Abstract
Epstein-Barr Virus (EBV) infects resting B cells, within which it establishes latency as a stable, circular episome with only two EBV components, the cis element oriP and the latently expressed protein EBNA1. It is believed that EBNA1's ability to tether oriP episomes to metaphase chromosomes is required for its stable replication. We created fusions between the DNA-binding domain (DBD) of EBNA1 and the cellular chromatin-binding proteins HMGA1a and HMG1 to determine the minimal requirements for stable maintenance of an oriP-based episome. These two proteins differ in that HMGA1a can associate with metaphase chromosomes but HMG1 cannot. Interestingly, coinciding with metaphase chromosome association, HMGA1a-DBD but not HMG1-DBD supported both the transient replication and stable maintenance of oriP plasmids, with efficiencies quantitatively similar to that of EBNA1. However, HMGA1a-DBD activated transcription from EBNA1-dependent episomal reporter to only 20% of the level of EBNA1. Furthermore, EBNA1 but not HMGA1a-DBD activated transcription from a chromosomally integrated EBNA1-dependent transcription reporter. This indicates that EBNA1 possesses functional domains that support transcription activation independent of its ability to tether episomal oriP plasmids to cellular chromosomes. We provide evidence that metaphase chromosome tethering is a fundamental requirement for maintenance of an oriP plasmid but is insufficient for EBNA1 to activate transcription.
Epstein-Barr virus (EBV) is a gammaherpesvirus that infects resting B cells and also epithelial cells. Upon infection, the double-stranded, linear EBV genome circularizes and persists as a stable episome, infrequently integrating into the human genome. In rapidly dividing cells, stable maintenance of the EBV episome at copy numbers between 1 and 100 (48) requires the replication and partitioning of its genome through ensuing cell cycles. During latency, EBV replicates only once per cell cycle semiconservatively (1, 47), utilizing the cellular replication machinery for its own DNA synthesis (2, 9, 41).
Stable maintenance of an EBV-based episomal plasmid requires only the origin of latent replication (in cis) oriP (46), and the latently expressed viral protein (in trans) EBNA1 (48). EBNA1 homotypically interacts with two clusters of imperfect, repetitive binding sites, the family of repeats (FR) and the dyad symmetry element (DS), found within the cis element oriP. Four binding sites within the DS, a formal origin of bidirectional replication, contribute to the initiation of DNA synthesis; the 20 binding sites comprising the FR support stable plasmid maintenance (2, 29) and transactivation by EBNA1 (39).
The amino terminus of EBNA1 has two positively charged regions, which we term A (amino acids 33 to 89) and B (amino acids 328 to 378), that are required for EBNA1 to attach to cellular chromosomes, activate transcription, support the stable replication of oriP plasmids, and link DNAs. EBNA1 participates in all these processes without possessing any known enzymatic activity (15, 34).
Within the carboxy terminus (DNA-binding domain [DBD]) of EBNA1 (amino acids 451 to 641) lies the region required for oriP binding and dimerization (3, 6, 8). Expression of the DBD alone causes the rapid loss of oriP plasmids from proliferating cells (27). This is because oriP fails to be replicated or to be segregated due to a loss of association with cellular chromosomes (25). In fact, the DBD serves as a potent dominant-negative inhibitor of oriP replication and maintenance when coexpressed with EBNA1 (27).
Episomal maintenance is thought to occur via tethering of oriP-containing episomes by EBNA1 to host cell chromosomes. Through this association, newly replicated episomes are segregated to both daughter cells upon each cell division. EBNA1 is associated with cellular chromosomes throughout the phases of the cell cycle, including mitosis (23, 24). Three regions that overlap the A and B domains of EBNA1 contribute to metaphase chromosomal localization: CBS-1 (amino acids 68 to 90), CBS-2 (amino acids 328 to 365), and CBS-3 (amino acids 8 to 67) (32). Extending these findings, recent studies have demonstrated that expression of an EBNA1 mutant lacking all three CBS domains failed to localize to metaphase chromosomes and was impaired in its ability to mediate transient replication of an oriP-based plasmid (25).
Functional mapping studies indicate much overlap in the regions that are required for functions mediated by EBNA1 (8, 27, 30, 45). We therefore tested fusions of EBNA1 that replace its functional amino terminus with cellular proteins possessing defined biochemical properties. By quantifying the biological function of a chimeric version of EBNA1, the role of the amino terminus of EBNA1 can be clarified; those fusions behaving biologically and quantitatively similarly to EBNA1 provide insight into the molecular mechanisms required for the replication and maintenance of an oriP replicon.
In this study we replaced EBNA1's amino terminus (amino acids 1 to 450) with either the HMGA1a or HMG1 cellular protein. The HMG family was attractive to use in a chimeric fusion analysis with EBNA1 for several reasons. First, previous findings have shown that a fusion between HMGA1a and the DBD of EBNA1 support maintenance of oriP plasmids in BJAB cells (20). Second, the HMG proteins serve as architectural transcription factors that remodel DNA through induction of specific bends within it (7, 37, 38). This, like the function of EBNA1, occurs without enzymatic activity from the HMG proteins (7, 37, 38). Third, and most importantly, HMGA1a and HMG1 possess quite distinct binding properties for different forms of cellular chromatin. Whereas HMGA1a associates with metaphase chromosomes (13, 33), HMG1 can be found associated with interphase chromatin but not with metaphase chromosomes (14, 22). Taken together, these properties of the HMG proteins provided the ability to directly test whether metaphase chromosome association is necessary for stable replication of oriP plasmids.
Here we present the first biochemical evidence that the stable replication of an oriP episome requires only its association with metaphase chromosomes. By utilizing EBNA1-DBD fusions with either HMGA1a or HMG1, we determined that quantitatively similar maintenance of an oriP-based episome could be recapitulated solely through its ability to be tethered to metaphase chromosomes. This is indicated by the observation that HMGA1a-DBD but not HMG1-DBD was sufficient to support transient replication of oriP plasmids and stably maintain oriP plasmids in long-term studies. Our data indicate that metaphase chromosome tethering is required not only for oriP plasmids to be maintained and partitioned but also for them to undergo DNA synthesis in the ensuing S phase.
As EBNA1 bound to oriP activates transcription from EBV promoters (16, 36), we tested whether metaphase chromosome tethering was sufficient for EBNA1 to activate transcription from episomal oriP-containing transcription reporters. We found that both HMGA1a-DBD and HMG1-DBD were impaired in their ability to activate transcription from an episomal reporter, suggesting that EBNA1 activates transcription independently of its ability to maintain oriP plasmids. Supporting this conclusion, EBNA1 but not HMGA1a-DBD activated transcription from a chromosomally integrated EBNA1-dependent transcription reporter.
MATERIALS AND METHODS
Bacterial strains and plasmid purification.
All plasmids were propagated in Escherichia coli strains DH5α, MC1061/P3, and STBL2 (Invitrogen, Carlsbad, Calif.). Plasmids used for transfection were purified on isopycnic CsCl gradients (31).
Oligonucleotides and plasmids.
Plasmid 1160 (27), which is an expression plasmid for the DNA-binding domain (DBD) (amino acids 451 to 641) of EBNA1, was used as the backbone hmg/EBNA1-DBD plasmid to create both fusion proteins. Oligonucleotides AGO107 and AGO108 were used for PCR amplification of hmg1. These oligonucleotides contain SbfI and HindIII recognition sites, respectively (italic): AGO107, 5′-GAATTCCCTGCAGGGGCCAGTCAGGCCCAACCATGGGCAAAGGAGATCCTAAGAAGC; and AGO108, 5′-GAATTCCTCGAGTCAAGCTTGTTCATCATCATCATCTTCTTCTTCATC. This PCR product was introduced into plasmid 1160 digested with SbfI and HindIII to create a fusion product between hmg1 and the DBD of EBNA1 to create plasmid AGP102.
Oligonucleotides AGO109 and AGO110 were used for PCR amplification of hmgA1a. These oligonucleotides contain SbfI and HindIII recognition sites, respectively (italic): AGO109, 5′-GAATTCCCTGCAGGGGCCAGTCAGGCCCAACCATGAGTGAGTCGAGCTCGAA GTCCA; and AGO110, 3′-GAATTCCTCGAGTCAAGCTTGCTGCTCCTCCTCCGAGGACTCCTG. This PCR product was introduced into plasmid 1160 digested with SbfI and HindIII to create a fusion between hmgA1a and the DBD of EBNA1 in plasmid AGP103.
To create derivatives of the retroviral vector pLXSN (AGP164) that express HMG1 or HMGA1a, the open reading frames comprising hmg1/EBNA1-DBD and hmgA1a/EBNA1-DBD were moved into AGP164 to create AGP176 and AGP177, respectively. oriP replication reporter plasmids AGP74 and AGP100 are derivatives of pPUR that were described previously (18). AGP74 contains a synthetic FR with 20 EBNA1 binding sites and is referred to as 20-FR in this study. AGP100 contains a synthetic FR with seven EBNA1 binding sites and is referred to as 7-FR in this study. AGP66 is a derivative of AGP74 that lacks DS and is referred to as DS− in this study. Plasmid 2380 is also a pPUR derivative that contains wild-type oriP. Plasmid 1033 is a transcription reporter containing oriP and expressing luciferase under the control of the BamHI-C promoter (27). It is referred to as oriP-BamHI-C-luciferase in this study. AGP51 is a transcription reporter plasmid that contains an FR with seven EBNA1 binding sites and the luciferase gene under the control of the herpes simplex virus type 1 thymidine kinase promoter (18). It is referred to as 7FR-HSV-1-TK-luciferase in this study. Construct 2145 is a cytomegalovirus-enhanced green fluorescent protein (EGFP) expression plasmid that was used to measure transcription efficiency and normalize for plasmid uptake (18). Sequences for all the plasmids used in this study can be found at http://ebv.mimnet.northwestern.edu/plasmids.
