Abstract
Flaviviruses assemble in the endoplasmic reticulum by a mechanism that appears to be driven by lateral interactions between heterodimers of the envelope glycoproteins E and prM. Immature intracellular virus particles are then transported through the secretory pathway and converted to their mature form by cleavage of the prM protein by the cellular protease furin. Earlier studies showed that when the prM and E proteins of tick-borne encephalitis virus are expressed together in mammalian cells, they assemble into membrane-containing, icosahedrally symmetrical recombinant subviral particles (RSPs), which are smaller than whole virions but retain functional properties and undergo cleavage maturation, yielding a mature form in which the E proteins are arranged in a regular T = 1 icosahedral lattice. In this study, we generated immature subviral particles by mutation of the furin recognition site in prM. The mutation resulted in the secretion of two distinct size classes of particles that could be separated by sucrose gradient centrifugation. Electron microscopy showed that the smaller particles were approximately the same size as the previously described mature RSPs, whereas the larger particles were approximately the same size as the virus. Particles of the larger size class were also detected with a wild-type construct that allowed prM cleavage, although in this case the smaller size class was far more prevalent. Subtle differences in endoglycosidase sensitivity patterns suggested that, in contrast to the small particles, the E glycoproteins in the large subviral particles and whole virions might be in nonequivalent structural environments during intracellular transport, with a portion of them inaccessible to cellular glycan processing enzymes. These proteins thus appear to have the intrinsic ability to form alternative assembly products that could provide important clues about the role of lateral envelope protein interactions in flavivirus assembly.
Flaviviruses are small enveloped viruses that are assembled intracellularly, apparently by budding into the endoplasmic reticulum (ER) (28). The entire virion, consisting of a nucleocapsid, lipid membrane, and two envelope glycoproteins, prM and E, is then transported through the secretory pathway (32) and, after undergoing posttranslational modifications that include the cleavage of the prM protein, is exported from the cell by exocytosis. Newly synthesized prM and E proteins rapidly associate to form heterodimers (6, 30, 46), which then further associate to organize the envelope into a regular icosahedral structure (8, 25, 48). The idea that lateral envelope protein interactions play a dominant role in assembly is supported by the observations that ordered membrane-containing subviral particles can be obtained by coexpressing prM and E in the absence of other viral components (4, 9, 19-23, 35, 36, 41) and that subviral particles containing only the envelope proteins are also secreted as a by-product of natural infections (39).
Shortly before being released from the cell, the prM proteins in the immature virus are cleaved by the trans-Golgi network resident proprotein convertase furin (43), resulting in the loss of approximately the N-terminal half of this protein and leaving the small (8 kDa) C-terminal portion, the M protein, anchored in the membrane. The cleavage of prM converts the viral envelope proteins to a metastable state (44), allowing them to undergo the further structural changes that are required for membrane fusion (3, 43).
Studies with recombinant subviral particles (RSPs) from tick-borne encephalitis (TBE) virus have shown that they are assembled in the ER and transported through the secretory pathway in the same manner as whole virus particles (31), that they undergo similar posttranslational modifications during transport (41), and that their envelope proteins are presented in a fully functional state (5, 41). The molecular organization of a mature RSP from TBE virus was previously determined by cryoelectron microscopy and icosahedral image reconstruction (8). It was demonstrated that the prevalent species secreted from transfected COS-1 cells expressing prM and E is an icosahedrally symmetrical particle with a diameter of 31.5 nm (compared to approximately 50 nm for the whole virus). Fitting of the X-ray crystal structure of the TBE virus E protein (38) into the experimental electron density revealed that the E proteins in the mature RSP make a regular lattice structure consisting of 30 E homodimers in a T = 1 arrangement. The lateral interactions that stabilize the E protein network are apparently identical at each position of the particle. Based on this structure, a model was proposed for the arrangement of the E proteins in the native mature TBE virion particle by using quasiequivalent interactions to make a T = 3 icosahedral lattice with a diameter of 50 nm (8). Since then, it has been shown by Kuhn et al. (25) that the E protein arrangement in another flavivirus, dengue virus, does not conform to this model but instead fits a herringbone model that is not governed by quasiequivalence and features different types of lateral interactions between the E proteins at different sites on the icosahedral lattice.
In this study, we generated immature subviral particles by introducing a mutation in the furin recognition site of prM. The mutation resulted in the secretion of two distinct, relatively homogeneous populations of prM-containing particles with diameters of about 30 and 55 nm. Further characterization showed that these two particle classes probably represent specific alternative assembly products with subtle differences in glycan modification that could indicate that, in contrast to the small particles, the E proteins in the large subviral particles and virions are in nonequivalent structural environments during intracellular transport.
MATERIALS AND METHODS
Construction of prM cleavage mutant.
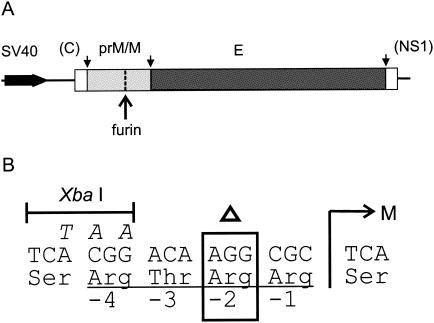
Plasmid SV-PEwt (1), which contains nucleotides 388 to 2550 from TBE virus strain Neudoerfl (34) (GenBank accession no. U27495) under the control of a simian virus 40 (SV40) early promoter, was the parent construct used for mutant construction and for wild-type controls. To construct the prM cleavage mutant SV-P(Δ88)E, a restriction fragment (SapI-AgeI, nucleotides 648 to 960) corresponding approximately to the prM coding region of SV-PEwt was replaced by the corresponding fragment from plasmid dR204, an intermediate construct used previously for constructing the full-length mutant clone pTNd/prM(Δ88) (7). This fragment contained the deletion of the codon for arginine 88 of prM (nucleotides 742 to 744) as well as silent mutations at nucleotides 735, 736, and 738 to produce a new XbaI site (Fig. 1). Plasmids were propagated in Escherichia coli HB101 and purified with a Qiagen plasmid mega kit. Sequence analysis of the prM and E coding regions was performed with an automated DNA sequencing system (ABI) to confirm that only the desired mutations were present.
