Abstract
Live attenuated RNA viruses make highly efficient vaccines. Among them, measles virus (MV) vaccine has been given to a very large number of children and has been shown to be highly efficacious and safe. Therefore, this vaccine might be a very promising vector to immunize children against both measles and other infectious agents, such as human immunodeficiency virus. A vector was previously derived from the Edmonston B strain of MV, a vaccine strain abandoned 25 years ago. Sequence analysis revealed that the genome of this vector diverges from Edmonston B by 10 amino acid substitutions not related to any Edmonston subgroup. Here we describe an infectious cDNA for the Schwarz/Moraten strain, a widely used MV vaccine. This cDNA was constructed from a batch of commercial vaccine. The extremities of the cDNA were engineered in order to maximize virus yield during rescue. A previously described helper cell-based rescue system was adapted by cocultivating transfected cells on primary chicken embryo fibroblasts, the cells used to produce the Schwarz/Moraten vaccine. After two passages the sequence of the rescued virus was identical to that of the cDNA and of the published Schwarz/Moraten sequence. Two additional transcription units were introduced in the cDNA for cloning foreign genetic material. The immunogenicity of rescued virus was studied in macaques and in mice transgenic for the CD46 MV receptor. Antibody titers and T-cell responses (ELISpot) in animals inoculated with low doses of rescued virus were identical to those obtained with commercial Schwarz MV vaccine. In contrast, the immunogenicity of the previously described Edmonston B strain-derived MV clone was much lower. This new molecular clone will allow for the production of MV vaccine without having to rely on seed stocks. The additional transcription units allow expressing heterologous antigens, thereby providing polyvalent vaccines based on an approved, safe, and efficient MV vaccine strain that is used worldwide.
Vaccines developed from live attenuated RNA viruses are very efficient. Mass vaccination with live attenuated polio, measles, or yellow fever vaccines has reduced the incidence of these infections and of the associated pathologies dramatically. Having been used on billions of people since the 1960s, these vaccines have a proven safety and efficacy record. They induce strong cellular and humoral immune responses and are particularly efficient at stimulating long-lasting memory B and T cells. Moreover, they are easy to produce and are cheap, and the means to deliver them worldwide already exist. All these characteristics make live attenuated RNA viruses excellent candidate vaccination vectors. As an example, poliovirus has been adapted to express foreign genes (1), and a simian immunodeficiency virus-recombinant Sabin poliovirus succeeded in protecting macaques from a virulent simian immunodeficiency virus challenge (6). However, the icosahedral symmetry of the viral capsid and its small volume impose stringent constraints on genome size. As a consequence, the poliovirus genome cannot be extended by more than 10% of its length to express foreign genes. Viruses with helical nucleocapsid and pleomorphic particles, such as paramyxoviruses, have fewer constraints. Their genomes can be adapted to accommodate long foreign genes.
Measles virus (MV) belongs to the genus Morbillivirus in the family Paramyxoviridae. The Edmonston strain of MV was isolated in 1954 (7), serially passaged on primary human kidney and amnion cells, and then adapted to chicken embryo fibroblasts (CEF) to produce Edmonston A and B seeds (see references 11 and 12 for reviews). Edmonston B was licensed in 1963 as the first MV vaccine. Further passages of Edmonston A and B on CEF produced the more attenuated Schwarz and Moraten viruses (27), whose sequences have recently been shown to be identical (23, 24). Being reactogenic, Edmonston B vaccine was abandoned in 1975 and was replaced by the Schwarz/Moraten vaccine. This is now the most commonly used measles vaccine (11, 12). By now, MV vaccine has been given to billions of people and is safe and efficacious. It induces a very efficient, life-long CD4, CD8, and humoral immunity after a single injection of 104 50% tissue culture infective doses (TCID50). Its safety is due to the fact that the genome is very stable, which explains that reversion to pathogenicity has never been observed, and that it cannot be integrated in host chromosomes, since viral replication is exclusively cytoplasmic. Thus, live attenuated MV could provide a safe and efficient pediatric vaccination vector.
In a noteworthy and pioneer work, members of the Billeter laboratory cloned an infectious cDNA corresponding to the antigenome of MV and established a reverse genetics procedure to rescue the corresponding virus (26). With this clone they developed a vector that can stably express as much as 5 kb of foreign genetic material (25, 28-30, 36). However, this vector was cloned from an Edmonston B strain of MV that had been propagated in HeLa cells (3). Its sequence diverges noticeably from that of Edmonston B and has 10 amino acid substitutions not related to any Edmonston subgroup. Moreover, despite the fact that this vector is immunogenic in mice expressing CD46 and lacking the interferon (IFN) type I receptor (29), we show in this article that it is not immunogenic in nonhuman primates when inoculated at the standard dose of 104 TCID50. Therefore, this vector does not appear to be suitable for human vaccination.
A recombinant MV vaccine might be an ideal vector to immunize children against measles and at the same time against another infectious disease, such as AIDS. However, such a vector will have to be derived from an approved and efficient strain of MV vaccine. Therefore, we cloned an infectious cDNA corresponding to the antigenome of the widely used Schwarz/Moraten vaccine strain. This cDNA allows the production of the Schwarz/Moraten vaccine without having to depend on the availability of seed stocks. Additional transcription units (ATU) were introduced in the viral genome to turn it into a vector expressing foreign proteins. This vector, which is very immunogenic in macaques, will allow constructing recombinant vaccines based on an approved, widely used, and efficient MV vaccine strain that can be grown on safe CEF.
MATERIALS AND METHODS
Cells.
Vero (African green monkey kidney) cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 5% fetal calf serum (FCS). Helper 293-3-46 cells used for viral rescue (26) (a kind gift from M. Billeter, Zurich University) were grown in DMEM supplemented with 10% FCS and 1.2 mg of G418 per ml. CEF were prepared as follows. Fertilized chicken eggs (Morizeau, Dangers, France) were incubated at 38°C for 9 days. Embryos were collected under sterile conditions. Head, limbs, and viscera were removed, and embryos were chopped and then trypsinized for 5 to 10 min at 37°C (Trypsine-EDTA, 2.5 g/liter). After filtration (70 μm) and several washes in DMEM-high glucose-10% FCS, cells were seeded (5 × 106 to 7 × 106 cells per petri dish) and were incubated overnight at 37°C before use for virus infection.
Plasmid constructions.
