Abstract
Analysis of the composition and regulation of the Borna disease virus (BDV) polymerase complex has so far been limited by the lack of a functional assay. To establish such an assay on the basis of an artificial minigenome, we constructed expression vectors encoding either nucleoprotein (N), phosphoprotein (P), X protein, or polymerase (L) of BDV under the control of the chicken β-actin promoter. A Flag-tagged version of L colocalized with virus-encoded N and P in characteristic nuclear dots of BDV-infected cells and increased viral N-protein levels in persistently infected Vero cells. Vector-driven expression of L, N, and P in BSR-T7 cells together with a negative-sense BDV minigenome carrying a chloramphenicol acetyltransferase (CAT) reporter gene resulted in efficient synthesis of CAT protein. Induction of CAT protein synthesis strongly depended on a 10- to 30-fold molar excess of the N-encoding plasmid over the P-encoding plasmid. Cotransfection of even small amounts of plasmid encoding the viral X protein reduced CAT synthesis to background levels. Thus, the N-to-P stoichiometry seems to play a central role in the regulation of the BDV polymerase complex. Our data further suggest a negative regulatory function for the X protein of BDV.
Borna disease virus (BDV) is a neurotropic, enveloped virus with a nonsegmented negative-strand RNA genome (4). In naturally and experimentally infected animals, BDV establishes a noncytolytic, persistent infection of the central nervous system that frequently results in a severe immune-mediated neurological disorder (10). Successful experimental infection of a broad range of warm-blooded animals has been reported (25). Serological evidence suggests that BDV or a BDV-like virus infects humans (2).
The compact BDV genome of approximately 8,900 nucleotides (nt) is transcribed and replicated in the nucleus and encodes at least six viral proteins organized in three transcription units (3, 4, 9). BDV employs a variety of strategies to regulate viral protein levels including overlapping open reading frames (ORFs), transcription units, and transcriptional signals, read-through of transcription termination signals, and alternative splicing of polycistronic mRNA (11, 21, 22). On the basis of these unique properties, BDV has been classified as the prototypic member of a new virus family, Bornaviridae, in the order Mononegavirales.
Negative-strand RNA viruses initiate infection by introducing their genetic material in the form of ribonucleoprotein complexes into the host cells. These complexes, which consist of the single-stranded RNA genome associated with the RNA-binding protein nucleoprotein (N), the polymerase cofactor phosphoprotein (P), and the RNA-dependent RNA polymerase L, are transcriptionally active and direct the synthesis of viral mRNAs (8, 17). Upon production of sufficient amounts of newly synthesized viral proteins, replication is initiated and new nucleocapsids can be formed. A consequence of this multiplication strategy is that, in contrast to the situation of positive-strand RNA viruses, naked genomic RNA of negative-strand viruses by itself is not competent to initiate infection. For the genetic manipulation of negative-strand viruses, novel systems in which cDNAs encoding viral proteins and cDNAs encoding artificial viral genomes are coexpressed had to be developed. Using such systems, in vivo reconstitution of active viral polymerase complexes and generation of recombinant viruses from cDNA was reported for a variety of negative-strand RNA viruses (8, 17). For the Bornaviridae, however, the reconstitution of functional nucleocapsid complexes has not yet been achieved.
The precise composition of the BDV nucleocapsid is still unknown. The p39 isoform of N and the p23 isoform of P are the main components of the BDV nucleocapsid. In contrast to other members of the order Mononegavirales, N-terminally truncated isoforms of N (p38) and P (p16) are present in infected cells (15, 22). These isoforms are produced by internal translation initiation at in-frame AUG codons. p38 lacks the nuclear localization signal located in the N-terminal 13 amino acids of p39 (14), but nuclear import of p38 in infected cells is still observed presumably due to interaction with p39 and P (14). A variety of p23 interaction partners have been described. Domains responsible for interaction with N and the viral X protein (p10) and for the formation of homo-oligomers were defined (24). The interaction of p23 with L was described but has not yet been characterized in detail (28). p23 becomes highly phosphorylated in the course of the viral infection cycle (23). It was proposed that differential phosphorylation regulates P-protein function. On the basis of the strong interaction of the X protein with p23 (24), it has been proposed that the X protein is an integral or regulatory component of the polymerase complex. Finally, by assuming that various conserved AUG codons in the L gene might be used for translation initiation, several isoforms of the L protein (p190/p180/p165) have been postulated (4). The impressive variety of BDV nucleocapsid components suggests that various forms of the BDV polymerase complex that exhibit differential transcriptional activity exist.
Several lines of evidence indicate that not only the nucleocapsid composition but also the stoichiometry of nucleocapsid components might be involved in the regulation of the BDV multiplication cycle. In tissue culture and in experimentally infected animals, major alterations of the N-to-P stoichiometry during the acute and persistent phases of the BDV infection were detected (31). Detailed analysis of a restriction phenomenon in BDV-infected cells, designated homologous interference, revealed that expression of single BDV nucleocapsid components can render cells resistant to subsequent infection with BDV (12). These studies further showed that transient expression of N-terminally Flag-tagged P inhibits BDV propagation in persistently infected Vero cells. Taken together, these data suggest that BDV employs a unique mechanism for the regulation of its polymerase.
We report here the establishment of a functional assay for the analysis of the BDV polymerase complex. The assay is based on the coexpression of viral nucleocapsid components with an artificial BDV minigenome harboring the chloramphenicol acetyltransferase (CAT) reporter gene. The BDV proteins L, N, and P, but not the X protein, were required for the induction of CAT synthesis. We further demonstrate that the BDV polymerase activity is tightly regulated by the N-to-P stoichiometry and strongly suppressed by the X protein. Our results thus provide the first insights into the composition and regulation of the BDV polymerase complex.
MATERIALS AND METHODS
Plasmid construction.
PCR was performed with proofreading Pwo DNA polymerase (Peqlab) and standard reaction conditions in the GeneAmp PCR cycler 9600 (Applied Biosystems). The integrity of all PCR-derived DNA fragments was verified by sequencing. All restriction digestions were performed with enzymes purchased from Fermentas or New England Biolabs. Ligation reactions were done using 2.5 Weiss units of the bacteriophage T4 DNA ligase (Fermentas) in a total volume of 5 μl. The ligation reaction mixtures were incubated at 16°C for at least 2 h and then used to transform 50 μl of competent Top10 bacteria (Invitrogen).