Cell culture, transfection, and generation of stable 293 cell lines.
The human EBNA1-expressing cell line 293/EBNA1 is a derivative of 293 cells that stably express wild-type EBNA1. 293/HMG1-DBD cells are a derivative of 293 cells which stably express, through retroviral integration, human wild-type HMG1 fused to the DNA-binding domain of EBNA1. 293/HMGA1a-DBD cells are a derivative of 293 cells which stably express, through retroviral integration, human wild-type HMGA1a fused to the DNA-binding domain of EBNA1. All cells were propagated in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics. G418 was added for 293/EBNA1 cells at a final concentration of 200 μg/ml and at 300 μg/ml for 293/HMG1-DBD and 293/HMGA1a-DBD cells. BJAB cells containing an integrated FR-HSV-1-TK-luciferase reporter were a gift from Bill Sugden and grown in RPMI 1640 supplemented with 10% fetal bovine serum and puromycin at 1 mg/liter. All cells were grown in 37°C in a humidified 5% CO2 atmosphere.
A derivative of 293 cells (17) stably expressing the retroviral Gag and Pol proteins (gp293) was used to package and propagate retroviral vectors. They were grown in the conditions described above to 50% cell density in a 10-cm dish and cotransfected by the calcium phosphate method with 10 μg of AGP176 or AGP177, 10 μg of vesicular stomatitis virus G expression plasmid (AGP154), and 100 ng of a simian virus 40 large-T-antigen expression plasmid (AGP5). After 16 h, the medium was replaced. After 72 h, the virus-containing medium was collected and centrifuged to pellet suspended cells, and 5 ml of virus-containing medium was used to infect 293 cells grown to 50% cell density in a 10-cm dish. To increase the efficiency of retroviral infection, Polybrene was added during the infection at a final concentration of 7.5 μg/ml. After 48 h, infected cells were split 1:10 and then placed under selection with 300 μg of G418 per ml. Protein expression was confirmed by immunoblotting and immunofluorescence.
Cells were transfected by the calcium phosphate method as described previously (31) unless otherwise noted. Transfections were normalized by the inclusion of 1 μg of a cytomegalovirus-EGFP expression plasmid, 2145, in each transfection. Upon harvest, a fraction of the cells were profiled with a Becton-Dickinson FACScan or FACSCalibur. Transfection efficiency was measured as the fraction of GFP-expressing, live cells quantified with CellQuest software from Becton-Dickinson (Franklin Lakes, N.J.).
Immunoblotting.
Immunoblotting was done by standard protocols (31). Briefly, extracts from 1.5 × 106 cells were separated on a sodium dodecyl sulfate-10% polyacrylamide gel and transferred to a polyvinylidene difluoride membrane. After blocking for 1 h with 1% fat-free milk and 0.05% Tween 20, the membrane was treated with a rabbit polyclonal anti-EBNA1 serum (30) diluted 1:1,000 in blocking solution for 1 h at 37°C. The secondary antibody was a horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin antibody diluted 1:10,000 in blocking solution for 1 h at 37°C. Membranes were briefly washed once with 0.5% Tween 20 in PBS and once briefly with deionized water. Detection was performed by conventional chemiluminescence methods.
Indirect immunofluorescence.
293, 293/EBNA1, 293/HMG1-DBD, and 293/HMGA1a-DBD log-phase cells were grown on cell culture-treated Thermanox plastic coverslips (Fisher, Hanover Park, Ill.) to 40 to 60% confluence. Cells were washed two times with phosphate-buffered saline (PBS), fixed in fresh 3.5% formaldehyde for 20 min at room temperature, and then blocked and permeabilized in PBS containing 3% bovine serum albumin and 0.5% Triton X-100 for an additional 20 min at room temperature. Alternatively, similar results were achieved by fixing and permeabilizing cells in −20°C methanol for 2 min. The permeabilized cells were then incubated with a rabbit polyclonal antibody against the EBNA1 DBD, K67.3 (1:1,000 dilution in PBS), for 30 min in a humidified chamber at. This was followed by incubation with a fluorescein isothiocyanate (FITC)-conjugated anti-rabbit immunoglobulin antibody (Amersham, Buckinghamshire, United Kingdom) (1:500 dilution in PBS) for 30 min in a humidified chamber at 37°C. The cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) for 1 min at room temperature in PBS and then mounted onto glass slides with 10 μl of antifade solution (Vectashield; Vector Laboratories, Burlingame, Calif.).
For cell imaging, an Olympus IX inverted fluorescence microscope and a Photometrix cooled charge-coupled device camera (CH350/LCCD) utilizing DeltaVision software from Applied Precision Inc. (Seattle, Wash.) was used for data collection. Twenty 200-nm optical sections were taken through the depth of the cell, and DeltaVision software (softWoRx) was used to deconvolve these images to individual layers. DeltaVision softWoRx uses a constrained iterative deconvolution algorithm to remove out-of-focus blur in fluorescence optical sections and was set for a minimum of 15 iterative cycles. Fluorescence emission was linear over the 100-fold range of emission signals, where FITC and DAPI signals were detected well within these ranges. Images were acquired with 100 NA 1.35 objectives with or without 1.5× enhancement.
Colony formation assays to assess plasmid maintenance and partitioning.
We cotransfected 10 μg of AGP74 or an equivalent number of moles of plasmid AGP100 or 2380 with 1 μg of 2145 into 107 293, 293/EBNA1, 293/HMG1-DBD, or 293/HMGA1a-DBD cells on a 10-cm dish. Medium was replaced 16 h posttransfection. After 48 h, cells were harvested, profiled by fluorescence-activated cell sorting (FACS) to measure GFP expression and viability, and replated in duplicate at 2 × 105 and 2 × 104 GFP-positive, live cells per 10-cm dish. Cells were placed under selection with 0.5 μg of puromycin per ml 4 days posttransfection. After 2 weeks of selection, the resulting puromycin-resistant colonies were fixed with formamide and subsequently stained with methylene blue. Colonies were counted with NIH Image. Briefly, scanned images of plates were cropped to the rectangular coordinate dimensions 1.370 by 0.915 arbitrary units with Adobe Photoshop. Colonies of the cropped image were counted with a macro within NIH Image. Because colony counts were calculated on a cropped image that comprised 60% of the total area of the scanned culture dish, total cell counts were normalized to represent colony counts on a full 10-cm culture dish (cell counts/0.6).
Southern hybridization analysis to assess plasmid replication.
We cotransfected 10 μg of AGP74 or an equivalent number of moles of plasmid AGP100 or 2380 with 1 μg of 2145 into 107 293, 293/EBNA1, 293/HMG1-DBD, or 293/HMGA1a-DBD cells on a 10-cm dish. Medium was replaced 16 h posttransfection. After 96 h, DNA was harvested from 2 × 107 cells, while 106 GFP-positive, live cells, as measured by FACS profiling, were plated per 10-cm dish for puromycin selection as described above. After 3 weeks of selection, DNA was extracted from 2 × 106 to 108 puromycin-resistant cells as described previously (18). Extracted DNAs were digested with 200 units of DpnI and/or an enzyme that linearizes each plasmid (as stated in the figure legends) in a final volume of 20 μl overnight at 37°C.
Restriction endonucleases were purchased from New England Biolabs (Beverly, Mass.) and used as per the manufacturer's instructions. Digestions were electrophoresed on a 0.8% agarose gel. DNAs were transferred from the gel to a Hybond membrane (Amersham, Buckinghamshire, United Kingdom) with an Appligene vacuum transfer apparatus (Boekel Scientific, Feasterville, Pa.). Radioactive probes were prepared by the incorporation of [α-32P]dCTP (6,000 Ci/mmol) (Amersham) during Klenow synthesis with random primers and XbaI-digested AGP83 (pPUR-DS) (or the indicated plasmid) as a template. Probe specific activities ranged from 109 cpm/μg to 3 × 109 cpm/μg. Southern hybridization was performed as described by Hubert and Laimins (19). Southern blots were visualized and quantified by phosphorimage analysis with a Molecular Dynamics Storm phosphorimager (Molecular Dynamics, Sunnyvale, Calif.).
Transcription reporter assays.
We cotransfected 1 μg of 1033 or an equivalent number of moles of AGP51 with 1 μg of 2145 into 293, 293/EBNA1, 293/HMG1-DBD, and 293/HMGA1a-DBD cells. The medium was replaced 16 h posttransfection. At 48 h, cells were harvested, counted twice with a Coulter counter, pelleted, and lysed in reporter lysis buffer (provided along with a luciferase assay kit from Promega, Madison, Wis.) at a concentration of 105 cells/μl. Lysates were spun for 5 min at 1,000 × g to remove nuclei and then frozen at −80°C until assay. For BJAB cells, 2.5 × 107 cells were resuspended in 4.5 ml of RPMI complete medium to a final concentration of 5.5 × 106 cells/ml. Cells were then electroporated (210 V, 25 Ω resistance, 950 μF capacitance) with 10 μg of expression plasmids pcDNA3.1, 1160, 1553, AGP102, and AGP103 along with 1 μg of 2145. Luminescence assays were performed as per the manufacturer's instructions with a Zylux FB 15 luminometer.