The Schwarz cDNA was cloned from viral particles purified from a batch of vaccine kindly provided by Aventis Pasteur (Marcy l'Etoile, France). This bulk vaccine preparation (50 ml, 3 × 104 TCID50/ml) was obtained by scraping infected CEF, freeze-thawing cells and medium, and filtering to remove cellular debris. Viral particles were concentrated by centrifugation through a 30% sucrose cushion. Viral RNA was extracted from lysed particles by using a silica gel-based membrane (QIAmp; Qiagen). The viral RNA was reverse-transcribed into cDNA by using a mixture of random hexamers as primers (pdN6; 1 μM) and a specific oligonucleotide complementary to the first 32 nucleotides of the MV genome (MVSchwRT1, 5′-ACCAAACAAAGTTGGGTAAGGATAGTTCAATC-3′; 10 μM). The SuperScript II DNA polymerase (GibcoBRL) was used to ensure accuracy and high yield.
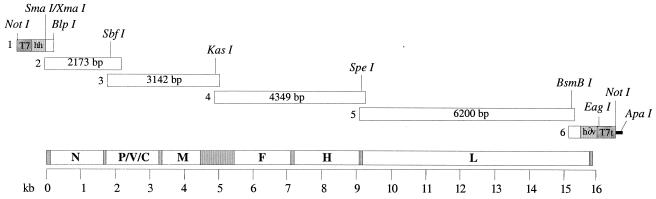
Six overlapping cDNA fragments covering the entire viral genome were generated by PCR using PfuTurbo DNA polymerase (Stratagene) and a set of specific primers (see Fig. 4). Fragment 1, at the 5′ end of the viral antigenome, was designed to fuse the first nucleotide of viral sequence with a hammerhead ribozyme sequence preceded by a T7 RNA polymerase promoter with the GGG motif necessary for full efficiency. To generate this fragment by PCR, the following two overlapping oligonucleotides were annealed: Leader 1 (5′-TATGCGGCCGCTAATACGACTCACTATAGGGCCAACTTTGTTTGGTCTGA-3′), which contains a NotI site, the T7 promoter (underlined), and the first 19 nucleotides of the hammerhead ribozyme sequence, and Leader 2 (5′-GGTGACCCGGGACTCCGGGTTTCGTCCTCACGGACTCATCAGACCAAACA-3′), which contains the hammerhead sequence with a SmaI/XmaI site (underlined). After PCR amplification, the resulting fragment was linked by PCR extension to a second fragment also generated by PCR from Schwarz cDNA using oligonucleotides MVSchw1 (5′-GAGTCCCGGGTCACCAAACAAAGTTGGGTAA G-3′), which overlaps the hammerhead sequence (underlined) and covers MV Schwarz genome 1 to 15, and MVSchw160 (5′-GGTTTGTCCTTGTTTCTTTT-3′, MV Schwarz genome 141 to 160). Fragments 2 to 5 were amplified by using specific oligonucleotides close to unique restriction sites (see Fig. 4). Fragment 6 at the 3′ end of the viral antigenome was designed to fuse the last nucleotide of the viral sequence with hepatitis delta virus (HDV) ribozyme sequence followed by the T7 terminator. Two overlapping fragments were generated by PCR and were annealed together. The first one was obtained by using the oligonucleotides MVSchw15155 (5′-GCAGCAGATAATTGAATCATCTGTGAGGACTTCAC-3′, MV Schwarz genome 15155 to 15190) and MVSchw15570 (5′-CCCGGAGTAAAGAAGAATGTGCCCCCAGAATTTGC-3′, MV Schwarz genome 15535 to 15570). The second one was obtained by PCR amplification of p(MV+) plasmid (26) (a kind gift from M. Billeter) by using oligonucleotides MVSchw15547 (5′-GGCACATTCTTCTTTACTCCGGGAACAAAAAGTTG-3′, MV Schwarz genome 15547 to 15581) and MVSchwEnd (5′-ATAGGGCCCGCGGCCGCATCCGGATATAGTTCCTCCTTTCA-3′, containing an ApaI restriction site [underlined]) linked to the last nucleotides of the T7 terminator. The six fragments thus generated were cloned in pCR2.1-TOPO vector (Invitrogen, Groningen, The Netherlands) and were sequenced.
FIG. 4.
Schematic map of the Schwarz MV cDNA. The six fragments generated to construct the pTM-MVSchw plasmid are shown in the upper part with the restriction sites used to assemble the complete cDNA. T7, T7 promoter; hh, hammerhead ribozyme; h∂v, hepatitis delta ribozyme; T7t, T7 RNA polymerase terminator (T7, hh, h∂v, and T7t are not represented at the same scale). A schematic map of MV genome is shown in the lower part (grayed portions represent the intergenic regions).
A modified pBlueScript KS(+) plasmid was constructed in order to assemble the full-length Schwarz MV cDNA. The pTM plasmid was previously derived from pBluescript KS(+) (Stratagene) by deletion of the T7 promoter (33). Two complementary oligonucleotides containing a NotI-KasI-NarI-SpeI-ApaI polylinker were annealed and inserted in the pTM plasmid after digestion with NotI/ApaI. The six MV Schwarz cDNA fragments were assembled together step by step using unique restriction sites (see Fig. 4). Fragments 1 and 2 were assembled together using the BlpI site in MV sequence and the BglII site in pCR2.1-TOPO vector backbone. The resulting plasmid was recombined with fragment 3 using the SbfI site in the MV sequence and the BglII site in the pCR2.1-TOPO vector backbone, giving plasmid pCR2.1-TOPO-MVSchw-1-2-3, which contained MV Schwarz fragments 1 to 3. After NotI/NarI digestion of this plasmid, the fragment containing the T7 promoter, hammerhead ribozyme, and the first 4,922 nucleotides of the MV Schwarz antigenome was inserted in NotI/NarI-digested pTM plasmid, yielding pTM-MVL. Fragments 5 and 6 were assembled together using the BsmBI site in MV sequence and the BssHII site in pCR2.1-TOPO vector backbone, giving plasmid pCR2.1-TOPO-MVSchw-5-6, which contained MV Schwarz fragments 5 and 6. After SpeI/ApaI digestion of this plasmid, the fragment containing the last 6,720 nucleotides of the MV Schwarz antigenome, the HDV ribozyme, and the T7 terminator sequence was inserted in SpeI/ApaI-digested pTM vector, giving pTM-MVT. The following four fragments were prepared and ligated together to generate the complete cDNA: (i) a SapI/SapI fragment of pTM-MVL (4,367 nucleotides long) containing a part of pTM backbone, the T7 promoter, the hammerhead ribozyme, and the first 1,813 nucleotides of the MV antigenome; (ii) a SapI/NarI fragment of pTM-MVL (3,110 nucleotides long) containing nucleotides 1813 to 4923 of the MV Schwarz antigenome; (iii) a NarI/SpeI fragment of pCR2.1-TOPO-MVSchw-3 (4,253 nucleotides long) containing nucleotides 4923 to 12157 of the MV Schwarz antigenome; and (iv) a SpeI/SapI fragment of pTM-MVT (7,235 nucleotides long) containing nucleotides 12157 to 15894 of the MV Schwarz antigenome, the HDV ribozyme, the T7 terminator, and a part of the pTM vector backbone. After ligation and cloning, several full-length constructs were obtained. The resulting plasmid was named pTM-MVSchw.