FIG. 1.
Mutation of the furin cleavage site in prM. (A) Schematic diagram of the portion of the TBE virus genome included in the wild-type expression plasmid SV-PEwt (1) and the SV-P(Δ88)E deletion mutant used in this study. These constructs contained the coding region for prM and E under the control of the SV40 early promoter as well as short flanking regions from the genes encoding the capsid protein (C) and nonstructural protein 1 (NS1). The sites where the polyprotein is cleaved by host cell signalase (28) are indicated by small arrows, and the furin cleavage site in prM is indicated by a large arrow. (B) Sequence changes at the furin recognition site. The position of the N terminus of the M protein, which is normally created by furin cleavage, is indicated by an M, and the Arg 88 codon at position −2 that was deleted in SV-P(Δ88)E is indicated by an open triangle. Additional silent mutations in SV-P(Δ88)E resulting in a new XbaI site are shown in italics above the wild-type sequence.
Production of subviral particles and virus.
COS-1 cells (ATCC CRL 1650) were grown in Dulbecco's modified Eagle medium (Life Technologies) supplemented with 10% fetal calf serum, 4 mM glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml at 37°C in 5% CO2. Cells were transfected with purified plasmid with a Bio-Rad gene pulser apparatus and allowed to grow for another 22 to 24 h in the normal growth medium. The medium was then replaced with serum-free bicarbonate- and HEPES-buffered Dulbecco's modified Eagle medium (Life Technologies), and incubation was continued for another 24 h.
The cell supernatants were cleared by centrifugation at 10,000 rpm for 30 min at 4°C in a Beckman JLA16.250 rotor, and particles were collected by polyethylene glycol (PEG) precipitation as described previously (8). In some cases, harvesting was done instead by pelleting in an ultracentrifuge (Beckman 45 Ti rotor, 44,000 rpm, 4°C, 120 min). The particles were then separated by rate-zonal sucrose density gradient centrifugation (41) and, when high purity was required, further purified by equilibrium density centrifugation on 20 to 50% sucrose gradients as described previously (41).
TBE virus strain Neudoerfl was grown in primary chicken embryo cells and purified by two cycles of sucrose gradient centrifugation as described previously (15). For production of secreted immature virus, ammonium chloride was added to the cell culture medium during virus growth to a final concentration of 20 mM to suppress prM cleavage (16).
Sedimentation analysis.
Subviral particles were analyzed by centrifugation in 5 to 20% (wt/wt) sucrose gradients made with TAN buffer (0.05 M triethanolamine [pH 8.0], 0.1 M NaCl). For membrane solubilization experiments, samples were treated first for 1 h at room temperature with Triton X-100 (final concentration, 0.5% [wt/wt]) and the corresponding sucrose gradients were made with 0.1% Triton X-100 to prevent aggregation. The gradients were then centrifuged in a Beckman SW 40 rotor at 38,000 rpm at 4°C for 90 min and fractionated from the top with a BioComp piston gradient fractionator. The amount of E protein in the fractions was quantified by a four-layer enzyme-linked immunosorbent assay (ELISA) after heating the samples and virus standard at 65°C for 30 min with 0.4% sodium dodecyl sulfate (SDS) (16).
Electron microscopy.
Sucrose gradient fractions were first diluted approximately threefold in TAN buffer plus 10 mM MgCl2. The particles were then concentrated by ultracentrifugation in a Beckman SW40 rotor at 43,000 rpm for 3 h at 4°C and resuspension in a small volume of buffer. Samples were adsorbed to glow-discharged carbon-coated copper grids, washed with two drops of deionized water, and stained with two drops of freshly prepared 0.75% uranyl formate. Images were taken with a Tecnai 12 electron microscope (Philips, Eindhoven, The Netherlands) at 120 kV, with a magnification of ×21,000 and a defocus of 2 to 3 μm.
Endoglycosidase treatment, gel electrophoresis, and immunoblotting.
Endoglycosidase Hf (endo Hf) and peptide N-glycosidase F (PNGase F) (New England Biolabs) were used according to the manufacturer's instructions with the provided buffers. Protein samples were precipitated with deoxycholate and trichloroacetic acid and separated by SDS-polyacrylamide gel electrophoresis (PAGE) as described by Laemmli and Favre (26). For analysis of protein composition, gels containing 15% acrylamide were used. For analysis of glycosidase-treated E and prM proteins, gels with 10% acrylamide were used.
Protein bands were visualized with Coomassie PhastGel Blue R (Pharmacia) or by immunoblotting onto a polyvinylidene difluoride membrane (Bio-Rad) with a Bio-Rad semidry transfer cell as described previously (41). In the latter case, a polyclonal rabbit serum (KVIII) recognizing the TBE virus structural proteins (43) was used together with a donkey anti-rabbit immunoglobulin horseradish peroxidase conjugate (Amersham) for detection.
Analysis of monoclonal antibody binding.
A four-layer ELISA (17) was used to measure the reactivity of the different preparations with a panel of E-protein-specific monoclonal antibodies (MAbs) (13, 18) as described previously by Schalich et al. (41). The antigen (0.5 μg of E protein per ml) was captured on the solid phase by a guinea pig anti-TBE virus immunoglobulin and tested with a single dilution of each MAb and a peroxidase-labeled rabbit anti-mouse immunoglobulin (Nordic) for detection.
RESULTS
Mutation of the furin cleavage site in prM.