Salt extraction of chromatin-associated proteins.
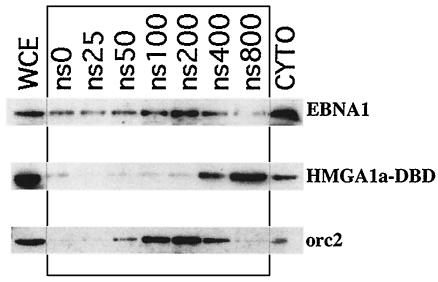
Cells were washed twice with PBS lacking Mg2+ and Ca2+ and resuspended in buffer A (10 mM HEPES [pH 7.6], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, and 10% glycerol). Triton X-100 was added to 0.1%, and the cells were incubated for 5 min at 4°C. Nuclei were pelleted at 4,000 rpm for 5 min. The supernatant was recovered and denoted the cytoplasm. The nuclei were washed once in buffer A before being divided between fresh aliquots of buffer A containing 0, 25, 50, 100, 200, 400, and 800 mM NaCl. Nuclei were extracted for 30 min at 4°C, followed by precipitation. The supernatants containing proteins released from the nuclei were recovered and are denoted NS 0 to 800. Immunoblots were performed as described with K67.3, a rabbit polyclonal antibody directed against amino acids 459 to 641 of EBNA1. The human orc2 protein was detected with a monoclonal antibody (Becton Dickinson, Franklin Lakes, N.J.).
RESULTS
Construction, expression, and localization of HMG-DBD fusions.
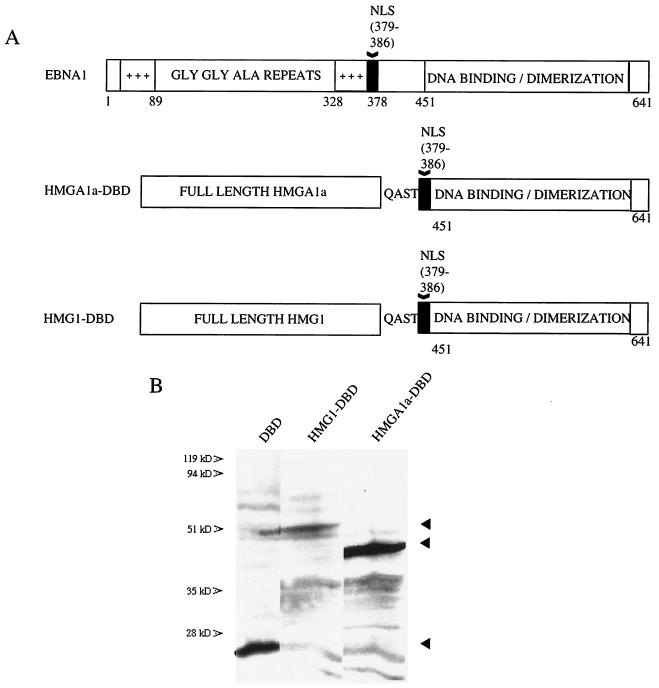
EBNA1 is detected in interphase nuclei and binds mitotic chromatin. To directly address whether these properties of EBNA1 were necessary and sufficient for stable maintenance of oriP plasmids, we created chimeras between the DBD of EBNA1 and the high-mobility group family members HMGA1a (formerly called HMGI) and HMG1. Full-length HMGs were fused by a short peptide linker of sequence QAST to the DBD of EBNA1 (amino acids 451 to 641) directly preceded by EBNA1's nuclear localization sequence (NLS, amino acids 379 to 386) (3) and are designated HMGA1a-DBD and HMG1-DBD throughout this report. Schematic representations of these fusions, compared to wild-type EBNA1 (EBNA1), are diagrammed in Fig. 1A.
FIG. 1.
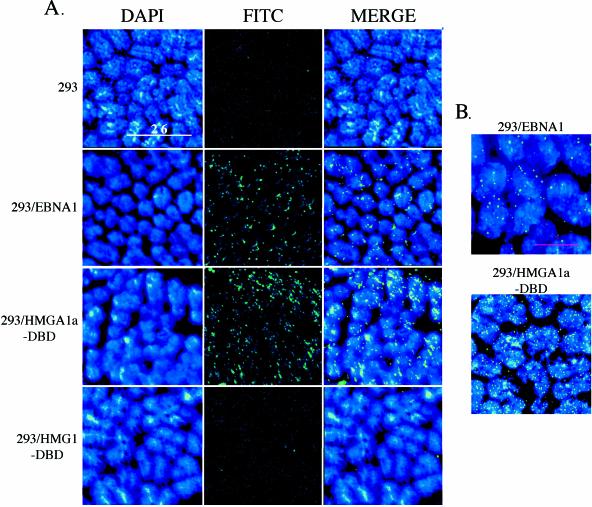
Schematic representation of EBNA1 and fusion proteins, their expression, and intracellular localization. (A) Wild-type EBNA1 from the prototype B95-8 strain of EBV is 641 amino acids (aa) long. The DNA binding and dimerization domain (DBD) lies between amino acids 451 and 641. Two positively charged regions implicated in oriP maintenance and transcription activation lie between amino acids 33 to 89 and 328 to 378 and are indicated. A nuclear localization sequence (NLS) has been identified between amino acids 379 and 386 and is also indicated. The HMGA1a-DBD fusion protein contains full-length human HMGA1a (GenBank accession number NM_145899) fused with a short peptide linker (QAST) to the NLS and DBD of EBNA1. The HMG1-DBD fusion protein contains full-length human HMG1 (GenBank accession number BC030981) fused to the NLS and DBD of EBNA1. (B) Immunoblots of extracts from 1.5 × 106 293 cells that stably express either the HMGA1a-DBD fusion or the HMG1-DBD fusion. The migration positions of the DBD of EBNA1 and fusion proteins are indicated by arrowheads. Immunoblots were probed with rabbit polyclonal antiserum raised against the DBD of EBNA1. (C) EBNA1, HMGA1a-DBD, and HMG1-DBD are detected in the nuclei of interphase, cycling cells stably expressing each of these proteins by indirect immunofluorescence. 293 derivative cell lines were grown on coverslips, fixed, and stained as described in Materials and Methods. EBNA1 was visualized with FITC-conjugated goat anti-rabbit IgG. DAPI-stained nuclei, FITC-stained cells, and merged images are indicated.
Immunoblots of extracts from 293 cells stably expressing the fusion proteins probed with anti-EBNA1 antiserum are shown in Fig. 1B. The electrophoretic mobilities of the HMG fusions were consistent with their predicted molecular weights.
The localization of EBNA1, HMGA1a-DBD, and HMG1-DBD in asynchronously growing cells was examined and is shown in Fig. 1C. Cells were fixed and probed with a rabbit polyclonal antibody against the EBNA1 DBD. Proteins were visualized with a FITC-conjugated goat anti-rabbit immunoglobulin secondary antibody, and the nucleus was counterstained with DAPI. As shown in the figure, all three proteins were predominantly nuclear in cycling cells.
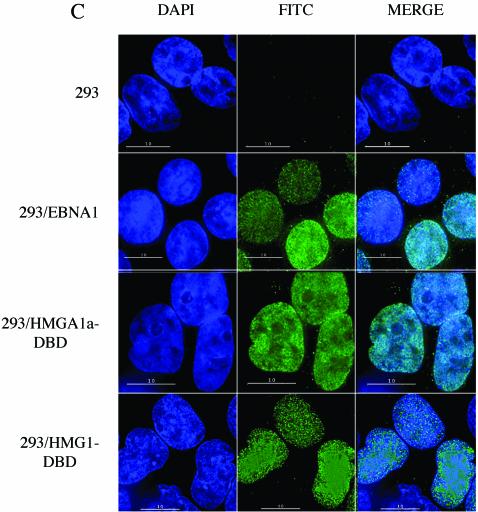
HMGA1a-DBD and EBNA1 but not HMG1-DBD are localized to mitotic chromosomes.
We next sought to determine the localization of the HMGA1a-DBD and HMG1-DBD fusions on cellular chromosomes during metaphase. Both endogenous HMGA1a and HMG1 have been shown previously to localize to interphase chromatin, as did our fusion proteins (Fig. 1C). However, HMGA1a, unlike HMG1, also associates with metaphase chromosomes (14, 22, 40). We wished to confirm that fusing the DBD of EBNA1 to either HMG protein would not disrupt its chromatin binding properties. To do this, indirect immunofluorescence with antiserum that recognizes the DBD of EBNA1 was performed on colcemid-blocked stable derivatives of 293 cells expressing the HMG fusions or EBNA1. Individual deconvolved layers of images were then analyzed for colocalization with metaphase chromosomes counter-stained with DAPI.