The ATU previously described for the EdB-tag vector (25) was amplified by PCR from p(+)MV2-GFP (a kind gift from M. Billeter, Zurich University). An 870-nucleotide-long fragment was generated with both ends overlapping with MV Schwarz sequence. This fragment consisted of a copy of the MV N-P intergenic sequence in which a multiple-site cassette containing the green fluorescent protein (GFP) sequence was introduced. This ATU was inserted in pTM-MVSchw plasmid by using site-directed mutagenesis in an SpeI site in position 3373 of the MW Schwarz genome. The new plasmid, named pTM-MVSchw-ATU2, contained the ATU between the P and M genes. The ATU was also inserted between the H and L genes by using another SpeI site in position 9175 of the MW Schwarz genome, giving rise to plasmid pTM-MVSchw-ATU3.
Rescue of Schwarz MV from the cloned cDNA.
Schwarz MV was rescued from the pTM-MVSchw cDNA with the helper-cell-based rescue system described by Radecke et al. (26) and modified by Parks et al. (22). Briefly, 293-3-46 cells (a kind gift of M. Billeter, Zurich University) were transfected by using the calcium phosphate procedure with pTM-MVSchw (5 μg) and a plasmid expressing the MV polymerase L gene (pEMC-La; 20 ng; a kind gift of M. A. Billeter). After overnight incubation at 37°C, the transfection medium was replaced by fresh medium and the cells were heat shocked at 43°C for 3 h and then returned to 37°C (22). After 2 days of incubation at 37°C, transfected cells were transferred onto a monolayer of CEF and incubated at 32°C or were transferred onto Vero cells and incubated at 37°C. Single syncytia were transferred to 35-mm wells of CEF or Vero cells and then were expanded to larger dishes. Virus was harvested from CEF after 5 to 7 days of infection and from Vero cells when syncytia involved 80 to 90% of the culture (usually after 2 days) by scraping infected cells, freeze-thawing cells and medium, and centrifuging them to remove cellular debris.
Growth curves and virus titers.
Monolayers of CEF or Vero cells in 6-well-plates were infected with viruses at different multiplicities of infection (MOI). At various times postinfection cells were scraped into culture medium. After freeze-thawing of cells and medium and clarification of cell debris, virus titers were determined on Vero cells. Vero cells were seeded into 96-well plates (7,500 cells/well) and were infected by serial 1:10 dilutions of virus sample in DMEM-5% FCS. After incubation at 37°C for 4 to 5 days (EdB-tag MV) or for 7 days (Schwarz MV), cells were stained with crystal violet and the TCID50 was calculated by the Kärber method (13).
Macaque immunization and characterization of humoral and cellular immune responses.
Colony-bred rhesus (Macaca mulatta) or cynomolgus (Macaca fascicularis) macaques that were seronegative for simian type D retrovirus, simian T-cell lymphotropic virus, simian immunodeficiency virus, and MV were housed in accordance with the American Association for Accreditation of Laboratory Animal Care. Monkeys were inoculated subcutaneously with different doses (103 to 105 TCID50) of EdB-tag or Schwarz MV diluted in OptiMEM (GibcoBRL) or with 104 TCID50 of the lyophilized Rouvax MV vaccine (Aventis Pasteur) diluted in the solution provided by the supplier. Blood samples were collected at different times after inoculation.
The presence of anti-MV antibodies in serum was looked for by enzyme-linked immunosorbent assay (ELISA) (Trinity Biotech) 1 month after vaccination. Each determination was done in triplicate on a 1/20 dilution of serum samples. A mixture of five samples from virus-negative monkeys was used as the negative control. To determine the immune status ratio (ISR) of each sample, the absorbance of the negative control was subtracted from the absorbance of the tested sample and the result was divided by the absorbance of a positive calibrator supplied in the ELISA kit, as recommended by the supplier. Only ISR values higher than 0.9 were considered positive in this test.
Cellular immune responses were determined by IFN-γ ELISpot assays. Frozen peripheral blood mononuclear cells (PBMC) were thawed and incubated overnight in RPMI-10% FCS and 4 U of recombinant human interleukin-2 (Boehringer Mannheim)/ml. Multiscreen hemagglutinin 96-well plates were coated overnight at 4°C with 4 μg of capture anti-IFN-γ (GZ-4; MAbTech)/ml in phosphate-buffered saline (PBS), washed, and then incubated with 100 μl of RPMI-10% FCS for 1 h at 37°C. The medium was replaced by 5 × 105 PBMC in suspension in 100 μl of RPMI-10% FCS and 100 μl of stimulating agent. The stimulating agent consisted of 107 PFU of recombinant modified vaccinia virus Ankara (MVA; a kind gift of Bernard Moss, National Institute of Allergy and Infectious Diseases); MVA-HMV or MVA-wt as a control. Cells were stimulated for 24 h at 37°C. Phytohemagglutinin A (2.5 μg/ml; Sigma) was used as positive control, and RPMI was used as negative control. The plates were washed twice with PBS, four times with PBS-0.05% Tween 20 (Sigma), and twice again with PBS. A biotinylated anti-IFN-γ antibody (7-B6-1; 100 μl; 1 μg/ml in PBS; MabTech) was added, and the plates were incubated for 2 to 4 h at 37°C. Streptavidin-alkaline phosphatase conjugate (100 μl; 1:2,000 dilution in PBS; Roche) was added, and spots were developed with 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium (Promega) in 1 M Tris (pH 9.5), 1.5 M NaCl, 0.05 M MgCl2. After drying overnight at room temperature, spots were counted by using an automated image analysis system (ELISpot Reader; Bio-Sys). The low background obtained after MVA-wt stimulation was subtracted, and the results were expressed as MVA-HMV-specific IFN-γ-producing cells per million PBMC.
Mice immunization and characterization of humoral immune responses.
FVB mice heterozygous for the CD46 transgene (37) (a kind gift from F. Grosveld, Erasmus University, Rotterdam, The Netherlands) were crossed with 129sv IFN-α/βR−/− mice which lack the type I IFN receptor (18) (a kind gift from M. Aguet, Swiss Institute for Experimental Cancer Research, Epalinges, Switzerland). The F1 progeny was screened by PCR, and the CD46+/− animals were crossed again with 129sv IFN-α/βR−/− mice. IFN-α/βR−/− CD46+/− animals were selected and used for immunization experiments. These mice are susceptible to MV infection (16, 17). Six-week-old female CD46+/− or CD46+/− IFN-α/βR−/− (CD46/IFNAR) mice were inoculated intraperitoneally with 104 TCID50 of the different vaccine preparations (four mice per group). The presence of anti-MV antibodies was looked for by ELISA (Trinity Biotech) in sera collected 1 month after vaccination. In this case, an anti-mouse immunoglobulin G monoclonal antibody (Amersham) was used as secondary antibody. Each determination was done in triplicate. The absorbance determined with a mixture of negative mice sera was subtracted from the absorbance measured in positive mice. Because it was not possible in this case to use the ISR to compare samples, serial dilutions of mice sera were tested to determine the end point limit positive dilution.
