contribution of any epitope present in the TMD of E2 on the strength of T-cell response, CT26-hghE2t cells or E2 peptide pools that were devoid of TMD were used for stimulation. At 5 weeks after the first immunization, pTV2-sE2 and pTV2-sΔST induced 152 and 200% of the number of IFN-γ spots induced by pTV2-sE2t (P < 0.001) and pTV2-sΔSTt (P < 0.001), an effect which was further enhanced by booster immunization to 250% (P < 0.005) and 256% (P < 0.004), respectively (Fig. 5). These results indicated that truncation of E2 TMD decreased E2-specific T-cell response no matter how E2 protein was expressed alone or as a portion of polyprotein, suggesting that E2 TMD is required for optimal induction of T-cell responses against E2.
FIG. 5.
Truncation of E2 TMD decreased the strength of E2-specific T-cell responses induced by DNA immunization. BALB/c mice were immunized intramuscularly with 100 μg of the indicated expression plasmids at 0 and 4 weeks. Splenocytes removed at 5 weeks after primary immunization or 3 weeks after boosting were used in IFN-γ ELISPOT assays after being stimulated either with a CT26-hghE2t or with an E2 peptide pool, respectively. Standard deviations are indicated as error bars. Data are representative of the results of three independent experiments.
DISCUSSION
In this study, we demonstrated that there is an inverse relationship between insert length and T-cell responses induced by the plasmids tested here. It is possible that a low level of antigen expression might have resulted in reduced T-cell responses, since long inserts led to decreased expression of DNA-encoded antigen. Although the expression level of an antigen has been shown to have a direct relationship with humoral responses in DNA immunization (5), its relationship with T-cell response is still controversial. Previous reports suggested a proportional relationship between expression level and the induced cellular immune responses in DNA immunization with a codon-optimized HIV-1 gag gene or a vaccinia virus infection model (44, 46). In contrast, no correlation was suggested in other reports of studies in which Venezuelan equine encephalitis virus containing HIV-1 gene or HBsAg and HBcAg DNA vaccine was tested (4, 21). Here, we demonstrated that the expression level appeared to affect (but was not directly proportional to) the level of T-cell responses induced by HCV structural gene immunization. Considering that each antigen has a different level of processing efficiency in the generation of immunogenic peptides which also have different binding affinities to MHC molecules, the nature of the antigen may be another determinant affecting the induction of T-cell responses. For example, when an antigen is efficiently processed into immunogenic peptides that bind to an MHC molecule with high affinity, a small amount of antigen is sufficient to induce a cellular immune response. Furthermore, it is known that the threshold for the induction of T-cell response is lower than that for antibody response (21). Thus, it is likely that the critical factor for inducing T-cell responses in DNA immunization is the nature of the antigen rather than the expression level.
Previous reports demonstrated that there is evidence of antigenic competition in infections with viruses such as lymphocytic choriomeningitis virus and influenza virus (41-43). However, in this study we showed that codelivery of separate plasmids expressing nonstructural proteins of HCV did not affect the induction of E2-specific T-cell response, indicating the absence of competition with coexpressed antigens. The level of antigen expression in DNA immunization in vivo is known to be far less than that in virus infection. This low level of expression may not reach to antigenic competition among dominant and/or subdominant epitopes if target cells provide a number of MHC molecules sufficient to accommodate all of the epitope peptides. Thus, a DNA immunization method may be basically different from virus infection regarding antigenic competition. Moreover, codelivery of plasmids expressing nonstructural proteins rather increased E2-specific T-cell response, presumably due to the presence of T-helper epitopes in the nonstructural region of HCV. It is worth noting that the immunization with a mixture of three DNA constructs (pTV2-sΔST, pTV2-NS34, and pTV2-NS5) was much more efficient at inducing E2-specific ELISPOT activity than immunization with pTV2-SN5 expressing whole HCV polyproteins (>26-fold). These results suggest that antigen processing and presentation induced by a single long polyprotein is inefficient compared to that induced by three separate short polyproteins. It remains to be determined whether the lack of antigenic competition for E2 is applicable to any other proteins within the polyprotein.