As shown in Fig. 2, 293/EBNA1 cells showed robust, punctate staining of EBNA1 on metaphase chromosomes, as expected from previous studies. A similar punctate staining of HMGA1a-DBD on metaphase chromosomes was observed. For EBNA1 and HMGA1a-DBD, nearly all the staining was metaphase chromosome associated, and little staining was observed in regions of the field where metaphase chromosomes were absent. In fact, the outlines of metaphase chromosomes can be detected simply by observing the fluorescence associated with EBNA1 or HMGA1a-DBD. In contrast, no specific metaphase chromosome-associated signal was detected with HMG1-DBD. When the FITC signal was set to be saturating, HMG1-DBD was observed to stain in a dispersed and diffuse manner. Little specific association with metaphase chromosomes was observed, and the staining was distributed throughout the field of view, mostly in areas that lacked metaphase chromosomes, indicating that this protein is not specifically associated with metaphase chromosomes. Furthermore, individual layers of deconvolved images indicated that the HMG1-DBD signal did not coincide with the DAPI signal for metaphase chromosomes. This confirmed that fusing the DBD of EBNA1 to either HMG protein had no effect on its localization in either mitotic or interphase cells (Fig. 1C and 2). In Fig. 2B, upon higher magnification, we noted that the specific staining pattern of HMGA1a-DBD on mitotic chromatin was quite similar to that of EBNA1 with respect to being both punctate and randomly distributed on metaphase chromosomes. We also observed more staining of metaphase chromosomes by HMGA1a-DBD, possibly due to higher expression levels.
FIG. 2.
Localization of EBNA1 and fusion proteins to metaphase chromosomes by indirect immunofluorescence. (A) 293, 293/EBNA1, 293/HMGA1a-DBD, and 293/HMG1-DBD cells were stalled in early mitosis with colcemid, swollen by hypotonic shock, and dropped on coverslips to release condensed metaphase chromosomes. Chromosomes were fixed and visualized by DAPI staining (deep blue) or probed with rabbit polyclonal antiserum against the EBNA1 DBD. EBNA1 and the fusion proteins were visualized with FITC-conjugated goat anti-rabbit IgG (bright green). The cell type used for the experiment is indicated to the left of each row of images, and the visualization used is indicated above each column of images. Size bars are indicated within some image panels. Images were taken at 60× magnification. The images indicate that EBNA1 and HMGA1a-DBD are localized to metaphase chromosomes. HMG1-DBD is not associated specifically with metaphase chromosomes but is dispersed throughout the field of view. (B) Merged images of metaphase chromosomes from 293/EBNA1 and 293/HMGA1a-DBD cells taken at 100× magnification with 1.5× enhancement.
Analysis of HMG fusion maintenance of oriP plasmids by colony formation.
Stable maintenance of oriP plasmids by EBNA1 requires its ability to replicate and faithfully partition an oriP-containing replicon through multiple, successive cell divisions. Drug-resistant colony formation has been used previously as a standard assay to evaluate the total maintenance function of EBNA1 (18). If a version of EBNA1 is capable of efficiently mediating both the replication and partitioning of an oriP plasmid containing a puromycin resistance gene, drug-resistant cells will form colonies under selection.
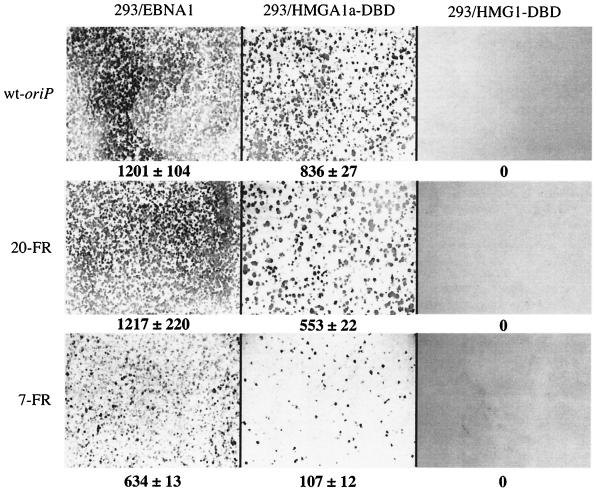
To determine the frequency at which HMGA1a-DBD and HMG1-DBD could establish colonies of oriP-containing cells, the 293 derivatives were transfected with a plasmid containing wild-type oriP and a puromycin resistance gene (2380). Briefly, 48 h posttransfection, equal numbers of live, transfected cells were plated and subsequently subjected to puromycin selection as described in the Materials and Methods section. Puromycin-resistant colonies were then stained, counted, and compared to colonies formed by 293/EBNA1 cells under the same conditions. As shown in Fig. 3A, cells expressing HMGA1a-DBD yielded colony counts (836 ± 27) close to those arising from cells expressing EBNA1 (1,201 ± 104). Strikingly we found that cells expressing HMG1-DBD, like 293 cells expressing no version of EBNA1, were completely impaired in their ability to maintain oriP plasmids, yielding no puromycin-resistant colonies.
FIG. 3.
oriP-puro replication reporter plasmids form colonies in 293/HMGA1a-DBD cells but not in 293/HMG1-DBD cells. 293/EBNA, 293/HMGA1a-DBD, and 293/HMG1-DBD cells were transfected with pPUR derivatives containing DS and either wild-type FR (wild-type oriP), 20 synthetic EBNA1 binding sites (20-FR), or seven synthetic EBNA1 binding sites (7-FR). Transfected cells were subjected to puromycin selection for 2 weeks as described in Materials and Methods, following which they were fixed and stained with methylene blue. Colonies were counted as described in Materials and Methods. The cell line used for the colony formation assay is indicated above each column of images, and the plasmid used in the assay is indicated to the left of each row of images. The numbers below each image indicate the number of colonies along with the standard deviation. The results indicate that 293/EBNA1 and 293/HMGA1a-DBD cells replicated stably and maintained all three oriP reporter plasmids. There was no stable replication and maintenance of oriP plasmids in 293 or 293/HMG1-DBD cells, indicated by a total absence of colonies.
We have observed previously that the maintenance function of EBNA1 is dependent on the number of EBNA1 binding sites within FR (18). To determine if a version of EBNA1 with its amino terminus replaced by HMGA1a was also sensitive to decreasing numbers of EBNA1 binding sites within FR, we transfected the 293 derivative cell lines with a puromycin-resistant replication reporter plasmid containing a synthetic FR with either seven (7-FR) or 20 (20-FR) EBNA1 binding sites. 20-FR gave results similar to those observed with wild-type oriP in all three 293 derivative cell lines. That is, 293/HMGA1a-DBD cells gave rise to colonies (553 ± 22), albeit to a lesser extent than 293/EBNA1 cells (1,217 ± 220), whereas 293/HMG1-DBD cells again gave rise to no puromycin-resistant colonies. Finally, we found that both 293/EBNA1 cells and 293/HMGA1a-DBD cells formed fewer colonies (634 ± 13 and 107 ± 12, respectively) when the replication reporter plasmid contained only seven EBNA1 binding sites in FR. From the colony formation assays, we conclude that cells expressing HMGA1a-DBD can maintain oriP plasmids at levels comparable to EBNA1, 293/HMGA1a-DBD cells, like 293/EBNA1 cells, show decreased efficiency in maintaining oriP plasmids with limiting numbers of EBNA1 binding sites, and HMG1-DBD fails to maintain oriP plasmids.
Under selection, HMGA1a-DBD maintains oriP plasmids at copy numbers similar to EBNA1.
To quantify the average copy number of an oriP plasmid maintained by HMGA1a-DBD relative to EBNA1, we performed Southern analysis on cells that stably maintained either the wild-type oriP or 7-FR replication reporter plasmid under selection. 293/EBNA1 and 293/HMGA1a-DBD cells were transfected with the wild-type oriP or 7-FR replication reporter plasmid and selected in puromycin (0.5 μg/ml) for up to 18 days posttransfection. At this time, DNA was isolated, DpnI digested to degrade unreplicated plasmid DNA, linearized with XbaI, and quantified by Southern blot. Average plasmid copy numbers were then calculated with DNA loading standards as shown in Fig. 4.
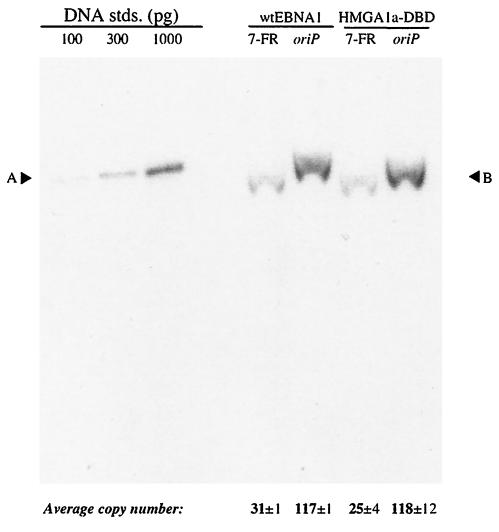
FIG. 4.
oriP-puro replication reporter plasmids are stably replicated as plasmids in the 293/EBNA1 and 293/HMGA1a-DBD cell lines. The 7-FR and wild-type oriP replication reporter plasmids described in the legend to Fig. 3 were transfected into 293/EBNA1 and 293/HMGA1a-DBD cells, which were subjected to 2 weeks of puromycin selection. At that time, DNA was isolated from the pool of puromycin-resistant cells and analyzed by Southern blotting with labeled probes made from plasmid pPUR-DS. The cell line used for transfection is indicated above each set of lanes, and the plasmid used for transfection is indicated above each lane. The amounts of standards loaded are indicated above each standard, and their electrophoretic mobility is indicated by A. B indicates the electrophoretic mobility of the test plasmids. The values below each lane indicate the average copy number per transfected cell and standard deviation. The experiment indicates that while under selection, both replication reporter plasmids were replicated equivalently in 293/EBNA1 and 293/HMGA1a-DBD cells.