RESULTS
Comparison of humoral immune responses after vaccination of macaques and mice with EdB-tag and Schwarz MV vaccines.
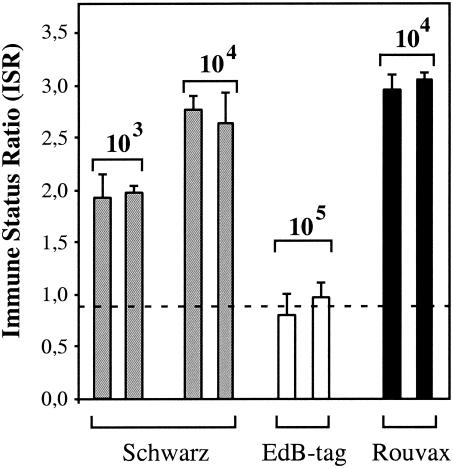
EdB-tag MV is a molecularly cloned MV derived from the Edmonston B strain (26). We compared its immunogenicity in macaques with that of the Schwarz commercial MV vaccine. The EdB-tag virus was prepared in Vero cells infected at an MOI of 0.05. When syncytia occupied 80 to 90% of the culture, the cells were scraped, cells and medium were freeze/thawed, and cell debris were eliminated by low-speed centrifugation. The Schwarz MV, obtained from Aventis Pasteur, was prepared in the same way from infected CEF grown at 32°C, the temperature at which this strain has been adapted to CEF. The titers of both vaccine preparations were determined by end point dilution assays in Vero cells and are expressed as TCID50. Different doses (103 to 105 TCID50) of EdB-tag and Schwarz MV were injected subcutaneously into rhesus macaques (two monkeys per dose). As a control, animals were also injected with 104 TCID50 of the lyophilized commercial Schwarz vaccine (Rouvax; Aventis Pasteur). Anti-MV antibody levels were determined by ELISA in macaque sera collected 1 month after vaccination (Fig. 1). Macaques inoculated with 103 and 104 TCID50 of the Schwarz MV had antibody levels similar to those induced by a standard dose of Rouvax vaccine. Macaques inoculated with 104 TCID50 of EdB-tag virus remained negative (data not shown). The injection of a 10-fold higher dose (105 TCID50) induced only a weak response that was lower than that observed with 103 TCID50 of Schwarz MV (Fig. 1). Vaccination with the commercial vaccine induced the best response, probably due to the adjuvant effect of lyophilization.
FIG. 1.
Detection of anti-MV antibodies in macaques immunized with different MV vaccine strains. Anti-MV antibodies were detected by ELISA 1 month after immunization of rhesus macaques (two monkeys per group) with Schwarz virus (gray bars), EdB-tag virus (white bars), and Rouvax vaccine (black bars) at the doses indicated. ISR were calculated as described in Materials and Methods. Only ISR values higher than 0.9 were considered positive (determinations were done in triplicate on 1/20 dilution of serum samples, and results are expressed as the mean values ± standard deviations).
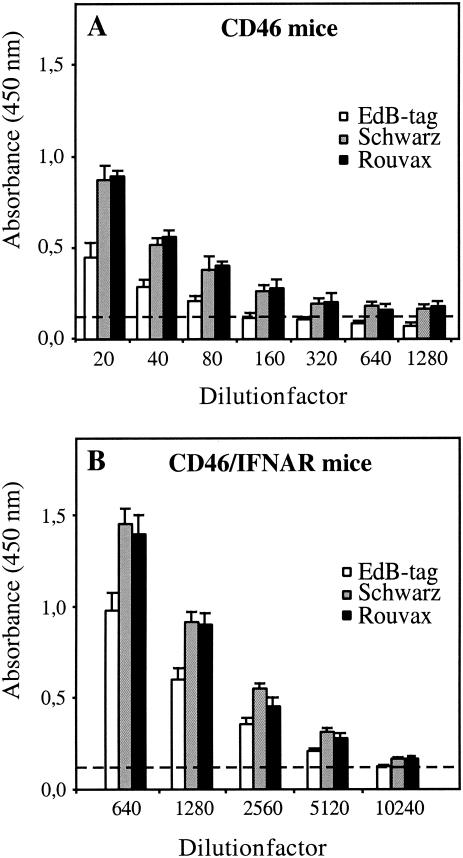
The different vaccine preparations were also tested in genetically modified mice obtained as described in Materials and Methods. Two types of mice were used: mice expressing CD46 (37), the human receptor for MV vaccine strains (19), and mice expressing CD46 and lacking the IFN type I receptor (CD46/IFNAR (17). Six-week-old mice were inoculated intraperitoneally with 104 TCID50 of the different vaccine preparations (four mice per group). Figure 2 shows the detection of anti-MV antibodies in sera of both types of mice collected 1 month after vaccination. In CD46 mice the EdB-tag virus was less immunogenic than the Schwarz vaccine. The average titer obtained with the former was 1/80, whereas it was 1/1,280 with the latter. The EdB-tag virus was also less immunogenic in CD46 mice lacking the IFN type I receptor, but the difference was less pronounced than in CD46 immunocompetent mice, possibly indicating a difference in sensitivity to IFN-α/β between the two viral strains.
FIG. 2.
Antibody titers to MV in mice immunized with different MV vaccine strains. Anti-MV antibodies were detected by ELISA 1 month after immunization of CD46 (A) and CD46/IFNAR (B) mice with 104 TCID50 of EdB-tag virus (white bars), Schwarz virus (gray bars), and Rouvax vaccine (black bars). Results are expressed as mean optical density values ± standard deviations (four mice per group) determined in serial dilutions of sera.
Comparison of the nucleotide sequence of various MV genomes.