To make subviral particles that are secreted from cells while still in their immature state, we altered the furin recognition site in prM to prevent cleavage during exocytosis. In an earlier study (7), it was shown with an infectious clone system that the deletion of arginine 88, two residues before the furin cleavage site in prM, prevented prM cleavage and eliminated the ability of the virus to be passaged in cell culture. This deletion disrupted the furin recognition motif R-X-R/K-R and rendered it uncleavable by furin. In the same study, it was also shown that infectivity could be restored by the addition of exogenous trypsin, indicating that the mutation had caused the envelope proteins to be held in a latent but nevertheless potentially functional state. We therefore introduced this same mutation into the corresponding site of plasmid SV-PEwt, which contains the coding region for prM and E under the control of an SV40 promoter and has been used in several previous studies for generating mature RSPs (2, 4, 5, 8, 41). The position of the deletion in the resulting plasmid construct, SV-P(Δ88)E, is shown in Fig. 1.
Expression and secretion of mutant subviral particles.
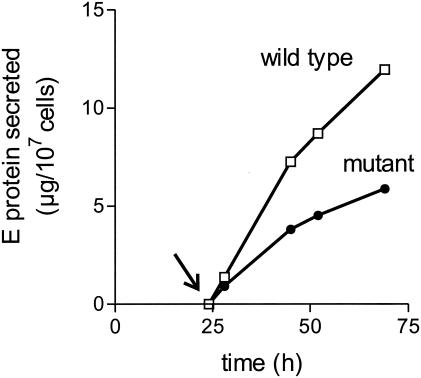
To investigate the effect of the mutation on the efficiency of particle assembly, transport, and secretion, COS-1 cells were transfected either with the wild-type plasmid or with the construct carrying the prM mutation, and the kinetics of secretion were monitored by quantifying the amount of E protein present in the cell culture medium at different time points. The transfection efficiency (approximately 50% for both the wild type and the mutant) was monitored by immunofluorescence staining (data not shown). As shown in Fig. 2, transfection with each of the plasmids resulted in a significant accumulation of extracellular E protein, but the amount secreted was somewhat less with the mutant than with the wild-type construct. In three separate experiments, the rate of extracellular E protein production by the mutant was 40 to 60% of that of the wild-type control, suggesting either that the mutation had caused a minor decrease in the overall rate of synthesis and particle assembly or that the mutant proteins required more time to pass through the secretory pathway.
FIG. 2.
Kinetics of E protein secretion from COS-1 cells transfected with plasmid SV-PEwt (wild-type) or SV-P(Δ88)E (mutant). The cell culture medium was replaced with fresh medium 24 h after transfection (arrow).
Two populations of subviral particles with different sedimentation properties.
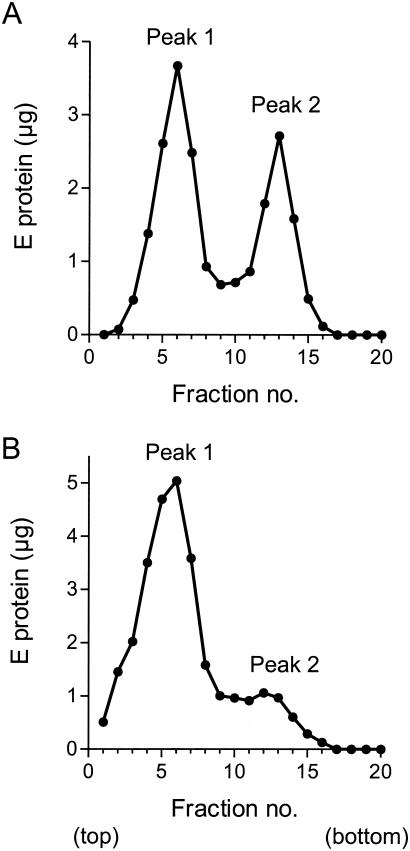
In order to characterize the mutant particles, larger-scale preparations were made. Particles were harvested from clarified cell culture supernatants by PEG precipitation and further purified by sucrose gradient centrifugation under conditions used previously for mature wild-type RSPs (8, 41). Unexpectedly, the harvested material from the prM mutant was found to contain two distinct major populations of particles that sedimented at different rates in rate-zonal sucrose density gradients (Fig. 3A). Peak 1 migrated to the same position as the previously reported mature RSP (41), whereas peak 2 sedimented much faster, suggesting a larger particle size. The ratio of the amount of E protein in peak 1 to that in peak 2 was approximately 1.5 to 1 and varied only slightly when samples were analyzed at different time points between 42 and 66 h posttransfection or when they were harvested by ultracentrifugation instead of PEG precipitation (data not shown).
FIG. 3.
Rate-zonal sucrose density gradient centrifugation of clarified cell culture supernatants. (A) Cells transfected with the prM mutant. (B) Cells transfected with wild-type plasmid. The sedimentation direction is left to right.
Similar analyses were performed to see whether cells transfected with the wild-type construct also secrete a faster-sedimenting particle form. As shown in Fig. 3B, the pattern of distribution in supernatants from wild-type-transfected cells was quite different from that of the mutant, and as expected, peak 1, which was known from earlier studies to correspond to the previously described mature RSP (8, 41), was by far the dominant form. Nevertheless, a second, minor population of particles with a sedimentation rate similar to that of the mutant peak 2 could also be consistently detected when sufficient amounts of starting material were analyzed (Fig. 3B).
Two different particle sizes shown by electron microscopy.
Particles from sucrose gradient peaks 1 and 2 were concentrated, stained with uranyl formate, and analyzed by electron microscopy. As shown in Fig. 4A, the predominant species in peak 1 from the mutant preparation was a spherical particle with a diameter of about 30 nm (n = 74, mean = 28.2 nm, standard deviation [SD] = 2.69 nm). This corresponds in size and shape to the mature wild-type RSP (n = 108, mean = 30.23 nm, SD = 2.35 nm) (Fig. 4B), which has been shown previously to have a T = 1 icosahedral arrangement of 30 E protein homodimers on its surface (8). In addition, the negatively stained images revealed a number of misshapen particles that probably represent damaged RSPs. Peak 2 from the prM mutant also contained mostly spherical particles (Fig. 4C), although these appeared somewhat more heterogeneous in shape than the peak 1 material. In contrast to peak 1, however, the most prevalent form in this population had a diameter of about 55 nm (n = 170, mean = 55.4 nm, SD = 3.22 nm), which corresponds approximately to the diameter of the whole virion. In addition, a few of the smaller 30-nm-diameter particles were visible in these preparations (Fig. 4C), probably due a slight overlap between the peaks or particle aggregation. The faster-sedimenting peak (peak 2) from the wild-type preparations also consisted primarily of round, approximately 55-nm-diameter particles (Fig. 4D). These two major size classes thus appear to be distinct assembly products.