The BDV minigenomes were assembled from three PCR fragments in plasmid p(+)MV (20). The BDV 5′ and 3′ noncoding regions (NCR) were fused to the T7 RNA polymerase promoter and the hepatitis delta virus ribozyme (δ), respectively, using the overlap extension PCR technique (13). To produce the fragment with 5′-proximal sequences of the BDV genome, the T7 promoter fragment was amplified from p(+)MV using primer 5′-CGAGGTGCCGTAAAGC-3′ that hybridizes to a sequence located 70 nt upstream of a unique SfiI restriction site and primer 5′-gttgtagcgcTATAGTGAGTCGTATTACA-3′ that binds to the T7 promoter and also contains a sequence that is complementary to the extreme 5′ end of the BDV genome. Hybridizing and nonhybridizing primer sequences are given in uppercase and lowercase letters, respectively. The sequence designed to overlap with the target sequence of the second PCR fragment is underlined. The viral 5′-NCR was amplified from a plasmid that contains 2,800 nt derived from the 5′-proximal part of strain He/80FR genome using primer 5′-ctcactataGCGCTACAACAAAGCAAC-3′ and primer 5′-GGCAGCATATCCTCGG-3′. The resulting PCR fragments were purified by agarose gel electrophoresis. The fragments were combined (1 ng of each fragment), and overlap extension PCR was performed using primers 5′-GAGCTTGACGGGGAAAG-3′ and 5′-gacctcGAGCCATCTACTGCCC-3′ to amplify a cDNA that contains the T7 promoter and the complete 5′-NCR of BDV. Primer-encoded restriction enzyme recognition sequences are indicated by bold type. The resulting fragment was digested with SfiI and XhoI and ligated into SfiI/XhoI-opened p(+)MV, resulting in the intermediate construct pT7-5′NCR/MV.
Two versions of the 3′-NCR of BDV were amplified from a plasmid containing a 2,000-nt fragment derived from the 3′ end of the genome of strain He/80FR using primer 5′-CGCAGCGTGCAGTCC-3′ in combination with either primer 5′-gggaccatgccggccTGTTGCGTTAACAACAAACC-3′ or primer 5′-gggaccatgccggccGTTGCGTTAACAACAAACC-3′. The ribozyme fragment was amplified from p(+)MV using primer 5′-GGCCGGCATGGTCCC-3′ and primer 5′-GTTGTGTGGAATTGTGAGC-3′ that hybridizes to a sequence located 100 nt downstream of an unique NotI restriction site. The final 3′-NCR-ribozyme fragments with or without the additional A residue were amplified by overlap extension PCR using primer 5′-gacggtaccTGCGTGTATTCATTTTGTTAG-3′ and primer 5′-GATTACGCCAAGCTCG-3′. The resulting product was digested with KpnI and NotI. Finally, the CAT gene was amplified using primers 5′-gaggtaccATGGAGAAAAAAATCACTG-3′ and 5′-gacctcgagTTACGCCCCGCCCTG-3′, and the resulting product was digested with KpnI and XhoI. The two fragments were joined and inserted into XhoI/NotI-opened pT7-5′NCR/MV in a three-way ligation reaction, yielding constructs pT7-gmgA and pT7-gmgC, respectively.
To create expression constructs for the various BDV nucleocapsid components, the multiple cloning site (MCS) of vector pCAGGS (pCA) (18) was first modified. The SacI/KpnI fragment of pCA was replaced by the corresponding MCS fragment of pBS-KS+ (Clontech). To eliminate several nonunique restriction sites, the resulting plasmid was cut with XmaI and religated, yielding a MCS with unique EcoRI, NotI, and NheI restriction sites in the desired order. The complete ORFs for N, P, and X of BDV strain He/80 were excised from existing plasmids (12) and inserted into the MCS of the modified pCA vector. To generate a suitable L expression plasmid, base changes unintentionally introduced into plasmid pBlue-L (19) during PCR were first corrected by site-directed mutagenesis. The corrected version of pBlue-L codes for the p190 L isoform of BDV strain He/80FR with arginine at position 451 and isoleucine at position 959. The complete L ORF of this plasmid was excised using EcoRI and NotI and inserted into the corresponding sites of the modified pCA vector.
A cDNA encoding Flag-tagged L was first assembled in pBlue-L and later transferred into pCA. Briefly, primer 5′-TCATGAGCGGATACATATTTG-3′ and primer 5′-atagcggccgctttatcgtcatcgtctttgtagtcCATTTTCCGGAATTCGATATC-3′ were used to amplify a 850-base-pair (bp) fragment of the vector backbone, thereby inserting the Flag tag (shown in italic type) and the NotI restriction site downstream of the EcoRI site. This DNA fragment was cut at the NotI site and at a unique NgoMIV site located 370 bp upstream of the EcoRI site. A 200-bp fragment encoding the 5′ end of the L ORF was amplified from pBlue-L using primers 5′-ccagcggccgctATGTCATTTCATGCGAGC-3′ and 5′-CAACTTGGCGGAATAGC-3′, thereby introducing a NotI restriction site and an additional T nucleotide to maintain the reading frame. The resulting PCR fragment was digested with NotI and BsiWI. Together with the NgoMIV/NotI fragment, it was then ligated into pBlue-L between the NgoMIV and BsiWI restriction sites. Due to the presence of a second NotI site at the 3′ end of the L expression cassette, a unique NheI site located in the L ORF was used to excise the complete flagL cDNA in the form of two distinct pieces. The EcoRI/NheI fragment encoding the N-terminal moiety and the NheI/NotI fragment encoding the C-terminal moiety were finally inserted between the EcoRI and NotI sites of the modified pCA vector in a three-way ligation.
The N, P, and X ORFs of BDV strain He/80FR were amplified from plasmids pTRE-N, pTRE-P, and pBI-X/GFP (12), respectively, using primers 5′-gcagcggccgctATGCCACCCAAGAGACG-3′ and 5′-gcatctagaCTAGTTTAGACCAGTCAC-3′ for N, primers 5′-gcagcggccgctATGGCAACGCGACCATC-3′ and 5′-gcatctagaTTATGGTATGATGTCCCAC-3′ for P, and primers 5′-gcagcggccgctATGAGTTCCGACCTCCGGC-3′ and 5′-gcatctagaTCATTCGATAGCTGCTCC-3′ for X. The resulting PCR fragments were digested with NotI and XbaI and ligated into modified vector pCA or pCA-flagL digested with NotI and NheI, creating constructs pCA-N, pCA-P, and pCA-X or constructs pCA-flagN, pCA-flagP, and pCA-flagX, respectively. The ligation of the XbaI-digested fragments into the NheI-opened vectors was possible, because both restriction enzymes create matching overhangs.
Cells and transfections.
Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) for Vero cells (African green monkey kidney) and with 10% FCS plus 0.5 mg of G418 per ml for BSR-T7 cells (baby hamster kidney) stably expressing the T7 RNA polymerase (5). Cells were kept at 37°C in a 5% CO2 humidified atmosphere.
Semiconfluent layers of Vero and BSR-T7 cells that were not infected or infected with BDV He/80FR were grown in 35-mm-diameter (6-well) or 25-mm-diameter (12-well) dishes before transfection with the various plasmids using Metafectene (Biontex). DNA dilutions were prepared in 100 μl of DMEM for transfection of the cells in 35-mm-diameter dishes and in 50 μl of DMEM for the transfection of the cells in 25-mm-diameter dishes. The DNA preparations were subsequently mixed with 100 μl of DMEM containing 10 μl of Metafectene and 50 μl of DMEM containing 5 μl of Metafectene, respectively. The transfection solutions were incubated for 30 min at room temperature and then applied directly to the cell supernatants. After overnight incubation, the transfection solution was removed from the cells, fresh medium was added, and incubation was continued for the indicated period of time.
RNA preparation.
For each RNA sample, five six-well cultures of BSR-T7 cells were transfected with the indicated amounts of plasmid DNA. Three days after transfection, the cells of one six-well culture were lysed in 500 μl of CAT lysis buffer (Roche) and CAT expression was determined. Total RNA from the remaining four six-well cultures was prepared using peqGOLD TriFast (PeqLab) as recommended by the manufacturer. To remove unwanted DNA, the RNA preparation was treated with 10 U of RNase-free DNase I (Ambion) for 1 h at 37°C.