The sequences of the genomes of the EdB-tag and Schwarz viruses were compared in an attempt to understand their differences in immunogenicity at the genetic level. The coding and noncoding sequences of Edmonston-derived vaccine strains have been previously compared to that of a low-passage isolate of the Edmonston wild-type MV (23, 24). The authors identified 10 amino acid substitutions shared by almost all the MV vaccine strains. We compared the genomic sequences of these Edmonston-derived vaccine strains and of two primary MV isolates (31, 32) with that of EdB-tag cDNA (26) (Fig. 3). The nucleotide sequence of EdB-tag (15,894 bp) differs from that of Rubeovax (Edmonston B vaccine) by 38 mutations (0.24%) and from that of the Schwarz/Moraten strain (Edmonston A vaccine) by 44 mutations (0.27%). The Schwarz/Moraten and Rubeovax sequences differ by only 16 mutations (0.1%). Among the 38 differences between EdB-tag and Rubeovax, 17 are amino acid substitutions in coding regions and 7 are located in noncoding regions. Among the 44 differences between EdB-tag and Schwarz/Moraten, 22 are amino acid substitutions and 9 are in noncoding regions. The 10 amino acids substitutions shared by almost all the Edmonston vaccine strains (23, 24) are conserved in EdB-tag cDNA. However, five of these substitutions are specific of the AIK-C and Zabreg subgroup, indicating that the virus from which EdB-tag was cloned diverged and did not correspond to any approved vaccine strain.
FIG. 3.
Sequence comparison of MV genomes. (A) Nucleotide changes for each coding region (capital letters in boxes) and in noncoding regions (lowercase letters) are shown in the lower part. Amino acid changes are shown in the upper part (one-letter amino acid symbol). The nucleotide (nt) and amino acid (aa) changes that are present only in the EdB-tag sequence are highlighted in gray. Nucleotide changes in positions 1805 and 1806 of EdB-tag correspond to the tag introduced. (B) Phylogenetic tree showing the EdB-tag among the Edmonston group (24) and two wild-type isolates (31, 32). The sequences were aligned using Clustal W (34). Nucleotide sequence distances were determined with Dnadist of the Phylip package, version 3.5 (9). The tree was derived by neighbor-joining analysis applied to pairwise sequence distances calculated by using a variety of methods, including the Kimura two-parameter method to generate unrooted trees. The final output was generated with Treeview (21).
Moreover, 10 amino acid substitutions in EdB-tag are not related to any Edmonston subgroup (highlighted in gray in Fig. 3), probably reflecting the adaptation to growth on HeLa and Vero cells and/or errors introduced during the cloning procedure. Among these specific changes, five are located in the P/V/C coding sequences and three are in the L polymerase gene, thus possibly affecting the replication capacity of the virus in vivo. These changes and others in cis-acting sequences may influence the immunogenicity or pathogenicity of the virus recovered from the EdB-tag cDNA. It has been shown that adapting MV to Vero cells causes a few amino acid changes in the polymerase (L) and accessory (P/V/C) proteins and that these changes are associated with loss of pathogenicity (14) most likely due to reduced transcription in lymphoid cells (31, 32). Because of these sequence differences and of the poor immunogenicity of the EdB-tag vector in immunocompetent animals, we decided to clone a cDNA from the Schwarz strain of MV.
Construction of a cDNA corresponding to the antigenome of the Schwarz vaccine strain of MV.
In order to clone a sequence corresponding precisely to that of the Schwarz vaccine, RNA was purified from viral particles present in a vaccine preparation kindly provided to us by Aventis Pasteur and which was not passaged in the laboratory. All the cloning methods used were optimized to ensure cloning fidelity (see Materials and Methods). A set of six overlapping cDNA fragments covering the entire viral genome (numbered 1 to 6 in Fig. 4) were generated. The 5′ end of the viral antigenome (fragment 1) was engineered by PCR with specific primers in order to contain a T7 RNA polymerase promoter with the GGG motif necessary for full efficiency, and a hammerhead ribozyme sequence was inserted between the T7 promoter and the first viral nucleotide. The HDV ribozyme followed by the T7 terminator were added to the 3′ end of the viral genome (fragment 6). The six fragments generated were sequenced and assembled step by step using unique restriction sites in a modified BlueScript plasmid (33) in which the T7 promoter has been deleted. The resulting plasmid, named pTM-MVSchw, was entirely sequenced. No mutation was found between this cDNA and the previously reported sequence of the Schwarz MV genome (23, 24).
Recovery of the Schwarz virus from pTM-MVSchw.
The Schwarz MV was rescued from the pTM-MVSchw plasmid with the helper-cell-based rescue system described by Radecke et al. (26) and modified by Parks et al. (22). In order to avoid adaptation to Vero cells, transfected 293-3-46 cells were cocultivated with CEF at 32°C, the temperature at which the Schwarz strain was adapted to chicken cells. Infectious virus was recovered between 3 and 7 days following cocultivation. The rescued Schwarz virus was passaged two times on CEF to prepare the virus used in all subsequent experiments. The cytopathic effect observed was identical to that of the parental Schwarz virus: only occasional syncytia developed in the CEF monolayer, and after 4 to 6 days of infection cells became refringent and began to detach from the plastic. To control for the genetic integrity of the rescued virus, viral particles were purified and viral RNA was reverse transcribed as described above with the primers used for cloning the cDNA. The genome of the rescued virus was entirely sequenced. The sequence was identical to that of the original Schwarz strain and of pTM-MVSchw.
The Schwarz virus was also rescued after cocultivation of transfected 293-3-46 helper cells at 37°C with primate Vero cells instead of CEF. In this case, syncytia were abundant after 2 days of coculture. Schwarz virus rescued on Vero cells was passaged two times on Vero cells. Viral particles were purified from the second passage, and viral RNA was reverse transcribed with the primers used for cloning. The viral genome was entirely sequenced. Two mutations out of 15,894 nucleotides were found when comparing the sequence with that of the cDNA used for transfection. Both mutations resulted in amino acid changes in the fusion protein (F): G→R in position 266 and Y→S in position 365. For each mutation, 10 randomly selected cDNA clones were sequenced. The mutations were found in 7 and 8 of the 10 clones, respectively, indicating a high percentage of mutation in the viral population. Therefore, changing the host cell of Schwarz virus leads to a rapid adaptation that might affect the properties of the vaccine.
Growth of the rescued virus on Vero cells and CEF.
The growth of the Schwarz virus rescued from the pTM-MVSchw cDNA was analyzed in Vero cells and in CEF and was compared to that of the industrial Schwarz vaccine from which it was derived and of the EdB-tag virus. Cells were infected using different MOI and were collected at different time points. Titers of cell-associated viruses were determined by end point dilution on Vero cells. Figure 5A shows that the growth kinetics of the pTM-MVSchw and EdB-tag viruses rescued from their respective cDNA were similar on Vero cells and were comparable to that of the commercial Schwarz virus. The yield of Schwarz virus on Vero cells was high (107 TCID50/ml). On CEF, the rescued Schwarz virus grew as well as the parental Schwarz virus at 37 or 32°C. However, growth was slower than that on Vero cells (Fig. 5B and C). On the other hand, the EdB-tag virus did not grow on CEF, confirming that the virus from which it was cloned diverged from any Edmonston A or B vaccine that have all been adapted to grow on CEF. The yield of Schwarz virus was lower on CEF than on Vero cells (106 TCID50/ml).
FIG. 5.