It is known that deletion of TMD usually increases the humoral responses by inducing secretion of antigen to the extracellular space, which facilitates antigen encounter with antigen-specific B cells (12, 39). Although this concept is generally accepted for humoral response, its influence on T-cell response is still controversial. While one group showed that the membrane-bound form of glycoprotein D from bovine herpesvirus induced a stronger CTL response than its secreted form in DNA immunization (29), another group reported there was no difference between the membrane-bound form and the secreted form of ovalbumin in DNA immunization (3). It was of interest to determine how the truncation of E2 TMD decreased IFN-γ ELISPOT responses for E2. Depending on the presence or absence of TMD, a major portion of E2 protein is either cell associated or secreted into the extracellular space (8, 26). One explanation may be the different efficiencies of E2 and TMD-truncated E2 proteins (E2t) in cross-presentation. Dendritic cells were known to process exogenous antigen to load on MHC class I molecules (cross-presentation) and thereby to stimulate CD8+ T cells (20). Several previous reports suggest that cross-presentation is a predominant mechanism for inducing CD8+ T-cell response in DNA immunization (6, 10, 11). During this process, the efficiency of cross-presentation depends on the form of antigen. The cell-associated form of antigen can be cross-presented 104 times more efficiently than the soluble form (30). Thus, the form of antigen provided by a transfected cell could be an important parameter affecting the induction of T-cell response. Also, the induction level of a T-cell response may be influenced by the period of retention time inside the cell before secretion. Thus, it would be disadvantageous for the induction of T-cell responses if secretion were to occur immediately after translation because the shorter retention time within the transfected cells would provide less chance for cross-presentation by dendritic cells.
The balance between T-cell responses and viral replication in HCV infection probably determines whether the infection will result in chronic progression or acute clearance. Thus, optimal induction of T-cell responses through customized modification of an antigen is likely to be necessary for eliminating virus infection. The present study provides information useful for the induction of strong multiepitope specific T-cell responses by tailored modification of HCV DNA in DNA immunization. Our results can be directly applied to the development of vaccine against HCV and other highly variable infectious agents, such as other RNA and retroviruses which contain polyprotein.
Acknowledgments
We thank S. C. Lee for devoted animal care and Mohamed Tarek Shata, Alfred M. Prince, and D. H. Lee for great help in preparing the manuscript. Especially, we appreciate Mohamed Tarek Shata for valuable advice in scientific discussions. We thank J. H. Byun for his kind donation of ACP vector.
This work was supported by a grant from the Korea Health 21 R&D Project, Ministry of Health & Welfare, Republic of Korea (02-PJ2-PG1-CH16-0002), a grant from the National Research Lab Program of the National S&T Program of the Ministry of S&T (2000-N-NL-01-202), and a grant from a consortium project (Genexine Co. Ltd., Daewoong Pharm. Co. Ltd., Dong-A Pharm. Co. Ltd., and POSCO).
REFERENCES
- 1.Andre, S., B. Seed, J. Eberle, W. Schraut, A. Bultmann, and J. Haas. 1998. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J. Virol. 72:1497-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berzofsky, J. A., J. D. Ahlers, and I. M. Belyakov. 2001. Strategies for designing and optimizing new generation vaccines. Nat. Rev. Immunol. 1:209-219. [DOI] [PubMed] [Google Scholar]