In support of the colony formation data (Fig. 3), we found that 293/HMGA1a-DBD cells (118 ± 12 copies per cell) maintained wild-type oriP with an efficiency very close to that found for 293/EBNA1 cells (117 ± 1 copies per cell) (Fig. 4). Furthermore, we found that the average copy number found in both 293/EBNA1 (31 ± 1 copies per cell) and 293/HMGA1a-DBD (25 ± 4 copies per cell) cells decreased substantially when FR contained only seven EBNA1 binding sites. These results indicate that under selective conditions, HMGA1a-DBD stably maintained oriP plasmids at efficiencies very close to EBNA1. It was observed under these same assay conditions that 293/HMG1-DBD did not survive puromycin selection, again confirming our colony formation data that oriP cannot be maintained stably by the HMG1-DBD fusion protein.
These results have shown that HMGA1a-DBD maintains oriP plasmids at copy numbers very close to those of EBNA1 under selective conditions. Additionally, as indicated by the colony formation data and plasmid copy number, 293/HMGA1a-DBD cells are also sensitive to the number of binding sites within FR. Taken together, these data provide strong support that HMGA1a-DBD mechanistically functions similarly to EBNA1 in its ability to stably maintain oriP plasmids.
293/HMGA1a-DBD cells but not 293/HMG1-DBD cells support the transient replication of oriP plasmids.
As stable maintenance requires an oriP plasmid to undergo successful DNA synthesis and faithful partitioning upon each cell generation, failure of HMG1-DBD to support colony formation results from the inability to support either replication or partitioning of an oriP plasmid. We therefore examined the ability of 293/HMGA1a-DBD and 293/HMG1-DBD cells to support DNA synthesis from an oriP plasmid in a transient assay. 293/EBNA1, 293/HMGA1a-DBD, and 293/HMG1-DBD cells were transfected with plasmid 1033, which contains wild-type oriP. Cells were harvested 4 days posttransfection, and DNA was isolated from them. This DNA was digested with DpnI to degrade unreplicated input plasmid and with HindIII to linearize the replicated plasmids. Linearized DNAs were quantified by Southern blot. This analysis is shown in Fig. 5.
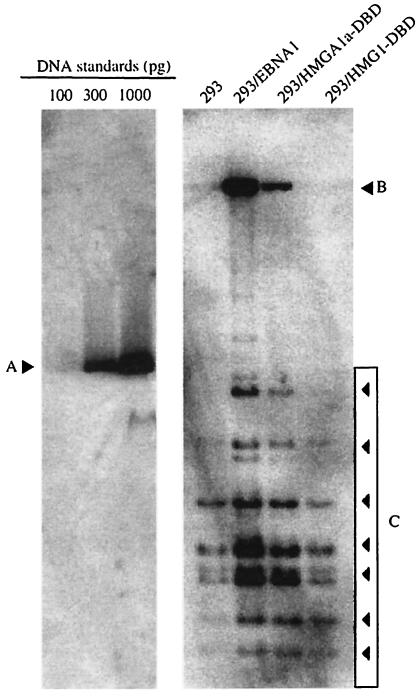
FIG. 5.
HMG1-DBD does not support transient replication of oriP plasmids. 293, 293/EBNA1, 293/HMGA1a-DBD, and 293/HMG1-DBD cells were transfected with the wild-type oriP plasmid described in the legend to Fig. 3. DNA was extracted from transfected cells 4 days posttransfection, digested with DpnI to degrade unreplicated, bacterially methylated input DNA, linearized with HindIII, and quantified by Southern blot. The blot was probed as described in the legend to Fig. 4. Quantification standards are shown on the left, with their migration position indicated by A, and the amount of the standard loaded is indicated above each lane. The identity of the cell line used for transient transfection is indicated above each lane on the right. The position of replicated DpnI-resistant, full-length linearized wild-type oriP plasmids is indicated by B, and the migration positions of DpnI-sensitive nonreplicated DNAs are indicated by the boxed arrowheads and C. No transient replication of wild-type oriP was detected in HMG1-DBD.
Plasmid 1033 was replicated efficiently in 293/EBNA1 cells but to a somewhat lesser extent in 293/HMGA1a-DBD cells by 4 days posttransfection. To our surprise, in multiple experiments, we failed to detect any replication of this reporter plasmid in 293/HMG1-DBD cells. In fact, even when Southern blots were overexposed, no replicated DNA could be detected in extracts prepared from transfected 293/HMG1-DBD cells, while small amounts of replicated 1033 DNA could be detected in 293 cells, confirming previous data that there is low-level nonspecific transient replication of oriP in the total absence of EBNA1 (Fig. 5, lane marked 293). To confirm the inability of HMG1-DBD to support replication of an oriP plasmid independent of the use of 293/HMG1-DBD cells, we transfected an expression plasmid for HMG1-DBD that also contained oriP (AGP111) into 293 cells and found that it had not undergone replication 2 days or 4 days posttransfection (data not shown). These data indicate that tethering by EBNA1 to metaphase chromosomes is required not only for oriP plasmid partitioning but also for replication of oriP plasmids.
Replication in 293/HMGA1a-DBD cells is DS dependent.
DS functions as the origin within oriP when EBNA1 is used as the replicator protein. EBNA1 binds four sites within DS, and this interaction is required for robust replication of oriP plasmids 4 days posttransfection. This may be because EBNA1 bound to DS mediates replication by either opening oriP through bending (5) or making DS accessible to the replication machinery by displacing nucleosomes (4). To determine whether HMGA1a-DBD still requires DS to mediate replication from oriP, we examined whether 293/HMGA1a-DBD cells could replicate an oriP plasmid that lacked DS. For this experiment we used the 20-FR plasmid used for the colony formation assays or a derivative of this plasmid that lacked DS. 293/EBNA1 and 293/HMGA1a-DBD cells were transfected with these plasmids, and DNAs were extracted from transfected cells 4 days posttransfection, DpnI digested to degrade unreplicated plasmids, linearized with XbaI, and examined by Southern blot.
As shown in Fig. 6, the DS-containing replication reporter plasmid was replicated in both cell types to similar average copies per cell (23.6 ± 5.4 for 293/EBNA1 and 25.1 ± 4.7 for 293/HMGA1a-DBD), despite differences in replication observed in transient assays with native oriP (Fig. 5); however, both 293/EBNA1 and 293/HGMA1a-DBD cells failed to replicate the reporter plasmid that lacked DS. The results in Fig. 6 support the notion that HMGA1a-DBD bound to DS possesses the same functional capabilities as EBNA1 for initiation of DNA synthesis from oriP. By extension, HMGA1a can replace any contributions made by the first 450 amino acids of EBNA1 toward the initiation of DNA synthesis from DS. That is, no specific protein-protein interactions are mediated by this large portion of EBNA1 required for the initiation of DNA synthesis from DS.
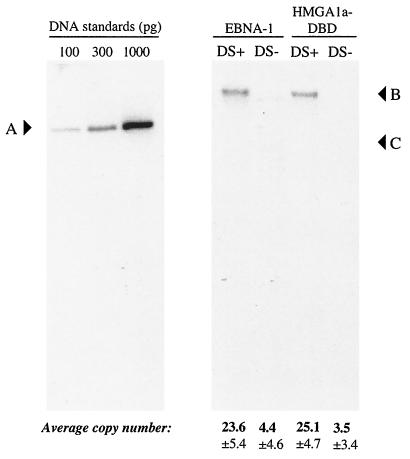
FIG. 6.
DS is required for the replication of oriP plasmids in 293/HMGA1a-DBD cells. 293/EBNA1 and 293/HMGA1a-DBD cells were transfected with the 20-FR plasmid described in the legend to Fig. 3 that either contained DS (DS+) or lacked DS (DS−). DNA was recovered from transfected cells 4 days posttransfection, digested exhaustively with DpnI to degrade input, bacterially methylated, unreplicated DNA, linearized with XbaI, and quantified by Southern blot. Quantification standards are shown on the left, with their migration position indicated by A, and the amount of the standard loaded is indicated above each lane. The identity of the transfected cell line is indicated above each set of lanes, and the identity of the transfected replication reporter is indicated above each lane. The migration position of replicated, DpnI-resistant, linearized replication reporter plasmid is indicated by B, while C indicates the position of the largest DpnI-sensitive fragment. The result indicates that replication of oriP plasmids in both cell lines occurred only in the presence of DS. The values below each lane indicate the copy number per cell along with the standard deviation.
oriP plasmids are lost at equivalent rates in 293/HMGA1a-DBD and 293/EBNA1 cells in the absence of selection.
In the presence of EBNA1, oriP is maintained efficiently, such that on average, only 2 to 6% of oriP plasmids are lost per cell per generation, depending on the cell type. Because of the short half-lives of the luciferase protein and mRNA (44), it has been shown that measuring luciferase activity from cells transfected with a luciferase reporter that contains oriP can also be used to follow plasmid maintenance in the presence or absence of EBNA1 (2, 44).
To determine if HMGA1a-DBD maintains oriP plasmids as efficiently as EBNA1, we calculated the rate at which oriP plasmids were lost in 293/HMGA1a-DBD cells in the absence of drug selection over a time course of 3 weeks. The protocol used for this experiment is diagrammed in Fig. 7. Briefly, 293/EBNA1 and 293/HMGA1a-DBD cells were cotransfected with plasmid 1033 and an EGFP expression plasmid. Plasmid 1033 contains oriP and expresses luciferase under the control of the EBV BamHI-C promoter. Beginning at 8 days posttransfection, the number of replicated oriP plasmids per cells was quantified by Southern blot, and the level of luciferase per transfected cell was measured. This process was repeated every 4 to 5 days under conditions in which the transfected cells were not permitted to reach confluence. The results of this assay, shown as the percentage of average copy number relative to the first time point (day 8), are summarized in Table 1.