FIG. 4.
Electron micrographs of mutant and wild-type subviral particles stained with formyl acetate and photographed at the same magnification. (A) Peak 1 (prM mutant); (B) peak 1 (wild type); (C) peak 2 (prM mutant); (D) peak 2 (wild type).
Biochemical and physical characteristics of subviral particles.
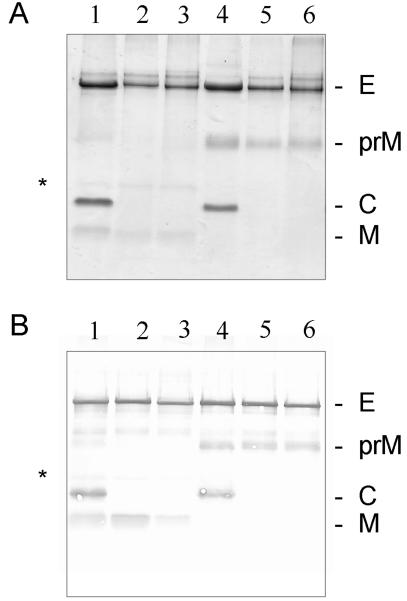
The four types of subviral particles were further purified and analyzed by SDS-PAGE (Fig. 5). Mature TBE virus (Fig. 5, lane 1) and immature TBE virus produced by treating infected cells with ammonium chloride (16) (Fig. 5, lane 4) were used as controls. In the Coomassie blue-stained gel shown in Fig. 5A, it can be seen that both the small (30-nm diameter) (Fig. 5, lane 5) and large (55-nm diameter) (Fig. 5, lane 6) particles produced by using the prM mutant construct contained E and uncleaved prM proteins, with no detectable M. In contrast, most of the prM protein had been cleaved in both the small (Fig. 5, lane 2) and large (Fig. 5, lane 3) particles produced by using the wild-type construct. These results were confirmed by immunoblotting, as shown in Fig. 5B, and the data together demonstrate that the deletion of arginine 88 in the furin recognition site of prM was indeed sufficient to abolish prM cleavage in COS-1 cells. Thus, both sizes of particles containing the mutation represent immature forms. Consistent with their immature state (43) and in contrast to both sizes of wild-type subviral particles and mature virus, they also lacked the ability to hemagglutinate goose erythrocytes at pH 6.4 (data not shown).
FIG. 5.
SDS-PAGE of proteins from purified virions and subviral particles. (A) Gel stained with Coomassie blue. (B) Immunoblot with polyclonal antiserum KVIII. Lanes: 1, mature TBE virus; 2, small wild-type subviral particle; 3, large wild-type subviral particle; 4, immature TBE virus; 5, small mutant subviral particle; 6, large mutant subviral particle. The positions of the bands for proteins E, prM, C (capsid protein), and M are indicated at the right. A minor band, corresponding to a previously identified SDS-resistant M dimer (43), is also indicated (*).
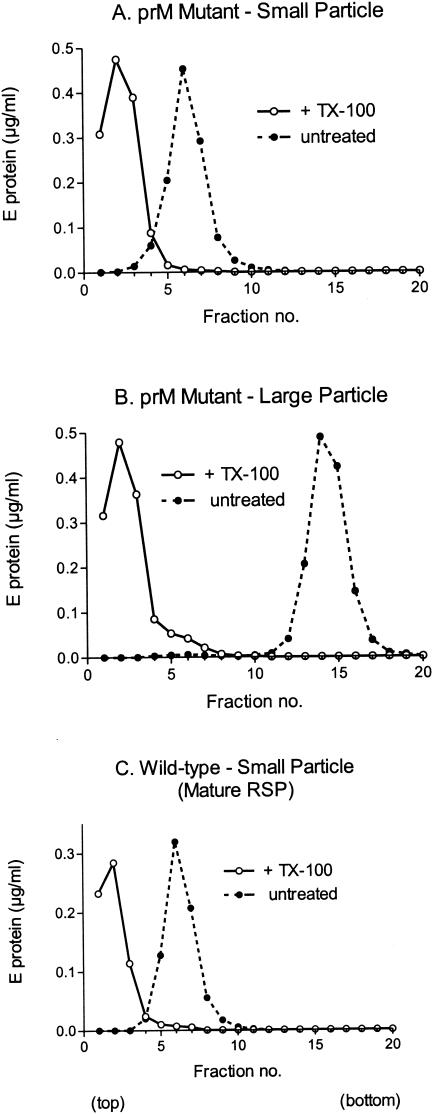
To get a first indication of whether both particle size classes contain a lipid membrane, samples from the peak fractions shown in Fig. 3A were treated with 0.5% Triton X-100 for 1 h and centrifuged in 5 to 20% sucrose gradients containing 0.1% Triton X-100. Untreated samples in gradients without detergent were used as controls. As shown in Fig. 6, both the small particles from peak 1 and the large particles from peak 2 were dispersed by the detergent treatment, causing the E protein peak to be shifted to the top of the gradient. This is consistent with the presence of a lipid membrane in both particle size classes. It is also significant that there was no change in the sedimentation behavior of the untreated controls between the first and second sedimentation analyses (compare Fig. 3 and 6), confirming that each peak represents a relatively stable and defined subpopulation.
FIG. 6.