Growth kinetics of rescued Schwarz and EdB-tag viruses on Vero and CEF cells. Cells on 35-mm-diameter dishes were infected with Schwarz MV rescued from pTM-MVSchw plasmid (▪), EdB-tag MV (○), and industrial Schwarz virus (▵) at different MOI (as indicated). At each time point cells were collected, and cell-associated virus titers were determined by using the TCID50 method on Vero cells. (A) Vero cells incubated at 37°C; (B) CEF incubated at 37°C; (C) CEF incubated at 32°C.
Immunogenicity of Schwarz MV recovered from cDNA.
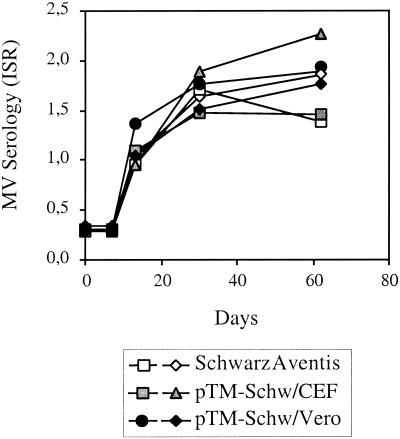
The immunogenicity for cynomolgus macaques of the virus rescued from pTM-MVSchw plasmid and passaged two times on CEF or Vero cells was compared to that of the industrial Schwarz vaccine. Cynomolgus macaques were used in this experiment because of the difficulty of obtaining rhesus macaques from China that were MV negative. These macaques are as sensitive to MV as rhesus macaques, as shown by several studies (15, 35). Monkeys (two animals per preparation) were injected subcutaneously with 104 TCID50 of Schwarz MV vaccine from Aventis or Schwarz MV rescued from pTM-MVSchw plasmid and grown either on CEF or Vero cells. The presence of anti-MV antibodies was determined in sera collected at different time points (Fig. 6). All the vaccinated macaques became positive. No statistically significant difference was observed, 1 or 2 months after immunization, between the different vaccine preparations tested. This result demonstrates that the virus rescued from the pTM-MVSchw plasmid has the same immunogenicity in nonhuman primates as the parental Schwarz vaccine. No difference was detected between the rescued viruses grown on CEF or Vero cells, indicating that the two mutations generated in the F protein by the passages on Vero cells did not affect the immunogenicity of the virus.
FIG. 6.
Detection of anti-MV antibodies in macaques immunized with different Schwarz MV preparations. Anti-MV antibodies were detected by ELISA at different time points after immunization of cynomolgus macaques (two monkeys per group) with 104 TCID50 of bulk industrial Schwarz virus (white marks) and Schwarz virus rescued from pTM-MVSchw plasmid and grown on CEF (gray bars) or Vero cells (black bars). ISR were calculated as described in Materials and Methods.
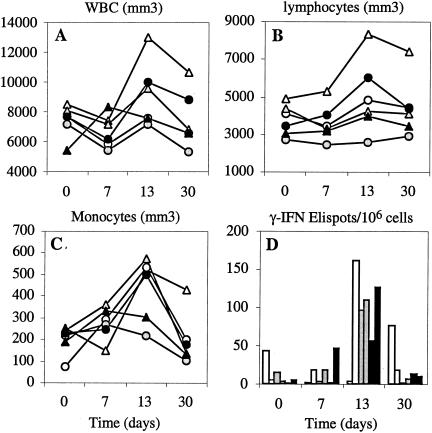
Changes in the number of total white blood cells (WBC), lymphocytes, and monocytes were observed during the first month following inoculation (Fig. 7A to C). There was a mild leukopenia during the first week, as previously observed after MV vaccination (2). During the second week a clear increase in the number of circulating lymphocytes and monocytes was observed. It coincided with a peak of the number of MV-specific T lymphocytes as detected by an IFN-γ ELISpot assay (Fig. 7D). No statistically significant difference was detected between the specific cellular immune responses induced by the Schwarz MV rescued from plasmid and the Schwarz vaccine prepared by Aventis.
FIG. 7.
Changes in the number of circulating leukocytes and MV-specific T-cell response in macaques immunized with different Schwarz MV preparations. Enumeration of white blood cells (A), lymphocytes (B), monocytes (C), and MV hemagglutinin-specific IFN-γ-ELISpots (D) in PBMC of cynomolgus macaques collected at different time points after immunization with 104 TCID50 of bulk industrial Schwarz virus (white marks) or Schwarz virus rescued from pTM-MVSchw plasmid and grown on CEF (gray bars) or Vero cells (black bars). IFN-γ-ELISpots were detected after stimulation of PBMC for 24 h with a recombinant MVA expressing the MV hemagglutinin. The background obtained with MVA-wt stimulation was subtracted, and the results are expressed as MVA-HMV-specific IFN-γ-producing cells per million PBMC. WBC, white blood cells.
Introduction of ATU in the Schwarz cDNA.
An ATU was introduced in the pTM-MVSchw cDNA in order to use the rescued Schwarz virus as a vector for foreign genes. A large number of transgenes have already been inserted in the EdB-tag vector in our and other laboratories. In order to facilitate the transfer of these transgenes into the Schwarz vector, the ATU was identical to that of the EdB-tag vector (25). This ATU is a multiple-cloning-site cassette inserted into a copy of the N-P intergenic region of the MV genome. This region contains the cis-acting sequences necessary for the transcription of the MV P gene. The GFP sequence was inserted in the cassette. The ATU was introduced into pTM-MVSchw in two different positions (between the P and M genes and between the H and L genes) (Fig. 8). The total number of antigenomic nucleotides was kept as a multiple of six (5). The resulting plasmids were named pTM-MVSchw-ATU2 and pTM-MVSchw-ATU3. Recombinant viruses were rescued from these plasmids and were used to infect Vero cells. As shown in Fig. 8, the GFP transgene was expressed in the syncytia induced in Vero cells.
FIG. 8.
Schematic representation of the pTM-MVSchw-ATU plasmids (A) and GFP expression in Vero cells infected by rescued recombinant viruses (B). Vero cells were infected with recombinant Schwarz MV-GFP either in position ATU2 (left side) or position ATU3 (right side), and the GFP fluorescence was observed in syncytia.