Northern blot analysis.
Total RNA (24 μg from each sample) was separated on a 1.2% agarose gel containing 3.7% formaldehyde (Fluka) and blotted overnight onto a positively charged nylon membrane (Schleicher & Schuell) in 10× SSC solution (1× SSC is 0.15 M NaCl plus 0.012 M sodium citrate adjusted to pH 7). The blot was dried for 2 h at 80°C. For prehybridization, the membrane was first incubated for 10 min at 68°C in 0.2× SSC containing 0.5% sodium dodecyl sulfate (SDS), then transferred to a hybridization bottle, and incubated for 3 h at 42°C in 10 ml of hybridization solution [25 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 6.8), 0.75 M NaCl, 25 mM EDTA, 1× Denhardt's solution, 50% formamide, 0.2% SDS, 0.2 mg of denatured, fragmented salmon sperm DNA per ml] under continuous rotation. After addition of the radiolabeled riboprobe (see below), hybridization was done overnight at 42°C. The blot was washed for 5 min at room temperature in 2× SSC plus 0.5% SDS and then washed twice for 30 min each time at 68°C in 2× SSC plus 0.5% SDS. The membrane was exposed for 3 days at −80°C to a Biomax MR film (Kodak).
Riboprobe synthesis.
To prepare the negative-sense riboprobe, 2 μg of pT7-gmgA was linearized with the restriction enzyme NotI, purified by agarose gel electrophoresis, and in vitro transcribed at 37°C for 90 min using 20 U of T7 RNA polymerase (MBI Fermentas) and 40 μCi of α-32P-labeled UTP (Amersham). To remove template DNA, RNase-free DNase I (Ambion) (2 U) was added, and incubation at 37°C was continued for another 60 min. The RNA was purified by phenol-chloroform treatment and precipitated with 0.4 M ammonium acetate in 75% ethanol solution supplemented with 10 μg of tRNA per ml. To improve the hybridization efficiency, we reduced the size of the riboprobe by alkali treatment. The RNA pellet was dissolved in 6.3 μl of diethyl pyrocarbonate (DEPC)-treated distilled water, 45 μl of ETS buffer (10 mM Tris [pH 7.4], 10 mM EDTA, 0.2% SDS), 1.7 μl of 5 M NaCl, and 1 μl of 1 M dithiothreitol. The mixture was incubated for 5 min on ice. For hydrolysis, 10 μl of 2 M NaOH was added to the probe, and the mixture was incubated on ice for 45 min. Hydrolysis was stopped by the addition of 20 μl of 2 M HEPES buffer. The riboprobe was precipitated by the addition of 200 μl of precipitation solution (90% ethanol, 0.4 M ammonium acetate, 30 μg of tRNA per ml), incubation for 1 h at −80°C, and centrifugation at 4°C for 30 min at 12,000 × g (Heraeus Biofuge 15). The pellet was washed in 80% ethanol, dried in a speed-vac centrifuge, dissolved in 200 μl of DEPC-treated distilled water, and added to the hybridization solution.
Fluorescence microscopy.
One day prior to analysis, transfected cells were trypsinized and seeded onto glass coverslips. The next day, the cells were fixed for 10 min in 3% paraformaldehyde, washed three times with 1 ml of phosphate-buffered saline (PBS), and then permeabilized by incubation for 5 min in PBS containing 0.5% Triton X-100. Transgene expression was detected using a 600-fold dilution of mouse Flag monoclonal antibody (MAb) M2 (Sigma). BDV-expressed N and P proteins were detected using a 1,000-fold dilution of antisera obtained from rabbits immunized with either recombinant N or P purified from Escherichia coli lysates. All antibody dilutions were prepared in PBS supplemented with 5% goat preimmune serum. Permeabilized cells were washed three times with 1 ml of PBS and incubated with 200 μl of diluted primary antibodies. Cells were washed three times for 5 min each time, and bound antibodies were detected using 200-μl samples of 200-fold dilutions of cytochrome 2-labeled goat anti-mouse and cytochrome 3-labeled goat anti-rabbit secondary antibodies (Dianova). After 30-min incubation, the cells were washed three times for 5 min each time, and fluorescence was analyzed using a Leica confocal microscope or a Reichert Polyvar 2 fluorescence microscope.
Quantification of CAT and luciferase gene expression.
BSR-T7 cells in 25-mm-diameter dishes were transfected with the indicated plasmids as described above. Three days after transfection, the cells were lysed in 250 μl of CAT lysis buffer, and CAT protein levels were determined by a CAT enzyme-linked immunosorbent assay (ELISA) (Roche), basically as described in the manufacturer's protocol. Briefly, upon addition of the lysis buffer, cells were incubated for 15 min on a shaking table. The lysate was then transferred to a 1.5-ml reaction tube and centrifuged at 4°C for 15 min at 12,000 × g (Heraeus Biofuge 15). Clarified supernatant (200 μl) was added to individual wells of 96-well plates coated with a CAT-specific MAb and incubated for 1 h at 37°C. The wells were washed extensively, and 200 μl of a 100-fold dilution of a second CAT-specific MAb labeled with digoxigenin (DIG) was added to each well, and the plate was incubated at 37°C for 1 h. The wells were washed extensively, and incubation at 37°C was continued with 200 μl of a 100-fold dilution of a peroxidase-coupled DIG-specific Fab fragment for 1 h. Finally, the wells were washed, and 200 μl of peroxidase substrate (without substrate enhancer) was added, resulting in a peroxidase-mediated color reaction. The colored product present after 20 min of incubation at room temperature was quantified on a Multiskan MCC/340 ELISA reader (Labsystems) at a wavelength of 405 nm. Background absorption was determined by measuring lysates from mock-transfected cells.
Luciferase expression was quantified by mixing 15-μl samples of the clarified CAT ELISA supernatant with 75 μl of luciferase substrate (Promega). Luciferase-mediated light emission was determined in a Lumat LB9501 luminometer (Berthold). We discarded the results of exceptional experiments in which the relative light unit (RLU) values of the individual samples differed by more than 50% from the average of all RLU values from one experiment. The ELISA absorption values were normalized for transfection efficiency by setting the average RLU value to 100% and by adjusting the individual absorption values in relation to their RLU values.
RESULTS
Altered cellular distribution of Flag-tagged L protein in BDV-infected cells.
To determine whether cellular RNA polymerase II can efficiently express a functional BDV polymerase from cDNA, we cloned the L ORF of strain He/80FR (19) into expression plasmid pCA controlled by the chicken β-actin promoter. The L ORF was introduced either alone (pCA-L) or fused to an N-terminal Flag tag (pCA-flagL), thereby creating a linker sequence composed of three alanine residues located between the Flag tag and the L sequence. To verify expression of Flag-tagged recombinant L, cells were transfected with pCA-flagL, and transgene expression was analyzed 48 h later using mouse monoclonal Flag antibody M2. Western blot analysis revealed the presence of a Flag-tagged protein of approximately 190 kDa (data not shown), presumably representing the BDV L polymerase.