- 3.Boyle, J. S., C. Koniaras, and A. M. Lew. 1997. Influence of cellular location of expressed antigen on the efficacy of DNA vaccination: cytotoxic T lymphocyte and antibody responses are suboptimal when antigen is cytoplasmic after intramuscular DNA immunization. Int. Immunol. 9:1897-1906. [DOI] [PubMed] [Google Scholar]
- 4.Caley, I. J., M. R. Betts, N. L. Davis, R. Swanstrom, J. A. Frelinger, and R. E. Johnston. 1999. Venezuelan equine encephalitis virus vectors expressing HIV-1 proteins: vector design strategies for improved vaccine efficacy. Vaccine 17:3124-3135. [DOI] [PubMed] [Google Scholar]
- 5.Chastain, M., A. J. Simon, K. A. Soper, D. J. Holder, D. L. Montgomery, S. L. Sagar, D. R. Casimiro, and C. R. Middaugh. 2001. Antigen levels and antibody titers after DNA vaccination. J. Pharm. Sci. 90:474-484. [DOI] [PubMed] [Google Scholar]
- 6.Cho, J. H., J. W. Youn, and Y. C. Sung. 2001. Cross-priming as a predominant mechanism for inducing CD8+ T-cell responses in gene gun DNA immunization. J. Immunol. 167:5549-5557. [DOI] [PubMed] [Google Scholar]
- 7.Cho, Y. G., J. W. Youn, K. L. Jang, C. M. Kim, and Y. C. Sung. 1993. Full genome cloning of hepatitis C virus from serum of Korean chronic hepatitis patients in Korea. Mol. Cells 355:195-202. [Google Scholar]
- 8.Cocquerel, L., C. Wychowski, F. Minner, F. Penin, and J. Dubuisson. 2000. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a major role in the processing, subcellular localization, and assembly of these envelope proteins. J. Virol. 74:3623-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper, S., A. L. Erickson, E. J. Adams, J. Kansopon, A. J. Weiner, D. Y. Chien, M. Houghton, P. Parham, and C. M. Walker. 1999. Analysis of a successful immune response against hepatitis C virus. Immunity 10:439-449. [DOI] [PubMed] [Google Scholar]
- 10.Corr, M., D. J. Lee, D. A. Carson, and H. Tighe. 1996. Gene vaccination with naked plasmid DNA: mechanism of CTL priming. J. Exp. Med. 184:1555-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doe, B., M. Selby, S. Barnett, J. Baenziger, and C. M. Walker. 1996. Induction of cytotoxic T lymphocytes by intramuscular immunization with plasmid DNA is facilitated by bone marrow-derived cells. Proc. Natl. Acad. Sci. USA 93:8578-8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drew, D. R., M. Lightowlers, and R. A. Strugnell. 2000. Humoral immune responses to DNA vaccines expressing secreted, membrane bound and non-secreted forms of the Tania ovis 45W antigen. Vaccine 18:2522-2532. [DOI] [PubMed] [Google Scholar]
- 13.Farci, P., A. Shimoda, A. Coiana, G. Diaz, G. Peddis, J. C. Melpolder, A. Strazzera, D. Y. Chien, S. J. Munoz, A. Balestrieri, R. H. Purcell, and H. J. Alter. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 14.Forns, X., S. U. Emerson, G. J. Tobin, I. K. Mushahwar, R. H. Purcell, and J. Bukh. 1999. DNA immunization of mice and macaques with plasmids encoding hepatitis C virus envelope E2 protein expressed intracellularly and on the cell surface. Vaccine 17:1992-2002. [DOI] [PubMed] [Google Scholar]
- 15.Geissler, M., K. Tokushige, T. Wakita, V. R. Zurawski, Jr., and J. R. Wands. 1998. Differential cellular and humoral immune responses to HCV core and HBV envelope proteins after genetic immunizations using chimeric constructs. Vaccine 16:857-867. [DOI] [PubMed] [Google Scholar]
- 16.Gordon, E. J., R. Bhat, Q. Liu, Y. F. Wang, C. Tackney, and A. M. Prince. 2000. Immune responses to hepatitis C virus structural and nonstructural proteins induced by plasmid DNA immunizations. J. Infect. Dis. 181:42-50. [DOI] [PubMed] [Google Scholar]