DISCUSSION
In the present work we describe cloning and rescuing the Schwarz/Moraten attenuated strain of MV, the constituent of two widely used measles vaccines, Attenuavax (Merck and Co. Inc., West Point, N.Y.) and Rouvax (Aventis Pasteur), and of the combined measles, mumps, and rubella vaccine (4). This cDNA was constructed with the aim of developing polyvalent MV vaccines expressing heterologous antigens. To be used in a pediatric clinical trial, recombinant live attenuated MV produced from a cDNA must be as safe and efficient as the parental vaccine. Assuming that safety and efficiency depend ultimately on the genomic sequence of the attenuated strain, we cloned the MV Schwarz cDNA from viral particles prepared from an industrial batch of vaccine using procedures optimized for fidelity of cloning. As a result, the sequence of the clone that we obtained was identical to that of the parental Schwarz MV genome. To maximize yield during rescue, the viral antigenomic cDNA was placed under the control of a T7 RNA polymerase promoter with the GGG motif necessary for full efficiency. A hammerhead ribozyme was inserted between this GGG motif and the first viral nucleotide to allow the exact cleavage of the viral RNA. In order to avoid adapting the Schwarz vaccine to noncertified cells during rescue, helper cells transfected with the engineered cDNA were cocultivated with CEF, the cells on which this vaccine was selected originally and is presently prepared. The rescued virus was passaged two times on CEF, and its genome was entirely sequenced. No mutation was found when the sequence was compared to that of the original virus. Moreover, the growth kinetics and the yield of the rescued virus and the original Schwarz virus on CEF were identical.
The Schwarz virus was also rescued after cocultivation of transfected helper cells with Vero cells, which are very permissive to MV. In this case, however, two mutations appeared in the viral fusion protein (F) after two passages on Vero cells. This rapid adaptation correlated with a much more fusogenic phenotype on Vero cells. In contrast, the rescued Schwarz MV was not fusogenic on CEF (only rare syncytia could be observed in infected CEF). The two mutations occurred in the F protein (G→R in position 266 and Y→S in position 365). These mutations are present in the EdB-tag virus (see Fig. 3) which is grown on Vero cells. They are also present in the Hallé strain, which is highly related to Edmonston strain and does not infect CEF (8). These two mutations thus appear to correlate with enhanced fusion in Vero cells. The rapid adaptation of the F protein after only two passages of the Schwarz virus on Vero cells shows that in order to keep its genetic integrity the vaccine must be grown on CEF.
The virus rescued from the pTM-MVSchw plasmid had the same immunogenicity in macaques as the parental Schwarz vaccine. It is important to emphasize that in these experiments macaques were inoculated with the low dose of virus used for human immunization. Therefore, it will be possible to conduct human clinical trials with this virus using standard vaccine doses (104 TCID50). In contrast, the previously cloned EdB-tag MV was not immunogenic in macaques and was poorly immunogenic in mice transgenic for CD46 when used at the same dose as that of the cloned Schwarz MV.
What could be the reason for the higher immunogenicity of the Schwarz MV strain? Inducing good immunogenicity with a live attenuated viral vaccine requires replication in tissues at a level high enough to prime the immune system adequately. Several of the mutations between the Schwarz and the EdB-tag MV genomes are located in the P/V/C and L genes, suggesting a difference in replication efficiency. It is possible that the Schwarz MV replicates in lymphoid cells in vivo more efficiently than the EdB-tag MV, even though they replicated at the same rate in Vero cells. Efficient replication in vivo requires some evasion mechanism from the IFN-α/β response. Vero cells, on which the EdB-tag virus was adapted, do not respond to IFN-α/β stimulation. Therefore, the EdB-tag MV was selected in the absence of an IFN-α/β response and might be particularly sensitive to this host defense mechanism. Indeed, it has been shown that passaging wild-type MV on Vero cells changes the phenotype of the virus from non-IFN inducer to IFN inducer (20). Also, the fact that the Ed-tag MV was immunogenic in mice transgenic for the CD46 receptor providing they were also knocked out for the IFN-α/β receptor suggests that this virus is particularly IFN sensitive. Interestingly, the IFN-α/β response helps priming the specific immune response against the vaccine. Therefore, a good live vaccine must at the same time induce an IFN-α/β response and evade it to some extent. For this reason selecting attenuated viral vaccines on primary cells with a strong IFN-α/β response, such as CEF, might be a good strategy.
The MV products which contribute to IFN resistance have not been identified. However, the nonstructural C protein of the closely related Sendai virus has been shown to counteract the IFN-induced antiviral state (10). The five mutations not related to any Edmonston subgroup that we found in the EdB-tag P/V/C gene might be responsible for its low immunogenicity in macaques. On the other hand, the two mutations generated in the F protein by passaging the Schwarz virus on Vero cells did not affect its immune potential, indicating that the fusogenic property of the viral envelope proteins may not play a significant role in immunogenicity.
The pTM-MVSchw plasmid was engineered for the expression of foreign genes by the introduction of two ATU at different positions of the genome. Rescued Schwarz recombinant MV expressed the GFP, thus showing that this new measles vaccine functions as a vector. In conclusion, this molecular clone will allow producing MV vaccine without having to rely on seed stocks. With its ATUs, it will be possible to use it as a vector to produce recombinant vaccines based on a vaccine strain that is approved, efficient, and used worldwide.
Acknowledgments
Chantal Combredet and Valérie Labrousse contributed equally to this work.
We are very grateful to Martin Billeter for his support during this work and for kindly providing plasmids and rescue helper cells. We thank Gudrun Christiansen for teaching us the rescue technique. We also thank Hussein Naim for helpful suggestions and positive collegiality.
C.L. was the recipient of a fellowship from the Agence Nationale pour la Recherche contre le SIDA (ANRS), L.M. was supported by a National Institutes of Health grant (NIH AI46007), and M.B.F. was supported by an Elizabeth Glaser Scientist Award from the Pediatric AIDS Foundation. This work was supported by institutional grants from the Pasteur Institute (PTR24) and CNRS and by NIH grant no. AI46007.
REFERENCES
- 1.Andino, R., D. Silvera, S. D. Suggett, P. L. Achacoso, C. J. Miller, D. Baltimore, and M. B. Feinberg. 1994. Engineering poliovirus as a vaccine vector for the expression of diverse antigens. Science 265:1448-1451. [DOI] [PubMed] [Google Scholar]
- 2.Auvaerter, P. G., P. A. Rota, W. R. Elkins, R. J. Adams, T. DeLozier, Y. Shi, W. J. Bellini, B. R. Murphy, and D. E. Griffin. 1999. Measles virus infection in rhesus macaques: altered immune responses and comparison of the virulence of six different virus strains. J. Infect. Dis. 180:950-958. [DOI] [PubMed] [Google Scholar]
- 3.Ballart, I., D. Eschle, R. Cattaneo, A. Schmid, M. Metzler, J. Chan, S. Pifko-Hirst, S. A. Udem, and M. A. Billeter. 1990. Infectious measles virus from cloned cDNA. EMBO J. 9:379-384. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Buynak, E., R. Weibel, W. J. Je, J. Stokes, Jr., and M. Hilleman. 1969. Combined live measles, mumps, and rubella virus vaccines. JAMA 207:2259-2262. [PubMed] [Google Scholar]
- 5.Calain, P., and L. Roux. 1993. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J. Virol. 67:4822-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crotty, S., C. J. Miller, B. L. Lohman, M. R. Neagu, L. Compton, D. Lu, F. X. Lu, L. Fritts, J. D. Lifson, and R. Andino. 2001. Protection against simian immunodeficiency virus vaginal challenge by using Sabin poliovirus vectors. J. Virol. 75:7435-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enders, J. F., and T. C. Peebles. 1954. Propagation in tissue cultures of cytopathogenic agents from patients with measles. Proc. Soc. Exp. Biol. Med. 86:277-286. [DOI] [PubMed] [Google Scholar]
- 8.Escoffier, C., and D. Gerlier. 1999. Infection of chicken embryonic fibroblasts by measles virus: adaptation at the virus entry level. J. Virol. 73:5220-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1989. PHYLIP: phylogeny interference package, ver. 3.2. Cladistics. 5:164-166.