Detergent sensitivity of small and large subviral particles. The peak fractions from the experiment shown in Fig. 3A were analyzed again by rate-zonal sucrose density gradient centrifugation after treatment with 0.5% Triton X-100 (open circles) or no detergent treatment (filled circles). Triton X-100 (0.1%) was included in the gradients of detergent-treated samples to prevent aggregation. (A) Small immature mutant subviral particle (peak 1); (B) large immature mutant subviral particle (peak 2); (C) small mature subviral particle (RSP control). The sedimentation direction is left to right.
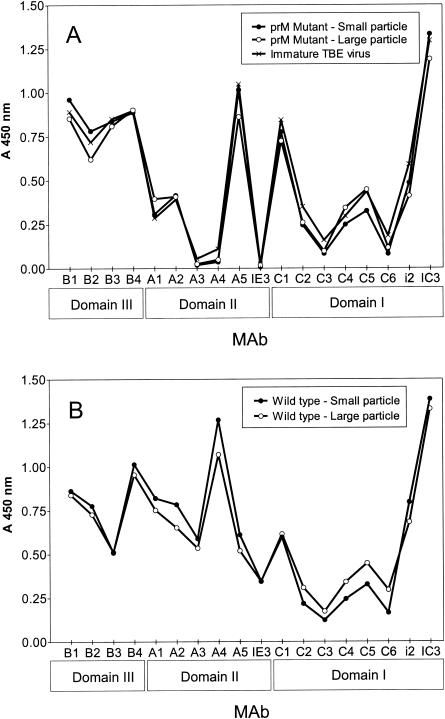
To assess the conformational state of the envelope glycoproteins in each of the particle types, we tested their reactivity in a four-layer ELISA, with a panel of 18 E-specific MAbs (13, 18), which has been used in several earlier studies for the analysis of the conformation of protein E. These MAbs have been shown to bind equally well to whole virions and RSPs (14, 41), but several of them react differently with the immature prM-containing forms, yielding different reactivity profiles in the ELISA (7, 16, 41). The data shown in Fig. 7 reveal that both populations of mutant, prM-containing, immature RSPs are very similar in their reactivity patterns to immature whole virions produced by suppressing prM cleavage by treating cells with ammonium chloride. This suggests that the E proteins in both the small and large immature subviral particles are in the same conformation as found on immature virions. The mature small and large wild-type particles yielded profiles that were very similar to each other but distinctly different from those of the immature forms (Fig. 7, compare panels A and B). The most pronounced differences between the mature and immature forms were observed with MAbs recognizing domain II (especially MAbs A3, A4, A5, and IE3), as has been observed previously (7, 16, 41). From these results, it therefore appears that the antigenic structure and, thus, the envelope protein conformation are independent of particle size.
FIG. 7.
Antibody binding profiles of immature (A) and mature (B) subviral particles. The binding of 18 E-protein-specific MAbs with each of the different subviral particle types was compared by four-layer ELISA. Immature TBE virus produced by treating cells with 20 mM NH4Cl was used as a control for the immature forms. The structural domain of the E protein to which each of the MAbs binds (38) is indicated at the bottom.
Differences in glycan processing.
The prM and E proteins of TBE virus each contain a single N-linked oligosaccharide moiety (30, 47). It has been shown previously that, in secreted mature 30-nm-diameter RSPs, all of the N-linked glycans attached to Asn 154 in the E protein are processed to an endo H-resistant form, whereas in secreted mature TBE virus particles produced in primary chicken embryo cells or COS-1 cells, a substantial portion consistently remains sensitive to endo H cleavage (41), indicating that only a subset of the E proteins possesses a fully processed glycan. It is not known, however, whether this difference is attributable to differences in accessibility to the processing enzymes or to other factors affecting the cellular machinery during infection. Therefore, to investigate potential differences in the degree of glycan processing, we treated all four subviral particle types as well as mature and immature virus controls with endo Hf, an enzyme that cleaves immature high-mannose-type glycans, and PNGase F, which removes both complex and high-mannose sugar chains (33).
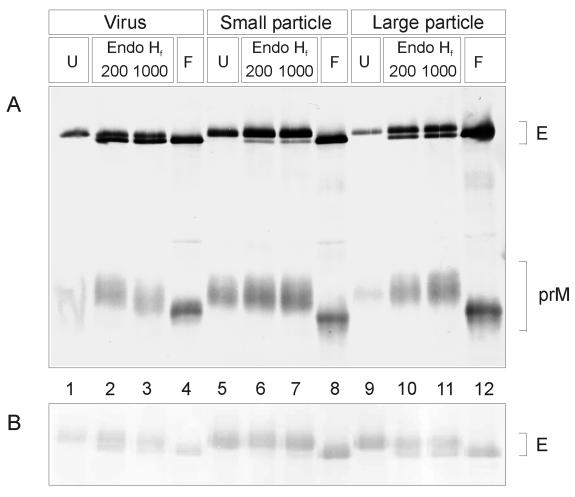
Figure 8A shows an immunoblot of a gel on which different glycosidase-treated samples were analyzed by electrophoresis. In this experiment, immature virus and the small and large immature subviral particles were treated with two different concentrations of endo Hf as well as with PNGase F. Treatment with PNGase F resulted in complete deglycosylation of E and prM for all particle types, causing a shift in the electrophoretic mobility of both proteins. However, with endo Hf, the E protein deglycosylation pattern was different for the small subviral particles than for the large subviral particles and whole immature virions. As expected from earlier work (41), nearly all of the glycans linked to the E proteins of the small immature subviral particles were endo Hf resistant, indicating that most of these were of the complex type. The large immature subviral particles, in contrast, displayed a pattern more similar to that of the immature virus, with an apparently larger proportion of the total E protein still in an endo Hf-sensitive form. An analysis of the E proteins of the corresponding mature forms yielded essentially identical results (Fig. 8B), as was expected because all of the sugar modifications occur while the particles are still in an immature state and are therefore independent of the subsequent prM cleavage step. The prM proteins, on the other hand, were in the fully processed endo Hf-resistant form, regardless of which immature particle type was analyzed (Fig. 8A). These data reveal differences in glycan processing patterns between the small and large particles that are possibly attributable to local differences in the accessibility of sugar chains to the processing enzymes of the secretory pathway. These data are consistent with a nonequivalent arrangement of E proteins in the large immature subviral particles and immature virions during intracellular transport.