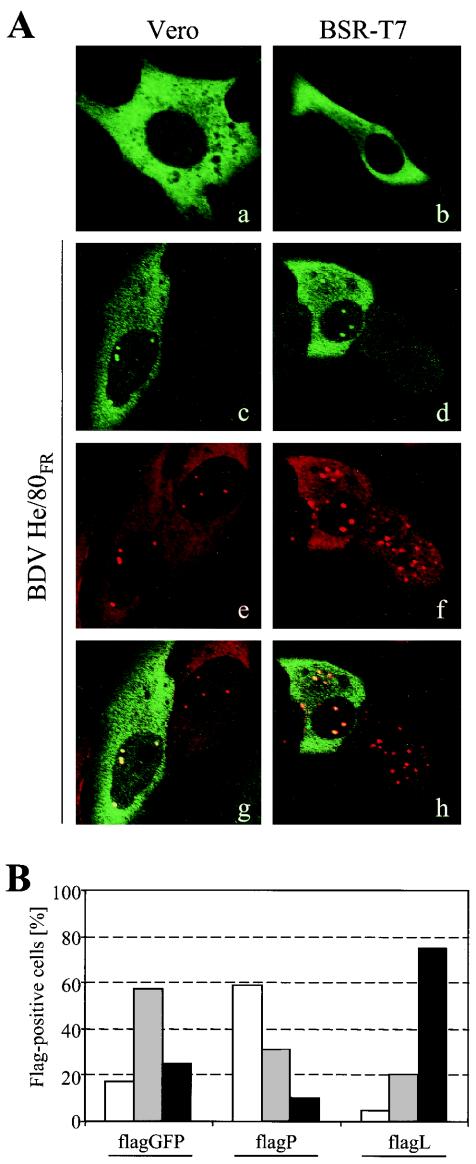
To investigate the cellular distribution of flagL, uninfected and BDV He/80FR-infected Vero and BSR-T7 cells were transfected with pCA-flagL (Fig. 1A). In uninfected cells (Fig. 1A, panels a and b), flagL showed a homogenous, almost exclusively cytoplasmic localization in both cell lines. In BDV-infected Vero and BSR-T7 cells, flagL was also mainly localized to the cytoplasm. In addition, it was efficiently incorporated into nuclear dots, which are thought to represent the sites of BDV replication and transcription (Fig. 1A, panels c and d). BDV P was also present in these nuclear dots (Fig. 1A, panels e and f). Picture overlays demonstrated that the nuclear signals of flagL and BDV P colocalized almost perfectly (Fig. 1A, panels g and h). Colocalization was also seen when an antiserum to BDV N was used instead of antiserum to BDV P (data not shown).
FIG. 1.
BDV L with an N-terminal Flag tag colocalizes with viral P protein and increases N expression levels in BDV-infected cells. (A) Uninfected (a and b) and BDV-infected (c to h) Vero and BSR-T7 cells were transfected with pCA-flagL and analyzed for the cellular localization of Flag-tagged BDV L by confocal immunofluorescence microscopy. Cells were stained with a mouse MAb to the Flag epitope and rabbit antiserum to BDV P. Bound antibodies were visualized by fluorescence-labeled goat anti-mouse (green) and goat anti-rabbit(red) antibodies. Panels g and h show the overlay of panels c and e and panels d and f, respectively. (B) BDV-infected Vero cells were transfected with either pCA-flagGFP, pCA-flagP, or pCA-flagL. Four days later, the cells were simultaneously stained with a mouse MAb to the Flag epitope and rabbit antiserum to BDV N and analyzed by -immunofluorescence microscopy with two immunofluorescent stains. The BDV N expression levels of approximately 200 individual Flag-positive cells from each transfection were evaluated. These cells were assigned into three categories, depending on whether they expressed low (white bars), medium (gray bars,) and high (black bars) levels of N protein.
The Flag-tagged L protein enhances viral antigen expression in BDV-infected cells.
We previously showed that transient overexpression of Flag-tagged BDV P in persistently infected Vero cells reduced the levels of virus-encoded antigen (12), suggesting an interference of the transgene product with the active viral polymerase complex. To test whether transient expression of flagL also affects BDV multiplication, persistently infected Vero cells were transfected with expression plasmids pCA-flagL and pCA-flagP or with the control construct pCA-flagGFP. Four days after transfection, the cells were stained twice for the flagged transgene product and for virus-encoded N antigen. The influence on the persisting virus was evaluated by determining the percentages of Flag-positive cells that expressed either low, medium, or high levels of BDV N (Fig. 1B). As described previously, expression of flagP reduced BDV N levels strongly. In this experiment, the number of flagP-positive cells expressing medium or high levels of BDV N dropped more than twofold from the number in the flagGFP control, while the number of cells expressing low levels of BDV N increased about fourfold (Fig. 1B). Interestingly, expression of flagL led to an increase rather than a decrease of BDV N levels in BDV-infected cells (Fig. 1B). Seventy-five percent of all flagL-positive cells showed high levels of BDV N, which were threefold more than the level in control transfections. This result indicated that in contrast to flagP, flagL promoted viral protein synthesis under these conditions, presumably by increasing the number of active BDV polymerase complexes.
L protein expressed from pCA-L supports in vivo reconstitution of the BDV polymerase complex.
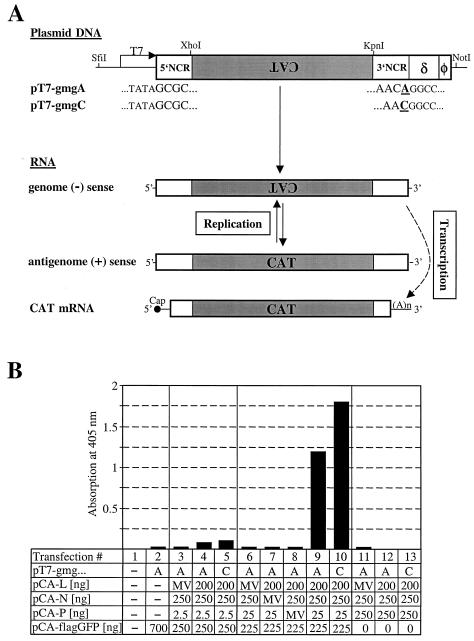
To analyze the functionality of the L-encoding cDNA clone in the absence of persistent BDV infection, we constructed plasmids from which BDV minigenomes of genomic orientation can be synthesized under the control of the T7 RNA polymerase promoter (Fig. 2A). These minigenomes encoded the CAT gene in the antisense orientation flanked by 78 nt of the 5′-NCR and 53 or 54 nt of the 3′-NCR of BDV strain He/80FR. These sequences are believed to harbor the antigenomic and genomic viral promoters, respectively (4). Since the existing data on the precise nature of the 3′ end of the BDV genome conflict, we constructed two versions of minigenomes, represented by plasmids pT7-gmgA and pT7-gmgC (Fig. 2A) which differed only in the number of terminal nucleotides in the 3′-NCR. Downstream of the minigenome cDNA, we inserted the autocatalytic hepatitis delta virus ribozyme sequence (δ) and the T7 terminator sequence (φ) which should create correct 3′ ends of the transcribed RNAs (Fig. 2A). pT7-gmgA and pT7-gmgC were cotransfected together with pCA-L, pCA-N, and pCA-P into BSR-T7 cells that stably express the bacteriophage T7 RNA polymerase (5). Upon successful in vivo reconstitution of the BDV polymerase complex, the genome negative-sense RNA should be copied into antigenome positive-sense RNA (Fig. 2A). It should further be transcribed into CAT mRNA.