- 10.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin, D. 2001. Measles virus, p. 1401-1441. In D. Knipe and P. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 12.Hilleman, M. 2002. Current overview of the pathogenesis and prophylaxis of measles with focus on practical implications. Vaccine 20:651-665. [DOI] [PubMed] [Google Scholar]
- 13.Karber, G. 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Arch. Exp. Pathol. Pharmak. 162:480-483. [Google Scholar]
- 14.Kobune, F., H. Sakata, and A. Sugiura. 1990. Marmoset lymphoblastoid cells as a sensitive host for isolation of measles virus. J. Virol. 64:700-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobune, F., H. Takahashi, K. Terao, T. Ohkawa, Y. Ami, Y. Suzaki, N. Nagata, H. Sakata, K. Yamanouchi, and C. Kai. 1996. Nonhuman primate models of measles. Lab. Anim. Sci. 46:315-320. [PubMed] [Google Scholar]
- 16.Mrkic, B., B. Odermatt, M. Klein, M. Billeter, J. Pavlovic, and R. Cattaneo. 1999. Lymphatic dissemination and comparative pathology of recombinant measles viruses in genetically modified mice. J. Virol. 74:1364-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mrkic, B., J. Pavlovic, T. Rulicke, P. Volpe, C. J. Buchholz, D. Hourcade, J. P. Atkinson, A. Aguzzi, and R. Cattaneo. 1998. Measles virus spread and pathogenesis in genetically modified mice. J. Virol. 72:7420-7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Müller, U., U. Steinhoff, L. F. L. Reis, S. Hemmi, J. Pavlovic, R. M. Zinkernagel, and M. Aguet. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918-1921. [DOI] [PubMed] [Google Scholar]
- 19.Naniche, D., G. Varior-Krishnan, F. Cervoni, T. F. Wild, B. Rossi, C. Rabourdin-Combe, and D. Gerlier. 1993. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 67:6025-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naniche, D., A. Yeh, D. Eto, M. Manchester, R. M. Friedman, and M. B. A. Oldstone. 2000. Evasion of host defenses by measles virus: wild-type measles virus infection interferes with induction of alpha/beta interferon production. J. Virol. 74:7478-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 22.Parks, C. L., R. A. Lerch, P. Walpita, M. S. Sidhu, and S. A. Udem. 1999. Enhanced measles virus cDNA rescue and gene expression after heat shock. J. Virol. 73:3560-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parks, C. L., R. A. Lerch, P. Walpita, H. P. Wang, M. S. Sidhu, and S. A. Udem. 2001. Analysis of the noncoding regions of measles virus strains in the Edmonston vaccine lineage. J. Virol. 75:921-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parks, C. L., R. A. Lerch, P. Walpita, H. P. Wang, M. S. Sidhu, and S. A. Udem. 2001. Comparison of predicted amino acid sequences of measles virus strains in the Edmonston vaccine lineage. J. Virol. 75:910-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radecke, F., and M. Billeter. 1997. Reverse genetics meets the nonsegmented negative-strand RNA viruses. Rev. Med. Virol. 7:49-63. [DOI] [PubMed] [Google Scholar]
- 26.Radecke, F., P. Spielhofer, H. Schneider, K. Kaelin, M. Huber, C. Dotsch, G. Christiansen, and M. A. Billeter. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwarz, A. 1962. Preliminary tests of a highly attenuated measles vaccine. Am. J. Dis. Child. 103:216-219. [DOI] [PubMed] [Google Scholar]
- 28.Singh, M., and M. Billeter. 1999. A recombinant measles virus expressing biologically active human interleukin-12. J. Gen. Virol. 80:101-106. [DOI] [PubMed] [Google Scholar]
- 29.Singh, M., R. Cattaneo, and M. A. Billeter. 1999. A recombinant measles virus expressing hepatitis B virus surface antigen induces humoral immune responses in genetically modified mice. J. Virol. 73:4823-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spielhofer, P., T. Bachi, T. Fehr, G. Christiansen, R. Cattaneo, K. Kaelin, M. Billeter, and H. Naim. 1998. Chimeric measles viruses with a foreign envelope. J. Virol. 72:2150-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeda, M., A. Kato, F. Kobune, H. Sakata, Y. Li, T. Shioda, Y. Sakai, M. Asakawa, and Y. Nagai. 1998. Measles virus attenuation associated with transcriptional impediment and a few amino acid changes in the polymerase and accessory proteins. J. Virol. 72:8690-8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeuchi, K., N. Miyajima, F. Kobune, and M. Tashiro. 2000. Comparative nucleotide sequence analyses of the entire genomes of B95a cell-isolated and vero cell-isolated measles viruses from the same patient. Virus Genes 20:253-257. [DOI] [PubMed] [Google Scholar]
- 33.Tangy, F., A. McAllister, and M. Brahic. 1989. Molecular cloning of the complete genome of Theiler's virus, strain GDVII, and production of infectious transcripts. J. Virol. 63:1101-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Binnendijk, R. S., R. W. J. van der Heijden, G. van Amerongen, F. UytdeHaag, and A. D. M. E. Osterhaus. 1994. Viral replication and development of specific immunity in macaques after infection with different measles virus strains. J. Infect. Dis. 170:443-448. [DOI] [PubMed] [Google Scholar]
- 36.Wang, Z., T. Hangartner, L. Cornu, A. Martin, M. Zuniga, M. Billeter, and H. Naim. 2001. Recombinant measles viruses expressing heterologous antigens of mumps and simian immunodeficiency viruses. Vaccine 19:2329-2336. [DOI] [PubMed] [Google Scholar]
- 37.Yannoutsos, N., J. N. Ijzermans, C. Harkes, F. Bonthuis, C. Y. Zhou, D. White, R. L. Marquet, and F. Grosveld. 1996. A membrane cofactor protein transgenic mouse model for the study of discordant xenograft rejection Genes Cells 1:409-419. [Erratum, 1: 785.] [DOI] [PubMed] [Google Scholar]