FIG. 8.
(A) Endoglycosidase treatment of glycoproteins from secreted immature TBE virus from infected cells treated with 20 mM NH4Cl (lanes 1 to 4), secreted small immature subviral particles (prM mutant) (lanes 5 to 8), and secreted large immature subviral particles (prM mutant) (lanes 9 to 12). (B) Same treatment as described for panel A but using the corresponding mature forms. Samples were treated with 200 or 1,000 U of endo Hf, as indicated, or 500 U of PNGase F (F) and compared to untreated controls (U) by SDS-PAGE and immunoblotting with polyclonal antiserum KVIII. The positions of the E and prM protein bands are indicated at the right.
DISCUSSION
It is an interesting feature of flaviviruses that their envelope proteins appear to play a dominant role in the assembly process. Their ability to form capsidless subviral particles suggests that lateral interactions between the heterodimers of prM and E in the ER membrane may be sufficient to induce budding into the lumen of this organelle, although it has not been ruled out that cellular components could also play at least a chaperone-like role in this process. Furthermore, the apparent scarcity of preformed core particles in the cytoplasm of infected cells (28) would suggest that an envelope protein lattice in the ER might even guide capsid assembly by ordering C protein capsomers at the budding site. A similar proposal has also been made in the case of the alphaviruses based on the properties of capsid mutants (10), but alphaviruses, in contrast to flaviviruses, bud at the plasma membrane and apparently do not make subviral particles (11). However, secreted subviral particles produced by expression of envelope proteins alone have been reported for hepatitis B virus (27) and the coronavirus mouse hepatitis virus (45), but also in these cases, the exact role of the envelope proteins in the budding mechanism is still unknown.
In this study, we found that coexpression of proteins E and prM leads to the secretion of two distinct classes of subviral particles, the first of which corresponds in size to the previously described RSPs (8, 41) and the second of which has approximately the same outer dimensions as a whole virus particle. Since both of these forms are assembled in the same cells from the same building blocks in the absence of capsid or other viral proteins, it appears that the ability to make particles of two specific sizes is a property that is encoded in the envelope proteins themselves. The intrinsic ability of viral structural proteins to form more than one type and size of closed icosahedral shell has been demonstrated previously with capsid proteins from several different viruses (24, 29, 42, 49). Although it is not yet known whether the immature subviral particles described in this study have icosahedral symmetry, the fact that they form two defined and stable subpopulations strongly suggests that the number of subunits per particle is not random and is most likely governed by specific lateral interactions that would favor an icosahedral arrangement.
Although the production and characterization of flavivirus subviral particles has been reported in a number of previous studies (4, 9, 19-23, 35-37, 40, 41), secreted particles of the larger size class had not yet been reported. In studies with wild-type RSPs, the larger particles were probably overlooked because of their low abundance relative to the small particles as well as the lack of sensitivity of certain assays. It is also possible that the ability to secrete the larger particles is a property that is not shared by all flaviviruses.
The successful use of mutations in the furin recognition site in prM to produce stable CHO cell lines secreting immature subviral particles has been reported recently for the Japanese encephalitis virus (21) and dengue virus type 2 (20) envelope proteins, but in each of these studies only one size class was reported. These authors showed that making prM uncleavable by furin improved the stability of the cell lines by eliminating the ability of the secreted particles to induce syncytium formation. For TBE virus, on the other hand, it has been shown recently that a CHO cell line constitutively producing wild-type RSPs could be established without mutating the prM cleavage site or suppressing fusogenic activity (12).
In this study, the mutation of the furin cleavage site in prM resulted in a substantial shift in the particle size ratio in favor of the larger size class. The reason for this phenomenon is not clear. It is possible that the structure of the mature envelope is relatively unstable in the absence of the viral core and that the presence of prM lends a degree of additional stability that favors an accumulation of the larger form. Alternatively, it is possible that the prM mutation causes a change in prM or E that is slightly unfavorable for the formation or release of the smaller particle type. COS-1 cells transfected with SV-PEwt (the same wild-type plasmid used in this study) appear to contain two different sizes of RSP-like particles in the ER and other compartments of the secretory pathway, including post-Golgi vesicles (31). These intracellular particles are approximately of the same sizes as the extracellular particles reported here, with the larger size accounting for about 25% of the total. In the present study, we found that with the prM mutant, the ratio of the amount of E protein in peak 1 to that in peak 2 was about 1.5 to 1. If we assume, based on the available structural studies with TBE virus RSPs (8) and dengue virus (25) that the large particles contain three times as many E protein molecules as the small ones (180 versus 60 monomers), the calculated particle number ratio for the prM mutant would be about 4.5 to 1, which is fairly consistent with what has been observed in cell sections by electron microscopy (31). In the case of the mature wild-type particles, however, we observed much higher small-to-large particle ratios in cell supernatants. The results of both studies together suggest that the large wild-type particles are probably efficiently synthesized and transported through the secretory pathway but that preventing prM cleavage somehow alters their relative rate of release from the cell or their ability to accumulate in the cell culture medium due to enhanced stability.
The large subviral particles and whole virions, in contrast to the small subviral particles, were found to contain a relatively large proportion of E proteins with sugar chains that had not been converted to the complex form during intracellular transport. Furthermore, because the two subviral particle types compared in this study came from the same cells, it can be concluded that these differences were not caused by any effects on the processing enzymes or cell machinery but instead were probably due to differences in the structural environment of the sugar chains themselves, although subtle differences in intracellular transport pathways have not yet been ruled out. The endo Hf digestion patterns therefore suggest that the E proteins in the virions and large subviral particles are in nonequivalent environments at the time that the particle is transported through the Golgi, consistent with the observed nonequivalence of the E protein contacts in mature dengue virus (25) and spike asymmetry in immature dengue and yellow fever viruses (48). The glycosylation pattern is a reflection of the state of the immature particle before transport to the trans-Golgi network, where prM cleavage and virus maturation presumably occur, and not of the E protein arrangement in the mature state, whose final geometry could be the result of further rearrangements.