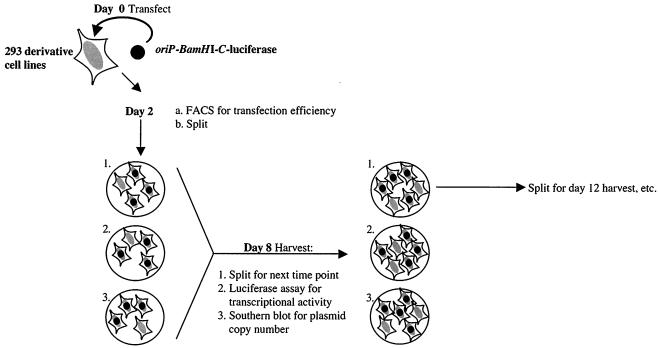
FIG. 7.
Schematic depiction of the rate of loss assay summarized in Table 1. On day 0, 293/EBNA1 and 293/HMGA1a-DBD cells were transfected with an oriP-BamHI-C-luciferase replication and transcription reporter plasmid. Two days posttransfection, cells were split onto three plates so that they would not reach confluence by 8 days posttransfection. On day 8, plasmid DNA was harvested from the cells on one plate and quantified by Southern blot, while the cells on another plate were used for FACS analysis and the luciferase assay. The third plate was split onto three additional plates for the next time of harvest, which was 4 days later. Subsequent harvests were carried out on days 12, 16, and 21 posttransfection.
TABLE 1.
Rate of loss of replicated oriP-BamHI-C-luciferase over time in the absence of selection in 293/EBNA1 and 293/HMGA1a-DBD cells
| Cell line | Avg copy no.a (% of total) on day posttransfection:
|
% Plasmid loss per cell generation | |||
|---|---|---|---|---|---|
| 8 | 12 | 16 | 21 | ||
| 293/EBNA1 | 100 | 85.4 ± 7.6 | 74.8 ± 11 | 60.9 ± 14 | 3.6 |
| 293/HMGA1a- DBD | 100 | 90 ± 23 | 85 ± 21 | 71.8 ± 7.2 | 3.5 |
Average copy number of DpnI-resistant, replicated plasmids maintained in the absence of selection, represented as a percentage the number of plasmid copies after 8 days.
In 293/EBNA1 cells oriP plasmids were lost at a rate of 3.6% per cell per generation, while in 293/HMGA1a-DBD cells, oriP plasmids were lost at a rate of 3.5% per cell per generation. We also determined the amount of luciferase activity for each cell type throughout the time course of the experiment. For both cell types, luciferase expression decreased at approximately 3.5% per cell generation, although the absolute levels of luciferase expressed in 293/EBNA1 cells were substantially higher than in 293/HMGA1a-DBD cells (data not shown).
These data show that even in the absence of selection, oriP plasmids are maintained with similar efficiencies by HMGA1a-DBD and EBNA1.
EBNA1 and HMGA1a-DBD bind interphase chromatin with substantially different affinities.
Although we postulate that our HMGA1a-DBD fusion functions like EBNA1 because both of these proteins associate with metaphase chromosomes, we decided to examine whether their affinities for interphase chromatin, as determined by salt extraction, also coincided. This would allow us to determine if a different property, the affinity of EBNA1 for chromatin, was related to its capacity to function in episomal tethering and maintenance. Nuclei were prepared from 293/EBNA1 and 293/HMGA1a-DBD cells and exposed to increasing concentrations of salt to determine the concentration that would liberate EBNA1 and HMGA1a-DBD from chromatin. The results of this analysis are shown in Fig. 8. Protein solubilized at each salt concentration was visualized by immunoblotting with rabbit polyclonal antiserum against the EBNA1 DBD. EBNA1 was extracted from interphase chromatin at between 100 mM and 200 mM salt, while HMGA1a-DBD was bound more tightly to interphase chromatin and extracted at 400 mM to 800 mM salt from intact nuclei. The results from this assay clearly indicate that a common affinity for interphase chromatin does not underlie the ability of EBNA1 and HMGA1a-DBD to maintain oriP plasmids.
FIG. 8.
EBNA1 and HMGA1a-DBD are differentially extracted from interphase nuclei. Nuclei were prepared from 293/EBNA1 and 293/HMGA1a-DBD cells and exposed to increasing concentrations of salt from 0 mM (ns 0) to 800 mM (ns800). Pelleted supernatants were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and immunoblotted with an antibody against the EBNA1 DBD. EBNA1 was maximally extracted from chromatin between 100 and 200 mM salt. HMGA1a-DBD was extracted from chromatin at between 400 and 800 mM salt. WCE, whole-cell extracts; CYTO, cytoplasmic extracts. A control extraction was performed with the chromatin-associated human protein orc2.
Metaphase chromosomal tethering of oriP plasmids is insufficient to activate transcription from oriP.
During latent infection, EBNA1 not only serves as a genome maintenance protein, but also functions as a potent transcriptional activator of several EBV promoters (16, 36). One such promoter is found in the BamHI-C EBV fragment that also contains oriP. Transcription from the BamHI-C promoter has been shown to be activated by EBNA1 bound to oriP (2, 36). We therefore tested whether HMGA1a-DBD could activate the BamHI-C promoter when bound to oriP. This would permit us to determine if tethering of oriP plasmids to metaphase chromosomes, which we have shown to be necessary for oriP maintenance, was sufficient for EBNA1 to activate transcription.
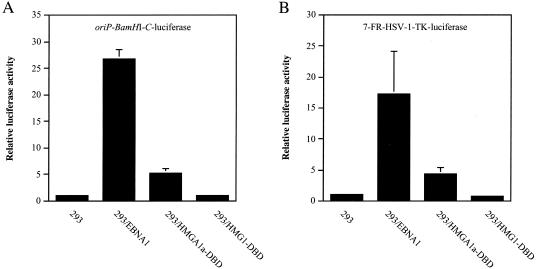
To examine this, 293, 293/EBNA1, 293/HMGA1a-DBD, and 293/HMG1-DBD cells were cotransfected with the transcription reporter plasmid oriP-BamHI-C-luciferase and an EGFP expression plasmid. At 48 h posttransfection, all cells were harvested and FACS profiled to normalize for plasmid uptake on the basis of GFP expression. Normalized extracts were then used to measure transactivation by EBNA1 and the HMG fusions. As shown in Fig. 9A, 293/EBNA1 cells readily transactivated the BamHI-C promoter approximately 27-fold over background levels. Surprisingly, we observed that despite the similar ability of EBNA1 and HMGA1a-DBD to maintain oriP plasmids, HMGA1a-DBD was severely impaired in the ability to activate transcription from the BamHI-C promoter, with only approximately a fivefold induction over background levels. Because HMG1-DBD was deficient in any replication or maintenance function, we expected and found 293/HMG1-DBD cells incapable of activating transcription from oriP-BamHI-C-luciferase. Although HMG1-DBD was unable to activate transcription from oriP-BamHI-C-luciferase, we did find that it interfered with the ability of EBNA1 to activate transcription from this reporter in a dominant negative manner (data not shown). This result indicates that this fusion protein retains the ability to recognize and bind oriP.
FIG. 9.
Transactivation of episomal transcription reporter plasmids by EBNA1, HMGA1a-DBD, and HMG1-DBD. 293, 293/EBNA1, 293/HMGA1a-DBD, and 293/HMG1-DBD cells were transfected with (A) oriP-BamHI-C-luciferase or (B) 7-FR-HSV1-TK-luciferase and an EGFP expression plasmid. Cells were harvested 48 h posttransfection and FACS profiled for EGFP expression to normalize for transfection efficiency, following which cytoplasmic extracts were prepared and examined for luciferase activity. The bars indicate the relative luciferase activity observed in each cell line over the activity observed in 293 cells, which was set at 1. The results indicate that HMG1-DBD did not activate transcription from either reporter plasmid, while HMGA1a-DBD activated transcription to about 20% of the level of wild-type EBNA1 for either transcription reporter plasmid.
Similar results were also found in luciferase assays from cells transfected with a FR-HSV-1-TK-luciferase reporter containing only seven EBNA1 binding sites. Figure 9B shows that EBNA1 was less efficient in activating transcription (17-fold versus 27-fold). As with transcription from the BamHI-C promoter, HMG1-DBD was unable to activate transcription and HMGA1a-DBD was able to activate transcription only weakly from a luciferase reporter containing seven EBNA1 binding sites.
Thus, these experiments indicate that tethering of oriP plasmids to metaphase chromosomes is insufficient to activate transcription by EBNA1 (Fig. 3 and 4 and Table 1).
HMGA1a-DBD cannot activate transcription from an integrated FR-TK-luciferase reporter to the same extent as EBNA1.
We observed that HMGA1a-DBD and EBNA1 functioned to similar levels with respect to maintenance but the former activated transcription from an episomal oriP plasmid very poorly. This led us to speculate that EBNA1's ability to transactivate requires properties besides its ability to tether oriP plasmids to cellular chromosomes. To determine this, we examined whether EBNA1 and HMGA1a-DBD could activate transcription from an FR-TK-luciferase reporter that was integrated in the host chromosome.