- 17.Gurunathan, S., D. M. Klinman, and R. A. Seder. 2000. DNA vaccines: immunology, application, and optimization. Annu. Rev. Immunol. 18:927-974. [DOI] [PubMed] [Google Scholar]
- 18.Ha, S. J., J. Chang, M. K. Song, Y. S. Suh, H. T. Jin, C. H. Lee, G. H. Nam, G. Choi, K. Y. Choi, S. H. Lee, W. B. Kim, and Y. C. Sung. 2002. Engineering N-glycosylation mutations in IL-12 enhances sustained cytotoxic T lymphocyte responses for DNA immunization. Nat. Biotechnol. 20:381-386. [DOI] [PubMed] [Google Scholar]
- 19.Ha, S. J., B. Y. Jeon, S. C. Kim, D. J. Kim, M. K. Song, Y. C. Sung, and S. N. Cho. Therapeutic effect of DNA vaccines combined with chemotherapy in a latent infection model after aerosol infection of mice with Mycobacterium tuberculosis. Gene Ther., in press. [DOI] [PubMed]
- 20.Heath, W. R., and F. R. Carbone. 2001. Cross-presentation in viral immunity and self-tolerance. Nat. Rev. Immunol. 1:126-134. [DOI] [PubMed] [Google Scholar]
- 21.Kwissa, M., J. Unsinger, R. Schirmbeck, H. Hauser, and J. Reimann. 2000. Polyvalent DNA vaccines with bidirectional promoters. J. Mol. Med. 78:495-506. [DOI] [PubMed] [Google Scholar]
- 22.Large, M. K., D. J. Kittlesen, and Y. S. Hahn. 1999. Suppression of host immune response by the core protein of hepatitis C virus: possible implications for hepatitis C virus persistence. J. Immunol. 162:931-938. [PubMed] [Google Scholar]
- 23.Lechmann, M., and T. J. Liang. 2000. Vaccine development for hepatitis C. Semin. Liver Dis. 20:211-226. [DOI] [PubMed] [Google Scholar]
- 24.Lee, C. H., Y. H. Choi, S. H. Yang, C. W. Lee, S. J. Ha, and Y. C. Sung. 2001. Hepatitis C virus core protein inhibits interleukin 12 and nitric oxide production from activated macrophages. Virology 279:271-279. [DOI] [PubMed] [Google Scholar]
- 25.Lee, J. W., K.-M. Kim, S.-H. Jung, K. J. Lee, E.-C. Choi, Y.-C. Sung, and C.-Y. Kang. 1999. Identification of a domain containing B-cell epitopes in hepatitis C virus E2 glycoprotein by using mouse monoclonal antibodies. J. Virol. 73:11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, K. J., Y. A. Suh, Y. G. Cho, Y. S. Cho, G. W. Ha, K. H. Chung, J. H. Hwang, Y. D. Yun, D. S. Lee, C. M. Kim, and Y. C. Sung. 1997. Hepatitis C virus E2 protein purified from mammalian cells is frequently recognized by E2-specific antibodies in patient sera. J. Biol. Chem. 272:30040-30046. [DOI] [PubMed] [Google Scholar]
- 27.Lee, S. W., J. H. Cho, and Y. C. Sung. 1998. Optimal induction of hepatitis C virus envelope-specific immunity by bicistronic plasmid DNA inoculation with the granulocyte-macrophage colony-stimulating factor gene. J. Virol. 72:8430-8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leitner, W. W., L. N. Hwang, M. J. DeVeer, A. Zhou, R. H. Silverman, B. R. Williams, T. W. Dubensky, H. Ying, and N. P. Restifo. 2003. Alphavirus-based DNA vaccine breaks immunological tolerance by activating innate antiviral pathways. Nat. Med. 9:33-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis, P. J., S. van Drunen Littel-van den Hurk, and L. A. Babiuk. 1999. Altering the cellular location of an antigen expressed by a DNA-based vaccine modulates the immune response. J. Virol. 73:10214-10223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, M., G. M. Davey, R. M. Sutherland, C. Kurts, A. M. Lew, C. Hirst, F. R. Carbone, and W. R. Heath. 2001. Cell-associated ovalbumin is cross-presented much more efficiently than soluble ovalbumin in vivo. J. Immunol. 166:6099-6103. [DOI] [PubMed] [Google Scholar]
- 31.Miller, C. L., C. Johnston, P. M. Spittal, K. Li, N. Laliberte, J. S. Montaner, and M. T. Schechter. 2002. Opportunities for prevention: hepatitis C prevalence and incidence in a cohort of young injection drug users. Hepatology 36:737-742. [DOI] [PubMed] [Google Scholar]