Although medium-resolution structures for both mature and immature dengue virus have recently been determined by cryoelectron microscopy (25, 48), the way in which the envelope proteins rearrange to the mature configuration upon prM cleavage remains enigmatic. This is because each of these forms has a complex geometry whose relationship to its counterpart is not obvious (48). It is likely that the two classes of subviral particles described here have spatially different packing arrangements but nevertheless undergo similar rearrangements upon prM cleavage. Their use in comparative structural studies might therefore simplify the process of identifying the underlying mechanisms that are important for flavivirus maturation.
Acknowledgments
We thank Angela Dohnal for excellent technical assistance throughout the course of this work. We are also grateful to Walter Holzer for help with virus production, to Elisabeth Krach for help with secretion time course experiments, and to Yifan Cheng for help with electron microscopy.
REFERENCES
- 1.Allison, S. L., C. W. Mandl, C. Kunz, and F. X. Heinz. 1994. Expression of cloned envelope protein genes from the flavivirus tick-borne encephalitis virus in mammalian cells and random mutagenesis by PCR. Virus Genes 8:187-198. [DOI] [PubMed] [Google Scholar]
- 2.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, and F. X. Heinz. 2001. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 75:4268-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 69:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allison, S. L., K. Stadler, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J. Virol. 69:5816-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corver, J., A. Ortiz, S. L. Allison, J. Schalich, F. X. Heinz, and J. Wilschut. 2000. Membrane fusion activity of tick-borne encephalitis virus and recombinant subviral particles in a liposomal model system. Virology 269:37-46. [DOI] [PubMed] [Google Scholar]
- 6.Courageot, M. P., M. P. Frenkiel, C. D. D. Santos, V. Deubel, and P. Desprès. 2000. α-Glucosidase inhibitors reduce dengue virus production by affecting the initial steps of virion morphogenesis in the endoplasmic reticulum. J. Virol. 74:564-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elshuber, S., S. L. Allison, F. X. Heinz, and C. W. Mandl. 2003. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J. Gen. Virol. 84:183-191. [DOI] [PubMed] [Google Scholar]
- 8.Ferlenghi, I., M. Clarke, T. Ruttan, S. L. Allison, J. Schalich, F. X. Heinz, S. C. Harrison, F. A. Rey, and S. D. Fuller. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis. Mol. Cell 7:593-602. [DOI] [PubMed] [Google Scholar]
- 9.Fonseca, B. A., S. Pincus, R. E. Shope, E. Paoletti, and P. W. Mason. 1994. Recombinant vaccinia viruses co-expressing dengue-1 glycoproteins prM and E induce neutralizing antibodies in mice. Vaccine 12:279-285. [DOI] [PubMed] [Google Scholar]
- 10.Forsell, K., L. Xing, T. Kozlovska, R. H. Cheng, and H. Garoff. 2000. Membrane proteins organize a symmetrical virus. EMBO J. 19:5081-5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garoff, H., R. Hewson, and D. E. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gehrke, R., M. Ecker, S. W. Aberle, S. L. Allison, F. X. Heinz, and C. W. Mandl. 2003. Incorporation of tick-borne encephalitis virus replicons into virus-like particles by a packaging cell line. 77:8924-8933. [DOI] [PMC free article] [PubMed]
- 13.Guirakhoo, F., F. X. Heinz, and C. Kunz. 1989. Epitope model of tick-borne encephalitis virus envelope glycoprotein E: analysis of structural properties, role of carbohydrate side chain, and conformational changes occurring at acidic pH. Virology 169:90-99. [DOI] [PubMed] [Google Scholar]
- 14.Heinz, F. X., S. L. Allison, K. Stiasny, J. Schalich, H. Holzmann, C. W. Mandl, and C. Kunz. 1995. Recombinant and virion-derived soluble and particulate immunogens for vaccination against tick-borne encephalitis. Vaccine 13:1636-1642. [DOI] [PubMed] [Google Scholar]
- 15.Heinz, F. X., and C. Kunz. 1981. Homogeneity of the structural glycoprotein from European isolates of tick-borne encephalitis virus: comparison with other flaviviruses. J. Gen. Virol. 57:263-274. [DOI] [PubMed] [Google Scholar]
- 16.Heinz, F. X., K. Stiasny, A. G. Puschner, H. Holzmann, S. L. Allison, C. W. Mandl, and C. Kunz. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198:109-117. [DOI] [PubMed] [Google Scholar]
- 17.Heinz, F. X., W. Tuma, F. Guirakhoo, and C. Kunz. 1986. A model study of the use of monoclonal antibodies in capture enzyme immunoassays for antigen quantification exploiting the epitope map of tick-borne encephalitis virus. J. Biol. Stand. 14:133-141. [DOI] [PubMed] [Google Scholar]
- 18.Holzmann, H., K. Stiasny, H. York, F. Dorner, C. Kunz, and F. X. Heinz. 1995. Tick-borne encephalitis virus envelope protein E-specific monoclonal antibodies for the study of low pH-induced conformational changes and immature virions. Arch. Virol. 140:213-221. [DOI] [PubMed] [Google Scholar]
- 19.Hunt, A. R., C. B. Cropp, and G. J. J. Chang. 2001. A recombinant particulate antigen of Japanese encephalitis virus produced in stably-transformed cells is an effective noninfectious antigen and subunit immunogen. J. Virol. Methods 97:133-149. [DOI] [PubMed] [Google Scholar]
- 20.Konishi, E., and A. Fujii. 2002. Dengue type 2 virus subviral extracellular particles produced by a stably transfected mammalian cell line and their evaluation for a subunit vaccine. Vaccine 20:1058-1067. [DOI] [PubMed] [Google Scholar]