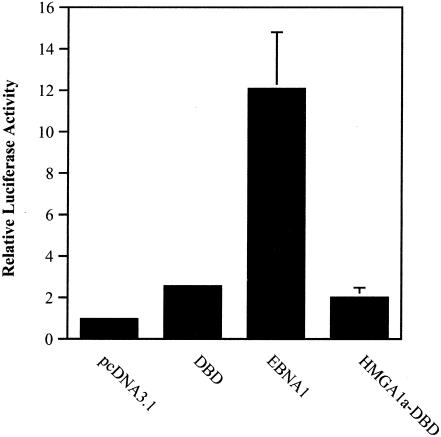
For this assay we used BJAB cells containing an integrated FR-TK-luciferase reporter. Cells were electroporated with EBNA1 or HMGA1a-DBD expression plasmids as well as an EGFP plasmid. Cells were normalized for transfection efficiency by FACS profiling and then examined for luciferase expression. This analysis is shown in Fig. 10. After 48 h, EBNA1 activated transcription from the integrated reporter 12-fold higher than electroporation with the pcDNA3.1 control. Interestingly, however, HMGA1a-DBD could activate transcription to levels no higher than the DBD of EBNA1. Because it could activate transcription, albeit poorly, from an episomal (Fig. 9) but not an integrated luciferase reporter (Fig. 10), we concluded that oriP retention (maintenance) is the major mechanism through which HMGA1a-DBD activates transcription. Furthermore, we concluded that the N terminus of EBNA1 possesses transactivation ability beyond its capacity to retain oriP plasmids in the nuclei of proliferating cells.
FIG. 10.
Transactivation of an integrated FR-HSV1-TK-luciferase reporter in BJAB cells by EBNA1 and HMGA1a-DBD. BJAB cells containing an integrated FR-HSV1-TK-luciferase reporter were electroporated with the control plasmid pcDNA3.1 or expression plasmids for the EBNA1 DBD, wild-type EBNA1 (EBNA1), or HMGA1a-DBD along with an EGFP expression plasmid. Two days after transfection, cells were FACS profiled for EGFP expression to normalize for transfection efficiency, following which cytoplasmic extracts were prepared and examined for luciferase activity. The bars indicate the relative luciferase activity observed over electroporation of the control plasmid pcDNA3.1. The results indicate that while EBNA1 could clearly transactivate an integrated transcription reporter with EBNA1 binding sites, HMGA1a-DBD did not transactivate this reporter over and above any effect of the DBD alone.
DISCUSSION
The latent genome of EBV is maintained as a stable episome within cells, a process that requires only the viral protein EBNA1 in trans and the viral element oriP in cis. It is postulated that EBNA1 maintains EBV genomes or oriP plasmids in proliferating cells by tethering them to host chromosomes. The genome or oriP maintenance activity of EBNA1 requires the amino-terminal domain of EBNA1 that is known to associate with chromatin. This association requires two positively charged regions within the amino terminus that we have termed A (amino acids 33 to 89) and B (amino acids 328 to 378). Our mutational analysis (M. Ujihara, J. Sears, and A. Aiyar, unpublished data) indicates that A and B are interchangeable in function, making it difficult to understand the contributions of these regions to maintenance in intact EBNA1.
In this study we used fusions between two host chromatin binding proteins and the DBD of EBNA1 to understand the nature of chromatin associations required for stable oriP replication and maintenance. EBNA1 has several properties that may enable it to mediate maintenance of oriP plasmids. First, it is a nuclear protein and is found in interphase nuclei (35). This property may be sufficient to retain oriP plasmids in the nuclei of proliferating cells. Second, EBNA1 is associated with interphase chromatin (23) and is salt extracted from interphase nuclei at 100 mM to 200 mM NaCl (Fig. 8). Third, EBNA1 associates with metaphase chromosomes (25, 32), and this property may permit tethered oriP plasmids to be retained in daughter nuclei over multiple cell cycles.
To understand which of these biological properties of EBNA1 is crucial for the long-term replication of oriP plasmids, we made fusions between two host cell chromatin binding proteins, HMGA1a and HMG1, to the DBD of EBNA1. Previously, work by Hung et al. showed that replacing most of EBNA1's amino terminus with either HMGI(Y) or histone H1 resulted in a fusion that could maintain oriP plasmids (20). On the basis of those findings, we used a similar approach to determine directly the relevance of metaphase chromosome association in contributing to EBNA1 function. Like EBNA1, both HMGA1a and HMG1 are nuclear proteins, associate with interphase chromatin, and stain interphase nuclei when examined by indirect immunofluorescence. However, a major difference between these proteins is that HMG1 does not associate specifically with metaphase chromosomes, whereas HMGA1a does.
Detailed characterization of the HMGA1a-DBD and HMG1-DBD fusion proteins indicates that they display the same biological properties as unfused HMGA1a and HMG1, respectively. Through our analyses, we were struck by the strong similarity between the mechanism of plasmid maintenance of HMGA1a-DBD and wild-type EBNA1: replication in 293/HMGA1a-DBD cells was dependent on the number of binding sites present within oriP and required DS, and plasmids were lost at a rate of 3.5% per cell generation, all features common to cells expressing wild-type EBNA1. These coincidences strongly suggest a similar mechanism of plasmid maintenance by both of these proteins.
HMGA1a binds chromosomes directly via AT hooks (21, 42, 43). We note that there is an AT hook sequence within EBNA1, and other stretches of sequence that strongly resemble AT hooks are peppered throughout the amino terminus of EBNA1. We are currently investigating whether such sequences can function as AT hooks and mediate an association between EBNA1 and host chromosomes.
Because metaphase chromosome association is a major difference between HMGA1a and HMG1, a lack of functionality in the chimeric fusion with HMG1 demonstrated that specific tethering to metaphase chromosomes is crucial for the stable long-term replication of oriP plasmids (Fig. 3 and 5). It is less obvious why oriP plasmids failed to replicate even transiently in 293/HMG1-DBD cells. HMG1 and this fusion protein are localized to interphase nuclei, especially S-phase nuclei, where DNA synthesis occurs. The results from the HMGA1a-DBD fusion make it clear that the amino terminus of EBNA1 makes no contribution to the synthesis of oriP DNA, so it cannot be argued that critical domains of EBNA1 are missing in the HMG1-DBD fusion protein. However, a more detailed consideration of the licensing model of eukaryotic DNA replication allows us to hypothesize a reason for the failure of the HMG1-DBD fusion to support even transient DNA replication of oriP plasmids.
Akin to chromosomal origins, oriP is replicated once per cell cycle. Elegant experiments by the groups of Hammerschmidt and Yates have demonstrated that the ORC proteins associate with DS (10, 41) and that replication from oriP is minichromosome maintenance (MCM) protein dependent (9). Association of MCM proteins with an origin is the licensing event that allows that origin to fire in the ensuing S phase. Recent work by Gilbert and coworkers has established that in mammalian cells, MCM2 and MCM3 (core components of the MCM complex) are loaded onto chromatin and therefore chromosomal origins in late telophase, a stage of mitosis immediately after metaphase (11, 12).
What relevance does this have for the replication of oriP? Although MCM proteins license replication from oriP, there has been no demonstration that they are loaded onto oriP via a direct association with EBNA1. We therefore propose that EBNA1 mediates replication from oriP by localizing oriP plasmids to metaphase chromosomes, the substrate on which the MCM complex is loaded; we hypothesize that the MCM complex is also loaded on the associated oriP replicons at the same time. The loaded MCM complexes allow replication firing from oriP in the ensuing S phase.
This hypothesis may underscore why HMGA1a-DBD permits efficient replication of oriP, although it is missing the first 450 amino acids of EBNA1. The minimal requirement may be that the fusion protein is localized to metaphase chromosomes — the appropriate time when MCM proteins are loaded onto chromosomes and, by our hypothesis, oriP plasmids. It may also explain why HMG1-DBD does not permit any oriP replication despite the fusion's localization to interphase chromatin. More critically, it does not associate with metaphase chromosomes, thereby preventing the licensed replication of oriP plasmids. Similarly, this hypothesis allows us to understand why deleting the chromosome binding domains of EBNA1 results in proteins that are unable to support even transient replication of oriP plasmids (25, 27). These proteins are likely crippled in their ability to provide oriP plasmids access to the licensing machinery.
Others have suggested that the strength with which a cellular protein binds chromatin will predict its ability to replace the amino terminus of EBNA1 in the maintenance function (20). Our salt extraction data in Fig. 8 indicate that although EBNA1 and HMGA1a-DBD have substantially different salt extraction profiles, they both support long-term replication of oriP plasmids. Taken together, our results suggest that metaphase chromosome association, not chromosome binding affinity, is critical to determining EBNA1's ability to support transient or stable replication of oriP plasmids.
Previous data suggest that the sole function of EBNA1 in transcriptional enhancement is to maintain oriP episomes within the nucleus, thus making templates for transcription readily available to the host cell transcriptional machinery (20, 26, 28). A corollary to this hypothesis is that EBNA1 has no intrinsic transcriptional enhancement ability beyond its capacity to maintain oriP in the nucleus. Our results support an alternative interpretation from the observation that while HMGA1a-DBD can maintain oriP episomes as efficiently as EBNA1, it activates transcription to approximately 20% of the level of EBNA1. This suggested to us that EBNA1 serves to activate transcription not only through tethering (maintenance) of oriP to cellular chromosomes but also through some form of intrinsic transactivation. One apparent discrepancy between our work and that conducted previously (20) is that the version of EBNA1 used in our studies to generate chimeric fusions lacks the region between amino acids 407 and 450, which may contribute to transcriptional activation by EBNA1.