FIG. 2.
In vivo reconstitution of the BDV polymerase complex. (A) Schematic drawing showing the T7 RNA polymerase-driven expression cassettes used to generate BDV minigenomes and the expected transcription and replication intermediates. The unique restriction sites used for cloning of the minigenome expression constructs are indicated. Transcription of two vectors, pT7-gmgA and pT7-gmgC, by T7 RNA polymerase in BSR-T7 cells should yield RNAs in the genome negative (−)-sense orientation that differ only in the last nucleotide at the 3′ end (shown in bold type and underlined) of the fully processed molecule. Transcription is terminated by a T7 terminator sequence (φ), and the correct 3′ terminus of the minigenome RNA is produced by self-cleavage of the hepatitis delta virus ribozyme (δ) sequence. Both minigenome constructs contain the CAT gene (gray box) flanked by BDV 5′- and 3′-NCR of 78 and 54 or 53 nt, respectively. In the presence of the BDV proteins L, N, and P, the minireplicon should be recognized and copied by the reconstituted BDV polymerase complex, resulting in complementary minigenome RNA in the antigenome positive (+)-sense orientation, which in turn can serve as the template for the synthesis of more negative-sense RNA. Transcription of the minigenome by the reconstituted BDV polymerase complex should further yield capped and polyadenylated CAT mRNA. (B) Analysis of reporter gene expression by CAT ELISA. BSR-T7 cells in 12-well plates were transfected with 400 ng of either pT7-gmgA or pT7-gmgC together with the various expression plasmids indicated below the graph. Constant amounts of pCA-L and pCA-N and variable amounts of pCA-P were used. To control for specificity, plasmids encoding the various BDV-derived nucleocapsid components were replaced by the same amounts of plasmids encoding the analogous proteins from MV. For an internal control for transfection efficiency, 100 ng of a plasmid encoding the firefly luciferase under the control of a T7 RNA polymerase promoter was cotransfected. The total amount of transfected DNA was kept constant by adding the indicated amounts of pCA-flagGFP. Seventy-two hours after transfection, the cells were lysed and analyzed for CAT protein levels and luciferase activity by ELISA and luciferase-mediated light emission, respectively. CAT values were normalized for transfection efficacy.
On the basis of the observation that transient overexpression of BDV P seemed to block the viral polymerase complex (Fig. 1B) (12), we reasoned that the amount of P protein might be critical for efficient replication and transcription of the minigenome. In an initial experiment, we tested this hypothesis by transfecting 10-fold serial dilutions (250, 25, and 2.5 ng) of pCA-P together with constant amounts of pCA plasmids encoding BDV L (200 ng), BDV N (250 ng), and one of the two versions of the BDV minigenome construct (400 ng). The total amount of transfected plasmid DNA was kept constant by adding pCA-flagGFP. Furthermore, we cotransfected a plasmid encoding the firefly luciferase under control of the T7 promoter (pBS-T7luc [100 ng]) to normalize the CAT signal for transfection efficiency. Three days after transfection, CAT and luciferase gene expression were determined. CAT protein was detected using a highly specific and sensitive CAT ELISA. Several controls were included in the experiment shown in Fig. 2B. Mock-transfected cells (Fig. 2B, lane 1) were used to determine the CAT ELISA background levels. Transfection of pT7-gmgA with pCA-flagGFP alone (Fig. 2B, lane 2) was done to detect unwanted expression of CAT protein from positive-sense RNA that might be synthesized by cryptic T7 promoters on the plasmid. We also replaced single BDV proteins with the analogous measles virus (MV) proteins to verify specificity (Fig. 2B, lanes 3, 6, 7, 8, and 11). In the presence of all BDV nucleocapsid components, specific induction of CAT protein expression was observed; this induction strongly depended on the amount of pCA-P applied. When 25 ng of pCA-P was used for the transfections, the CAT signals increased 54- and 82-fold for constructs pT7-gmgA and T7-gmgC, respectively (Fig. 2B, lanes 9 and 10) over the MV polymerase control. Thus, both versions of the minigenome were active. The use of 10-fold-reduced amounts of plasmid pCA-P resulted in more than 10-fold-reduced CAT synthesis (Fig. 2B, lanes 4 and 5). Interestingly, when the amount of pCA-P was increased 10-fold to 250 ng, CAT protein levels were reduced to background levels (Fig. 2B, lanes 12 and 13).
The N-to-P ratio determines the activity of the BDV polymerase complex.
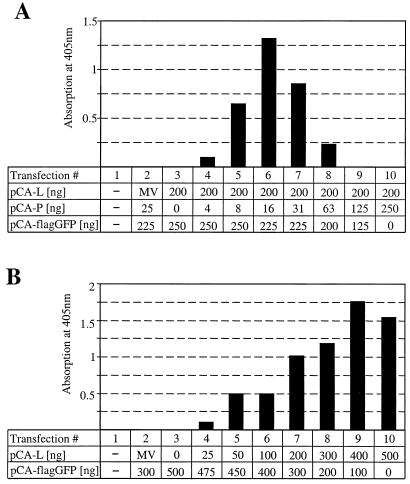
To further analyze the effects of variation of the N-to-P and L-to-P plasmid ratios on the efficiency of CAT expression, we tested serial dilutions of pCA-P (Fig. 3A) and pCA-L (Fig. 3B). We first cotransfected twofold serial dilutions of pCA-P ranging from 250 to 4 ng with constant amounts of pCA-L (200 ng) and pCA-N (250 ng). CAT protein synthesis was most efficient when 8 to 31 ng (Fig. 3A, lanes 5 to 7) of pCA-P were applied, with the signal peaking at 16 ng and decreasing sharply at higher and lower dilutions. The narrow range of efficient CAT expression suggested that a roughly 10- to 30-fold excess of L- and/or N-encoding plasmid over pCA-P is crucial for optimal activity of the minireplicon. To determine the importance of the L plasmid concentration, we next tested a serial dilution of pCA-L ranging from 25 to 500 ng, together with a fixed 10:1 ratio of pCA-N (250 ng) and pCA-P (25 ng). Increasing amounts of L plasmids correlated well with an increase in CAT protein synthesis in an almost linear fashion (Fig. 3B). An activity plateau was reached only with the two highest pCA-L amounts applied. This result suggests that up to 400 ng of pCA-L, suboptimal amounts of L protein were present in our assay. It argues against a direct regulatory effect of the L-to-P ratio.
FIG. 3.
Inhibitory effect of increasing concentrations of BDV P in the minireplicon assay. (A) Serial twofold dilutions of pCA-P were cotransfected with 400 ng of pT7-gmgA, 250 ng of pCA-N, 200 ng of pCA-L, and 100 ng of pBST7-luc. The total amount of transfected DNA was kept constant by adding the indicated amounts of pCA-flagGFP. To control for specificity, plasmid pCA-L was replaced by a plasmid that codes for the L protein of MV. CAT values were normalized for transfection efficacy. (B) Serial twofold dilutions of pCA-L were cotransfected with 400 ng of pT7-gmgA, 250 ng of pCA-N, 25 ng of pCA-P, and 100 ng of pBST7-luc. The total amount of transfected DNA was kept constant by adding the indicated amounts of pCA-flagGFP. To control for specificity, plasmid pCA-L was replaced by 200 ng of a plasmid that codes for the L protein of MV. CAT values were normalized for transfection efficacy.