- 21.Konishi, E., A. Fujii, and P. W. Mason. 2001. Generation and characterization of a mammalian cell line continuously expressing Japanese encephalitis virus subviral particles. J. Virol. 75:2204-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konishi, E., S. Pincus, E. Paoletti, R. E. Shope, T. Burrage, and P. W. Mason. 1992. Mice immunized with a subviral particle containing the Japanese encephalitis virus prM/M and E proteins are protected from lethal JEV infection. Virology 188:714-720. [DOI] [PubMed] [Google Scholar]
- 23.Kroeger, M. A., and P. C. McMinn. 2002. Murray Valley encephalitis virus recombinant subviral particles protect mice from lethal challenge with virulent wild-type virus. Arch. Virol. 147:1155-1172. [DOI] [PubMed] [Google Scholar]
- 24.Krol, M. A., N. H. Olson, J. Tate, J. E. Johnson, T. S. Baker, and P. Ahlquist. 1999. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc. Natl. Acad. Sci. USA 96:13650-13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli, U. K., and M. Favre. 1973. Maturation of the head of bacteriophage T4. I. DNA packaging events. J. Mol. Biol. 80:575-599. [DOI] [PubMed] [Google Scholar]
- 27.Laub, O., L. B. Rall, M. Truett, Y. Shaul, D. N. Standring, P. Valenzuela, and W. J. Rutter. 1983. Synthesis of hepatitis B surface antigen in mammalian cells: expression of the entire gene and the coding region. J. Virol. 48:271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae, p. 991-1041. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 29.Lokesh, G. L., T. D. S. Gowri, P. S. Satheshkumar, M. R. N. Murthy, and H. S. Savithri. 2002. A molecular switch in the capsid protein controls the particle polymorphism in an icosahedral virus. Virology 292:211-223. [DOI] [PubMed] [Google Scholar]
- 30.Lorenz, I. C., S. L. Allison, F. X. Heinz, and A. Helenius. 2002. Folding and dimerization of tick-borne encephalitis virus envelope proteins prM and E in the endoplasmic reticulum. J. Virol. 76:5480-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorenz, I. C., J. Kartenbeck, A. Mezzacasa, S. L. Allison, F. X. Heinz, and A. Helenius. 2003. Intracellular assembly and secretion of recombinant subviral particles from tick-borne encephalitis virus. J. Virol. 77:4370-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 75:10787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maley, F., R. B. Trimble, A. L. Tarentino, and T. H. Plummer, Jr. 1989. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180:195-204. [DOI] [PubMed] [Google Scholar]
- 34.Mandl, C. W., F. X. Heinz, and C. Kunz. 1988. Sequence of the structural proteins of tick-borne encephalitis virus (western subtype) and comparative analysis with other flaviviruses. Virology 166:197-205. [DOI] [PubMed] [Google Scholar]
- 35.Mason, P. W., S. Pincus, M. J. Fournier, T. L. Mason, R. E. Shope, and E. Paoletti. 1991. Japanese encephalitis virus-vaccinia recombinants produce particulate forms of the structural membrane proteins and induce high levels of protection against lethal JEV infection. Virology 180:294-305. [DOI] [PubMed] [Google Scholar]
- 36.Pincus, S., P. W. Mason, E. Konishi, B. A. Fonseca, R. E. Shope, C. M. Rice, and E. Paoletti. 1992. Recombinant vaccinia virus producing the prM and E proteins of yellow fever virus protects mice from lethal yellow fever encephalitis. Virology 187:290-297. [DOI] [PubMed] [Google Scholar]
- 37.Pugachev, K. V., P. W. Mason, and T. K. Frey. 1995. Sindbis vectors suppress secretion of subviral particles of Japanese encephalitis virus from mammalian cells infected with SIN-JEV recombinants. Virology 209:155-166. [DOI] [PubMed] [Google Scholar]
- 38.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 39.Russell, P. K., W. E. Brandt, and J. Dalrymple. 1980. Chemical and antigenic structure of flaviviruses, p. 503-529. In R. W. Schlesinger (ed.), The togaviruses. Academic Press, New York, N.Y.
- 40.Sato, T., C. Takamura, A. Yasuda, M. Miyamoto, K. Kamogawa, and K. Yasui. 1993. High-level expression of the Japanese encephalitis virus E protein by recombinant vaccinia virus and enhancement of its extracellular release by the NS3 gene product. Virology 192:483-490. [DOI] [PubMed] [Google Scholar]
- 41.Schalich, J., S. L. Allison, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1996. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J. Virol. 70:4549-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorger, P. K., P. G. Stockley, and S. C. Harrison. 1986. Structure and assembly of turnip crinkle virus. II. Mechanism of reassembly in vitro. J. Mol. Biol. 191:639-658. [DOI] [PubMed] [Google Scholar]
- 43.Stadler, K., S. L. Allison, J. Schalich, and F. X. Heinz. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71:8475-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stiasny, K., S. L. Allison, C. W. Mandl, and F. X. Heinz. 2001. Role of metastability and acidic pH in membrane fusion by tick-borne encephalitis virus. J. Virol. 75:7392-7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vennema, H., G. J. Godeke, J. W. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. Opstelten, and P. J. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, S., R. He, and R. Anderson. 1999. PrM- and cell-binding domains of the dengue virus E protein. J. Virol. 73:2547-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winkler, G., F. X. Heinz, and C. Kunz. 1987. Studies on the glycosylation of flavivirus E proteins and the role of carbohydrate in antigenic structure. Virology 159:237-243. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, Y., J. Corver, P. R. Chipman, W. Zhang, S. V. Pletnev, D. Sedlak, T. S. Baker, J. H. Strauss, R. J. Kuhn, and M. G. Rossmann. 2003. Structures of immature flavivirus particles. EMBO J. 22:2604-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zlotnick, A., N. Cheng, J. F. Conway, F. P. Booy, A. C. Steven, S. J. Stahl, and P. T. Wingfield. 1996. Dimorphism of hepatitis B virus capsids is strongly influenced by the C terminus of the capsid protein. Biochemistry 35:7412-7421. [DOI] [PubMed] [Google Scholar]