The HMGA1a-DBD fusion constructed by us poorly activates transcription from episomal reporter plasmids, especially compared to the activation ability of wild-type EBNA1. We suspected that this residual activation resulted from retention of episomal transcription templates in the nuclei of transfected cells. To investigate this further, we examined the ability of EBNA1 and HMGA1a-DBD to activate transcription from an integrated EBNA1-dependent transcription reporter. In contrast to the results of Kang and Kieff (26), we found that EBNA1 could activate transcription from an integrated transcription reporter, coinciding with the results of Kennedy and Sugden (personal communication). In this assay, which does not depend on episomal reporter retention in the nucleus of a transfected cell, we found that HMGA1a-DBD transactivated an integrated FR-TK-luciferase reporter to levels that were indistinguishable from those with the DBD alone. Our findings provide evidence that the N terminus of EBNA1 possesses transcriptional activation ability that is independent of its ability to maintain oriP plasmids.
.
Acknowledgments
We thank Leslie Elbert and Scott Battle for constructing some of the plasmids used in this study and Maki Ujihara for help in conducting some of the experiments described here. We thank Ray Reeves (Washington State University, Pullman) for providing cDNA clones of HMGA1a and HMG1 and for generous advice. We thank George Kennedy and Bill Sugden for generously providing the BJAB cell line containing an integrated EBNA1-responsive transcription reporter and for other plasmid reagents. We thank Lou Laimins, Kasturi Haldar, and members of their laboratories for assistance during conduct of the experiments described here. We thank Lou Laimins, Kathy Rundell, and Pat Spear for critically reviewing the manuscript. FACS analyses were performed at the Immunobiology Center of Northwestern University.
J.S. is a predoctoral trainee supported by Carcinogenesis Training Grant T32 CA09560. A.A. is supported by awards from the Leukemia Research Foundation and the National Cancer Institute.
REFERENCES
- 1.Adams, A. 1987. Replication of latent Epstein-Barr virus genomes in Raji cells. J. Virol. 61:1743-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiyar, A., C. Tyree, and B. Sugden. 1998. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 17:6394-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambinder, R. F., M. A. Mullen, Y. N. Chang, G. S. Hayward, and S. D. Hayward. 1991. Functional domains of Epstein-Barr virus nuclear antigen EBNA1. J. Virol. 65:1466-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avolio-Hunter, T. M., P. N. Lewis, and L. Frappier. 2001. Epstein-Barr nuclear antigen 1 binds and destabilizes nucleosomes at the viral origin of latent DNA replication. Nucleic Acids Res. 29:3520-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bashaw, J. M., and J. L. Yates. 2001. Replication from oriP of Epstein-Barr virus requires exact spacing of two bound dimers of EBNA1 which bend DNA. J. Virol. 75:10603-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bochkarev, A., J. A. Barwell, R. A. Pfuetzner, W. Furey, Jr., A. M. Edwards, and L. Frappier. 1995. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein EBNA 1. Cell 83:39-46. [DOI] [PubMed] [Google Scholar]
- 7.Bustin, M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19:5237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ceccarelli, D. F., and L. Frappier. 2000. Functional analyses of the EBNA1 origin DNA binding protein of Epstein-Barr virus. J. Virol. 74:4939-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 98:10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 11.Dimitrova, D. S., T. A. Prokhorova, J. J. Blow, I. T. Todorov, and D. M. Gilbert. 2002. Mammalian nuclei become licensed for DNA replication during late telophase. J. Cell Sci. 115:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimitrova, D. S., I. T. Todorov, T. Melendy, and D. M. Gilbert. 1999. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol. 146:709-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Disney, J. E., K. R. Johnson, N. S. Magnuson, S. R. Sylvester, and R. Reeves. 1989. High-mobility group protein HMG-I localizes to G/Q- and C-bands of human and mouse chromosomes. J. Cell Biol. 109:1975-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falciola, L., F. Spada, S. Calogero, G. Langst, R. Voit, I. Grummt, and M. E. Bianchi. 1997. High mobility group 1 protein is not stably associated with the chromosomes of somatic cells. J. Cell Biol. 137:19-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frappier, L., and M. O'Donnell. 1991. Epstein-Barr nuclear antigen 1 mediates a DNA loop within the latent replication origin of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 88:10875-10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gahn, T. A., and B. Sugden. 1995. An EBNA1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J. Virol. 69:2633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 18.Hebner, C., J. Lasanen, S. Battle, and A. Aiyar. 2003. The spacing between adjacent binding sites in the family of repeats affects the functions of Epstein-Barr nuclear antigen 1 in transcription activation and stable plasmid maintenance. Virology, 311:263-274. [DOI] [PMC free article] [PubMed]
- 19.Hubert, W. G., T. Kanaya, and L. A. Laimins. 1999. DNA replication of human papillomavirus type 31 is modulated by elements of the upstream regulatory region that lie 5′ of the minimal origin. J. Virol. 73:1835-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung, S. C., M. S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huth, J. R., C. A. Bewley, M. S. Nissen, J. N. Evans, R. Reeves, A. M. Gronenborn, and G. M. Clore. 1997. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 4:657-665. [DOI] [PubMed] [Google Scholar]
- 22.Isackson, P. J., D. L. Bidney, G. R. Reeck, N. K. Neihart, and M. Bustin. 1980. High mobility group chromosomal proteins isolated from muclei and cytosol of cultured hepatoma cells are similar. Biochemistry 19:4466-4471. [DOI] [PubMed] [Google Scholar]
- 23.Ito, S., E. Gotoh, S. Ozawa, and K. Yanagi. 2002. Epstein-Barr virus nuclear antigen-1 is highly colocalized with interphase chromatin and its newly replicated regions in particular. J. Gen. Virol. 83:2377-2383. [DOI] [PubMed] [Google Scholar]
- 24.Ito, S., and K. Yanagi. 2003. Epstein-Barr virus (EBV) Nuclear antigen 1 colocalizes with cellular replication foci in the absence of EBV plasmids. J. Virol. 77:3824-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanda, T., M. Otter, and G. M. Wahl. 2001. Coupling of mitotic chromosome tethering and replication competence in Epstein-barr virus-based plasmids. Mol. Cell. Biol. 21:3576-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang, M. S., S. C. Hung, and E. Kieff. 2001. Epstein-Barr virus nuclear antigen 1 activates transcription from episomal but not integrated DNA and does not alter lymphocyte growth. Proc. Natl. Acad. Sci. 98:15233-15238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirchmaier, A. L., and B. Sugden. 1997. Dominant-negative inhibitors of EBNA1 of Epstein-Barr virus. J. Virol. 71:1766-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langle-Rouault, F., V. Patzel, A. Benavente, M. Taillez, N. Silvestre, A. Bompard, G. Sczakiel, E. Jacobs, and K. Rittner. 1998. Up to 100-fold increase of apparent gene expression in the presence of Epstein-Barr virus oriP sequences and EBNA1: implications of the nuclear import of plasmids. J. Virol. 72:6181-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lupton, S., and A. J. Levine. 1985. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol. Cell. Biol. 5:2533-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackey, D., and B. Sugden. 1999. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol. Cell. Biol. 19:3349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 32.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, and J. C. Nicolas. 1999. Mapping EBNA1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martelli, A. M., M. Riccio, R. Bareggi, G. Manfioletti, G. Tabellini, G. Baldini, P. Narducci, and V. Giancotti. 1998. Intranuclear distribution of HMGI/Y proteins. An immunocytochemical study. J. Histochem. Cytochem. 46:863-864. [DOI] [PubMed] [Google Scholar]
- 34.Middleton, T., and B. Sugden. 1992. EBNA1 can link the enhancer element to the initiator element of the Epstein-Barr virus plasmid origin of DNA replication. J. Virol. 66:489-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Middleton, T., and B. Sugden. 1994. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein-Barr virus replication protein EBNA1. J. Virol. 68:4067-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puglielli, M. T., M. Woisetschlaeger, and S. H. Speck. 1996. oriP is essential for EBNA gene promoter activity in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J. Virol. 70:5758-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reeves, R. 2001. Molecular biology of HMGA proteins: hubs of nuclear function. Gene 277:63-81. [DOI] [PubMed] [Google Scholar]
- 38.Reeves, R. 2000. Structure and function of the HMGI(Y) family of architectural transcription factors. Environ. Health Perspect. 108(Suppl. 5):803-809. [DOI] [PubMed] [Google Scholar]
- 39.Reisman, D., and B. Sugden. 1986. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol. Cell. Biol. 6:3838-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saitoh, Y., and U. K. Laemmli. 1994. Metaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell 76:609-622. [DOI] [PubMed] [Google Scholar]
- 41.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solomon, M. J., F. Strauss, and A. Varshavsky. 1986. A mammalian high mobility group protein recognizes any stretch of six A. T base pairs in duplex DNA. Proc. Natl. Acad. Sci. USA 83:1276-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strauss, F., and A. Varshavsky. 1984. A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell 37:889-901. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, J. F., L. S. Hayes, and D. B. Lloyd. 1991. Modulation of firefly luciferase stability and impact on studies of gene regulation. Gene 103:171-177. [DOI] [PubMed] [Google Scholar]
- 45.Wu, H., P. Kapoor, and L. Frappier. 2002. Separation of the DNA replication, segregation, and transcriptional activation functions of Epstein-Barr nuclear antigen 1. J. Virol. 76:2480-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yates, J., N. Warren, D. Reisman, and B. Sugden. 1984. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. USA 81:3806-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yates, J. L., and N. Guan. 1991. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J. Virol. 65:483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]