BDV X protein strongly inhibits viral polymerase activity.
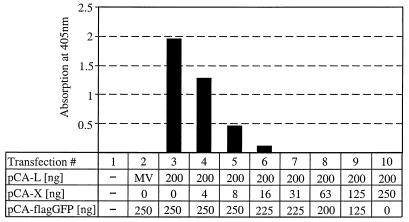
In the above experiments, no plasmid for BDV X was included, clearly demonstrating that the X protein is not required for reconstitution of the active BDV polymerase complex. We therefore looked at whether the X protein has regulatory functions in BDV transcription and/or replication. To test this hypothesis, we prepared a twofold dilution series of pCA-X and cotransfected the individual dilutions together with minigenome plasmid and constant amounts of pCA-N (250 ng), pCA-L (200 ng), and pCA-P (25 ng) into BSR-T7 cells (Fig. 4). The BDV polymerase complex was highly sensitive to the addition of plasmid encoding X. Upon the addition of only 4 ng of pCA-X (Fig. 4, lane 3), the CAT signal was reduced by about 30%, and it reached background levels when more than 16 ng of pCA-X was used.
FIG. 4.
The X protein of BDV exhibits potent inhibitory activity in the minireplicon assay. Serial twofold dilutions of pCA-X were cotransfected with 400 ng of pT7-gmgA, 200 ng of pCA-L, 250 ng of pCA-N, 25 ng of pCA-P, and 100 ng of pBST7-luc. The total amount of transfected DNA was kept constant by adding the indicated amounts of pCA-flagGFP. To control for specificity, plasmid pCA-L was replaced by 200 ng of a plasmid that codes for the L protein of MV. CAT values were normalized for transfection efficacy.
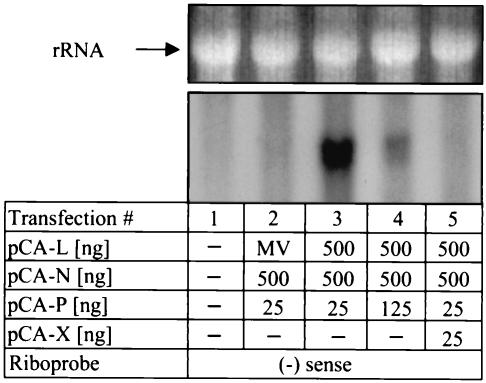
To determine whether the X protein indeed blocked the activity of the BDV polymerase complex, we performed Northern blot analysis with RNA from cells that were transfected with the various minireplicon components in the presence or absence of the X protein. Under experimental conditions that yielded high CAT protein levels (e.g., when the N-to-P ratio was 20:1), we obtained a strong hybridization signal with the minus-sense RNA probe (Fig. 5, lane 3). The observed double band presumably represents the combined signal resulting from the presence of minigenome cRNA and CAT mRNA, which have similar sizes. The CAT minireplicon cRNA is expected to consist of 804 nt, whereas CAT mRNA contains 695 template-encoded nucleotides plus a poly(A) tail of unknown length. Under experimental conditions that yielded low CAT protein levels (e.g., when the N-to-P ratio was 4:1), the Northern hybridization signal was much weaker (Fig. 5, lane 4), as would be expected if the CAT values were proportional to BDV polymerase activity. When the reconstitution of the BDV polymerase complex was done in cells that expressed X protein, no plus-strand minigenome RNA could be detected (Fig. 5, lane 5), suggesting that the X protein inhibits both viral transcription and replication.
FIG. 5.
Analysis of BDV minireplicon-derived RNA by Northern blotting. BSR-T7 cells in six-well plates were either mock transfected (lane 1) or transfected with 1.5 μg of pT7-gmgA and the indicated amounts of pCA expression vectors (lanes 2 to 5). To control for specificity, plasmid pCA-L was replaced by 500 ng of a plasmid that codes for the L protein of MV. Ethidium bromide staining for rRNA verified that comparable amounts of RNA were loaded (top panel). Radiolabeled minus-strand minigenome RNA was used as the hybridization probe. (−), negative.
DISCUSSION
We describe a novel BDV minireplicon assay which allows us to study the composition and functional regulation of the BDV polymerase complex. The assay is based on T7 promoter-driven expression of a BDV minigenome combined with expression of the BDV nucleocapsid components L, N, and P from vectors containing the chicken β-actin promoter. Cotransfection of these plasmids into BSR-T7 cells stably expressing the T7 RNA polymerase resulted in efficient CAT synthesis, provided a narrowly defined ratio of N and P plasmids was used. Addition of plasmid encoding BDV X protein had a strong inhibitory effect on CAT gene expression, suggesting a negative regulatory function for this viral protein.
Efficient expression of L was the major obstacle in the establishment of a functional BDV minireplicon assay. We first generated a BDV L cDNA encoding the previously deduced consensus sequence for the L protein (p190) of strain He/80FR (19) and engineered a Flag epitope into its N terminus to facilitate detection of the transgene product. We tried to express the BDV L cDNA using various RNA polymerase II promoters and the T7 RNA polymerase promoter, but easily detectable amounts of Flag-tagged protein were found only when the chicken β-actin promoter was used to control expression of L (unpublished results). Immunofluorescence analysis revealed that flagL predominantly localized to the cytoplasm of transfected Vero and BSR-T7 cells, and almost no nuclear staining was observed. In BDV-infected cells, however, a fraction of the Flag-tagged L protein was transported to the nucleus and incorporated into dot-like structures. Our analysis showed that flagL colocalized with viral P and N in the nucleus (Fig. 1A). Thus, it appears that these dots represent the nuclear sites of active viral genome transcription and replication. The fact that efficient nuclear import of flagL was observed only in BDV-infected cells suggests that L translocation into the nucleus depends on additional viral factors. Since coimmunoprecipitation experiments have demonstrated a direct interaction of L with P in BDV-infected cells but not with other viral proteins (28), it seems likely that the P protein mediates the nuclear import of L. Our L localization data are not in agreement with recently published work. Using T7 RNA polymerase-driven expression of an L cDNA derived from BDV strain V, Walker and coworkers observed a predominantly nuclear localization of Flag-tagged L in noninfected BSR-T7 cells (28). Using this expression system, Walker and Lipkin further defined a nuclear localization signal within the L protein (29). The reason for this discrepancy is unclear. The differences in cellular localization may be due to the different expression systems used or might reflect strain-specific differences in the amino acid sequence of the L protein. We noted, however, that the proposed nuclear localization signal of L is completely conserved between the two strains.
We observed a strong enhancement in viral N-protein expression in persistently infected Vero cells that overexpressed flagL (Fig. 1B), suggesting that L might be a rate-limiting factor which determines the extent of BDV propagation. Similar experiments in which flagP was overexpressed yielded remarkably different results. Excess P protein strongly inhibited viral N expression (Fig. 1B) (12). Although the mechanistic details of this phenomenon are currently not fully understood, it is interesting that the BDV polymerase seemed to follow the same rules in the newly established minireplicon system (Fig. 3).