- 32.Neumann, A. U., N. P. Lam, H. Dahari, D. R. Gretch, T. E. Wiley, T. J. Layden, and A. S. Perelson. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103-107. [DOI] [PubMed] [Google Scholar]
- 33.Power, C. A., C. L. Grand, N. Ismail, N. C. Peters, D. P. Yurkowski, and P. A. Bretscher. 1999. A valid ELISPOT assay for enumeration of ex vivo, antigen-specific, IFNγ-producing T cells. J. Immunol. Methods 227:99-107. [DOI] [PubMed] [Google Scholar]
- 34.Prince, A. M., and M. T. Shata. 2001. Immunoprophylaxis of hepatitis C virus infection. Clin. Liver Dis. 5:1091-1103. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez, F., and J. L. Whitton. 2000. Enhancing DNA immunization. Virology 268:233-238. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Shimizu, Y. K., A. J. Weiner, J. Rosenblatt, D. C. Wong, M. Shapiro, T. Popkin, M. Houghton, H. J. Alter, and R. H. Purcell. 1990. Early events in hepatitis C virus infection of chimpanzees. Proc. Natl. Acad. Sci. USA 87:6441-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugiyama, K., N. Kato, T. Mizutani, M. Ikeda, T. Tanaka, and K. Shimotohno. 1997. Genetic analysis of the hepatitis C virus (HCV) genome from HCV-infected human T cells. J. Gen. Virol. 78:329-336. [DOI] [PubMed] [Google Scholar]
- 39.Svanholm, C., L. Bandholtz, A. Lobell, and H. Wigzell. 1999. Enhancement of antibody responses by DNA immunization using expression vectors mediating efficient antigen secretion. J. Immunol. Methods 228:121-130. [DOI] [PubMed] [Google Scholar]
- 40.Uchijima, M., A. Yoshida, T. Nagata, and Y. Koide. 1998. Optimization of codon usage of plasmid DNA vaccine is required for the effective MHC class I-restricted T-cell responses against an intracellular bacterium. J. Immunol. 161:5594-5599. [PubMed] [Google Scholar]
- 41.van der Most, R. G., K. Murali-Krishna, J. L. Whitton, C. Oseroff, J. Alexander, S. Southwood, J. Sidney, R. W. Chesnut, A. Sette, and R. Ahmed. 1998. Identification of Db- and Kb-restricted subdominant cytotoxic T-cell responses in lymphocytic choriomeningitis virus-infected mice. Virology 240:158-167. [DOI] [PubMed] [Google Scholar]
- 42.van der Most, R. G., A. Sette, C. Oseroff, J. Alexander, K. Murali-Krishna, L. L. Lau, S. Southwood, J. Sidney, R. W. Chesnut, M. Matloubian, and R. Ahmed. 1996. Analysis of cytotoxic T-cell responses to dominant and subdominant epitopes during acute and chronic lymphocytic choriomeningitis virus infection. J. Immunol. 157:5543-5554. [PubMed] [Google Scholar]
- 43.Vitiello, A., L. Yuan, R. W. Chesnut, J. Sidney, S. Southwood, P. Farness, M. R. Jackson, P. A. Peterson, and A. Sette. 1996. Immunodominance analysis of CTL responses to influenza PR8 virus reveals two new dominant and subdominant Kb-restricted epitopes. J. Immunol. 157:5555-5562. [PubMed] [Google Scholar]
- 44.Wherry, E. J., K. A. Puorro, A. Porgador, and L. C. Eisenlohr. 1999. The induction of virus-specific CTL as a function of increasing epitope expression: responses rise steadily until excessively high levels of epitope are attained. J. Immunol. 163:3735-3745. [PubMed] [Google Scholar]
- 45.Wolff, J. A., R. W. Malone, P. Williams, W. Chong, G. Acsadi, A. Jani, and P. L. Felgner. 1990. Direct gene transfer into mouse muscle in vivo. Science 247:1465-1468. [DOI] [PubMed] [Google Scholar]
- 46.zur Megede, J., M.-C. Chen, B. Doe, M. Schaefer, C. E. Greer, M. Selby, G. R. Otten, and S. W. Barnett. 2000. Increased expression and immunogenicity of sequence-modified human immunodeficiency virus type 1 gag gene. J. Virol. 74:2628-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]