Expression of our L cDNA clone mediated efficient replication and transcription of negative-sense BDV minigenome RNA in transfected cells, resulting in up to 80-fold induction of the CAT reporter gene signal over background levels. CAT synthesis depended on the presence of the BDV L, N, and P proteins and was highly specific, since omission or replacement of one of the BDV proteins by its functional analogue from MV reduced the CAT signal to background levels. We observed a strong correlation between the ratio of plasmids encoding the various nucleocapsid components and the efficiency of CAT synthesis. Optimal activity was observed only if plasmids encoding L and N were used in amounts approximately 10- to 30-fold over that of plasmid encoding P. Reduced CAT synthesis at lower concentrations of P plasmid can easily be explained by limiting P-protein concentrations. The lack of detectable CAT expression at higher concentrations of P, however, suggests that increasing amounts of P inhibited the BDV polymerase complex due to unfavorable stoichiometry of the various nucleocapsid components. Since increasing the quantity of L plasmid led to a continuous and almost linear increase of the CAT signal until a plateau was reached, we conclude that the L-to-P ratio is far less critical for polymerase activity than the N-to-P ratio. We are aware of the fact that the amount of input plasmid DNA must not necessarily correlate with the amount of protein synthesized. Because of the lack of suitable ELISAs, the resulting protein ratios could not be determined precisely. However, it should be noted that the N, P, and L cDNAs were all cloned into the same vector with the initiation codons of N and P embedded in the same sequence context. Furthermore, when comparing expression of N and P proteins carrying N-terminal Flag tags using the Flag MAb M2, we found that upon transfection of identical amounts of plasmid DNA, similar quantities of products were present (unpublished results). This suggests that the accumulation of untagged proteins might similarly be proportional to the amount of plasmid used for transfection. Alterations of the N-to-P ratio seem to regulate the BDV polymerase. This interpretation is in accordance with recent data which showed that the amount of N exceeds that of P in the acute phase of the BDV infection, while P is present in an approximately eightfold molar excess over N in the persistent phase (31). Since N and P are both located in the cytoplasm and in the nucleus of infected cells, the possibility remains that the N-to-P ratio in the nuclear compartment might oscillate even more strongly. In any case, our results can readily explain our previous observation that expression of single viral nucleocapsid components (12) can strongly interfere with BDV replication.
The X protein was not required for replication of the BDV minireplicon. Rather, addition of minimal amounts of X-encoding plasmid had a strong inhibitory effect on CAT protein synthesis (Fig. 4) and minireplicon-derived RNA products (Fig. 5), indicating that X is a negative regulator of the BDV polymerase complex. Previous studies revealed that the BDV X protein strongly interacts with BDV P and promotes its homo-oligomerization (33). This suggests two possible mechanisms for the inhibitory effect of X. The X protein could either sequester the P protein by direct binding or could induce the formation of P oligomers. It is conceivable that complexes of either type no longer interact with N and/or L. Similarly, increasing the amount of P above a certain threshold could favor its multimerization, thereby reducing the level of monomeric P available for interaction with N and/or L. However, without further information on the precise nature of the interaction of P with N and L, this view remains highly speculative. It also has to be kept in mind that not all BDV proteins are present in our minireplicon assay and that the interactions of viral proteins might be more complex. It will be of interest to determine whether the matrix proteins and glycoproteins of BDV can modulate the activity of the BDV polymerase complex in our minireplicon assay or affect the function of X as a negative regulator. Other negative-strand RNA viruses also code for proteins that act as negative regulatory factors. The M1 protein of influenza A virus and the Z protein of lymphocytic choriomeningitis virus exhibit an inhibitory effect on virus polymerase activity (16, 30). The NS1 and M2-2 proteins of respiratory syncytial virus are potent inhibitors of viral transcription and replication (1, 7). Similarly, the V and C proteins of Sendai virus, the V protein of MV, and the NSs protein of bunyamwera virus were shown to interfere with viral genome replication (6, 26, 32).
The correct structures of the viral genome ends are thought to be critical parameters that determine the functionality of minireplicons. Extensive analysis of the genome ends of BDV strain He/80 in different laboratories yielded no concise result for the 3′ end of the viral genome. In contrast to originally published sequences (4), recent work emphasized the presence of an additional A residue at the extreme 3′ end of the genome of strain BDV He/80 (19). Taking these conflicting results into account, we tested two versions of the BDV minigenome differing only in the 3′-terminal nucleotide. We found that both variants of the minigenome were able to support specific and efficient induction of CAT synthesis (Fig. 2B). The presence of an additional A residue at the 3′ end of the fully processed minigenome RNA led to a slight decrease of the CAT signal under the conditions tested. A possible interpretation of this result is that both versions reflect replication-competent, naturally occurring BDV genome ends. The presence of an A residue at the 3′ end of the BDV genome would imply that the BDV polymerase is able to initiate transcription with an UTP. If true, this would represent a unique feature of BDV because the polymerases of other viruses from the order Mononegavirales all seem to employ ATP or GTP for initiation of transcription. Our results might alternatively be explained by inaccurate cleavage of the minigenome RNA by the hepatitis delta virus ribozyme upstream of the 3′-terminal A residue. Finally, it is possible that internal initiation of replication by the BDV polymerase resulted in the elimination of the additional 3′-terminal A residue. Internal initiation of replication has been described for the Sendai virus polymerase (27). To distinguish between these possibilities, the 5′ end of the positive-sense minigenome RNA synthesized by the BDV polymerase will need to be characterized extensively. In this context, it is noteworthy that the 5′ and 3′ ends of our minigenomes have the potential to form an imperfect panhandle structure with an overhanging 3′ end of 3 and 4 nt, respectively. In the study performed by Pleschka and coworkers, several intermediate sequences and a single clone with a 5′ end that perfectly matched the deduced 3′ end of the BDV genome were detected (19). This sequence would allow the formation of a perfect panhandle structure, as is the case for many other negative-strand RNA viruses. These results support the assumption that BDV might employ microheterogeneity at the genome ends to regulate replication and/or transcription.
The analysis of the BDV polymerase complex using the novel BDV minireplicon assay has already provided important insights into the replication strategy of this intriguing virus. We were able to identify the N-to-P stoichiometry and the X protein as key regulating factors of BDV replication. Studies involving naturally occurring isoforms and mutated forms of BDV proteins will further elucidate the roles of the single nucleocapsid components in replication and transcription of the BDV genome. Finally, this information will provide the basis for the rescue of recombinant BDV from cDNA.
Acknowledgments
We thank Stephan Pleschka for help with the construction of the first BDV L cDNA clone, Christel Hässler for technical assistance, Klaus Conzelmann for providing BSR-T7 cells, and Friedemann Weber, Georg Kochs, Otto Haller, and Jürgen Hausmann for helpful comments on the manuscript.
This work was supported in part by a grant from the Deutsche Forschungsgemeinschaft. U.S. was supported by a fellowship from the Swiss National Science Foundation.
REFERENCES
- 1.Atreya, P. L., M. E. Peeples, and P. L. Collins. 1998. The NS1 protein of human respiratory syncytial virus is a potent inhibitor of minigenome transcription and RNA replication. J. Virol. 72:1452-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billich, C., C. Sauder, R. Frank, S. Herzog, K. Bechter, K. Takahashi, H. Peters, P. Staeheli, and M. Schwemmle. 2002. High-avidity human serum antibodies recognizing linear epitopes of Borna disease virus proteins. Biol. Psychiatry 51:979-987. [DOI] [PubMed] [Google Scholar]
- 3.Briese, T., J. C. de la Torre, A. Lewis, H. Ludwig, and W. I. Lipkin. 1992. Borna disease virus, a negative-strand RNA virus, transcribes in the nucleus of infected cells. Proc. Natl. Acad. Sci. USA 89:11486-11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briese, T., A. Schneemann, A. J. Lewis, Y. S. Park, S. Kim, H. Ludwig, and W. I. Lipkin. 1994. Genomic organization of Borna disease virus. Proc. Natl. Acad. Sci. USA 91:4362-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cadd, T., D. Garcin, C. Tapparel, M. Itoh, M. Homma, L. Roux, J. Curran, and D. Kolakofsky. 1996. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J. Virol. 70:5067-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, P. L., M. G. Hill, J. Cristina, and H. Grosfeld. 1996. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc. Natl. Acad. Sci. USA 93:81-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conzelmann, K. K. 1998. Nonsegmented negative-strand RNA viruses: genetics and manipulation of viral genomes. Annu. Rev. Genet. 32:123-162. [DOI] [PubMed] [Google Scholar]
- 9.Cubitt, B., and J. C. de la Torre. 1994. Borna disease virus (BDV), a nonsegmented RNA virus, replicates in the nuclei of infected cells where infectious BDV ribonucleoproteins are present. J. Virol. 68:1371-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Torre, J. C. 2002. Bornavirus and the brain. J. Infect. Dis. 186(Suppl. 2):S241-S247. [DOI] [PubMed] [Google Scholar]
- 11.de la Torre, J. C. 2002. Molecular biology of Borna disease virus and persistence. Front. Biosci. 7:569-579. [DOI] [PubMed] [Google Scholar]
- 12.Geib, T., C. Sauder, S. Venturelli, C. Hassler, P. Staeheli, and M. Schwemmle. 2003. Selective virus resistance conferred by expression of Borna disease virus nucleocapsid components. J. Virol. 77:4283-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi, T., Y. Shoya, T. Koda, I. Takashima, P. K. Lai, K. Ikuta, M. Kakinuma, and M. Kishi. 1998. Nuclear targeting activity associated with the amino terminal region of the Borna disease virus nucleoprotein. Virology 243:188-197. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi, T., M. Watanabe, W. Kamitani, K. Tomonaga, and K. Ikuta. 2000. Translation initiation of a bicistronic mRNA of Borna disease virus: a 16-kDa phosphoprotein is initiated at an internal start codon. Virology 277:296-305. [DOI] [PubMed] [Google Scholar]
- 16.Lee, K. J., I. S. Novella, M. N. Teng, M. B. Oldstone, and J. C. de La Torre. 2000. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J. Virol. 74:3470-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neumann, G., M. A. Whitt, and Y. Kawaoka. 2002. A decade after the generation of a negative-sense RNA virus from cloned cDNA—what have we learned? J. Gen. Virol. 83:2635-2662. [DOI] [PubMed] [Google Scholar]
- 18.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 19.Pleschka, S., P. Staeheli, J. Kolodziejek, J. A. Richt, N. Nowotny, and M. Schwemmle. 2001. Conservation of coding potential and terminal sequences in four different isolates of Borna disease virus. J. Gen. Virol. 82:2681-2690. [DOI] [PubMed] [Google Scholar]
- 20.Radecke, F., P. Spielhofer, H. Schneider, K. Kaelin, M. Huber, C. Dotsch, G. Christiansen, and M. A. Billeter. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneemann, A., P. A. Schneider, S. Kim, and W. I. Lipkin. 1994. Identification of signal sequences that control transcription of Borna disease virus, a nonsegmented, negative-strand RNA virus. J. Virol. 68:6514-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneemann, A., P. A. Schneider, R. A. Lamb, and W. I. Lipkin. 1995. The remarkable coding strategy of Borna disease virus: a new member of the nonsegmented negative strand RNA viruses. Virology 210:1-8. [DOI] [PubMed] [Google Scholar]
- 23.Schwemmle, M., B. De, L. Shi, A. Banerjee, and W. I. Lipkin. 1997. Borna disease virus P-protein is phosphorylated by protein kinase C epsilon and casein kinase II. J. Biol. Chem. 272:21818-21823. [DOI] [PubMed] [Google Scholar]
- 24.Schwemmle, M., M. Salvatore, L. Shi, J. Richt, C. H. Lee, and W. I. Lipkin. 1998. Interactions of the Borna disease virus P, N, and X proteins and their functional implications. J. Biol. Chem. 273:9007-9012. [DOI] [PubMed] [Google Scholar]
- 25.Staeheli, P., C. Sauder, J. Hausmann, F. Ehrensberger, and M. Schwemmle. 2000. Epidemiology of Borna disease virus. J. Gen. Virol. 81:2123-2135. [DOI] [PubMed] [Google Scholar]
- 26.Tober, C., M. Seufert, H. Schneider, M. A. Billeter, I. C. Johnston, S. Niewiesk, V. ter Meulen, and S. Schneider-Schaulies. 1998. Expression of measles virus V protein is associated with pathogenicity and control of viral RNA synthesis. J. Virol. 72:8124-8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vulliemoz, D., and L. Roux. 2002. Given the opportunity, the Sendai virus RNA-dependent RNA polymerase could as well enter its template internally. J. Virol. 76:7987-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker, M. P., I. Jordan, T. Briese, N. Fischer, and W. I. Lipkin. 2000. Expression and characterization of the Borna disease virus polymerase. J. Virol. 74:4425-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker, M. P., and W. I. Lipkin. 2002. Characterization of the nuclear localization signal of the Borna disease virus polymerase. J. Virol. 76:8460-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe, K., H. Handa, K. Mizumoto, and K. Nagata. 1996. Mechanism for inhibition of influenza virus RNA polymerase activity by matrix protein. J. Virol. 70:241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe, M., Q. Zhong, T. Kobayashi, W. Kamitani, K. Tomonaga, and K. Ikuta. 2000. Molecular ratio between Borna disease viral-p40 and -p24 proteins in infected cells determined by quantitative antigen capture ELISA. Microbiol. Immunol. 44:765-772. [DOI] [PubMed] [Google Scholar]
- 32.Weber, F., E. F. Dunn, A. Bridgen, and R. M. Elliott. 2001. The Bunyamwera virus nonstructural protein NSs inhibits viral RNA synthesis in a minireplicon system. Virology 281:67-74. [DOI] [PubMed] [Google Scholar]
- 33.Wolff, T., R. Pfleger, T. Wehner, J. Reinhardt, and J. A. Richt. 2000. A short leucine-rich sequence in the Borna disease virus p10 protein mediates association with the viral phospho- and nucleoproteins. J. Gen. Virol. 81:939-947. [DOI] [PubMed] [Google Scholar]