Abstract
ETnII elements are mobile members of the repetitive early transposon family of mouse long terminal repeat (LTR) retroelements and have caused a number of mutations by inserting into genes. ETnII sequences lack retroviral genes, but the recent discovery of related MusD retroviral elements with regions similar to gag, pro, and pol suggests that MusD provides the proteins necessary for ETnII transposition in trans. For this study, we analyzed all ETnII elements in the draft sequence of the C57BL/6J genome and classified them into three subtypes (α, β, and γ) based on structural differences. We then used database searches and quantitative real-time PCR to determine the copy number and expression of ETnII and MusD elements in various mouse strains. In 7.5-day-old embryos of a mouse strain in which two mutations due to ETnII-β insertions have been identified (SELH/Bc), we detected a three- to sixfold higher level of ETnII-β and MusD transcripts than in control strains (C57BL/6J and LM/Bc). The increased ETnII transcription level can in part be attributed to a higher number of ETnII-β elements, but 70% of the MusD transcripts appear to have been derived from one or a few MusD elements that are not detectable in C57BL/6J mice. This element belongs to a young MusD subgroup with intact open reading frames and identical LTRs, suggesting that the overexpressed element(s) in SELH/Bc mice might provide the proteins for the retrotransposition of ETnII and MusD elements. We also show that ETnII is expressed up to 30-fold more than MusD, which could explain why only ETnII, but not MusD, elements have been positively identified as new insertions.
Early transposon (ETn) elements are middle repetitive sequences first discovered and characterized in the 1980s (2, 3). They possess long terminal repeats (LTRs) and are presumed to transpose via reverse transcription of an RNA intermediate. ETn elements were originally divided into two groups (ETnI and ETnII) which differ only in the 3′ half of the LTR and in the 5′ end of the internal region (17, 36, 39). The rest of the internal region contains mainly nonretroviral, noncoding sequences of unknown origin, except for a short stretch of a retroviral pol gene (24). Since their initial characterization, 15 new germ line mutations and 4 somatic mutations in mouse cell lines have been reported to be caused by insertions of ETn elements into genes (for a list, see reference 1). In most of these cases the loss of gene function was due to aberrant transcripts as a result of ETn-induced alternative splicing and/or premature termination. Four of the 19 mutations were found in A/J mice and two were found in SELH/Bc mice, suggesting that ETn retrotransposition occurs relatively frequently in these strains. In all cases with sufficient sequence data to distinguish between the two ETn groups, ETnII, but never ETnI, elements were found at the insertion site. Recently we identified a third ETn-related subfamily, MusD, with gag, pro, and pol genes homologous to those in type D viruses or betaretroviruses (24), suggesting that MusD elements may provide the proteins necessary for ETnII retrotransposition in trans. The LTRs of MusD and ETnII are almost identical, and both elements share the 5′ internal region upstream of the gag gene, including an intact primer binding site for tRNALys and a possible retroviral packaging signal. Like ETn elements, MusD is present in multiple copies in the genome (2, 24). The evolutionarily younger ETnII elements are probably descended from MusD elements by recombinatory replacement of the retroviral genes with sequences of unknown origin (24). MusD elements, too, might still be retrotranspositionally active. Only seven of the characterized ETnII elements found as new mouse mutations could be unequivocally distinguished from MusD. In eight other cases, the sequence data are insufficient to determine if the inserted element is an ETnII or MusD element. Indeed, MusD as well as ETnII elements display insertional site polymorphisms in different mouse strains, indicating that both types of elements still possess mobile capacity (1).
Transposition of these elements should be tightly controlled at the transcriptional level since they presumably transpose in a replicative manner via an RNA intermediate. However, little is known about the expression levels or copy numbers of MusD and the different ETn groups. ETn transcripts were found in plasmacytoma and embryonic carcinoma cells, as well as in normal cells of the early embryo, where the expression peaks between day 3.5 and 7.5 (3, 35). Tanaka and Ishihara (40) described high levels of RNA in acute myeloid leukemia cells from C3H/He inbred mice but only a small amount in hepatomas, lymphomas, and normal cells (liver, thymus, and spleen cells). Brûlet et al. (2) also reported low expression in the livers of 129 mice. All of these data were primarily obtained by Northern blot analyses and in situ hybridization using molecular probes that did not differentiate between the three ETn groups, and therefore the relative expression patterns of these groups are unknown. It is also unclear why ETnII elements currently seem to be the most active and why some mouse strains, such as A/J and SELH/Bc, appear more prone to ETnII insertions than other strains. To address these questions, we have performed a detailed analysis of the copy number, structure, and embryonic expression of ETn and MusD elements in various mouse strains. Our results identify an active subtype of ETnII elements and a young MusD subgroup with intact retroviral genes and identical LTRs. These findings provide insight into the relationships between ETn and MusD elements and their activities in the mouse.
MATERIALS AND METHODS
Mice and cells.
The inbred strain C57BL/6J (B6), obtained from Jackson Laboratory, and Crl:CD-1 (ICR)BR outbred mice (CD-1), obtained from Charles River Laboratories, were kept under standard conditions in the animal facility of the Terry Fox Laboratory (B. C. Cancer Agency, Vancouver, British Columbia, Canada). Inbred SELH/Bc (SELH) and LM/Bc (LM) mice were maintained in the animal unit of the Department of Medical Genetics at the University of British Columbia under standard conditions described elsewhere (14), including Purina Laboratory rodent diet no. 5001 administered ad libitum. SELH/Bc mice show a high incidence of the neural tube closure defect exencephaly, a large number of spontaneous heritable mutations (9, 14), and an unusually high incidence of ovarian teratomas and thymic tumors (unpublished data). LM/Bc is a distinct strain, probably related to the C3H strain, with normal neural tube closure, that is used as a control in studies with SELH mice (15, 16). The embryonic stem cell line R1, derived from 129 mice, was kindly provided by Pamela Hoodless.
Preparation of RNA, DNase treatment, and reverse transcription.
Embryos (7.5 and 9.5 days postconception) of naturally mated inbred female mice (C57BL/6J, SELH/Bc, and LM/Bc) or superovulated CD-1 female mice (for superovulation protocol, see reference 10) were dissected free from their extra-embryonic membranes in ice-cold 1× phosphate-buffered saline. Until final dissection, deciduae and embryos were kept in M2 medium (Sigma-Aldrich Canada Ltd.) on ice. The entire litter of a pregnant mouse (approximately 8 to 12 embryos for naturally mated females and 20 to 25 embryos for CD-1 females) was combined and immediately lysed in 200 μl of TRIzol (Invitrogen Life Technologies, Buro Ltd., Montevideo, Uruguay), and two deciduae from each mouse were lysed in 500 μl of TRIzol. Tissue samples and embryonic stem (ES) cells were kept at −70°C or in RNAlater (Ambion RNA Diagnostics, Austin, Tex.) and were then homogenized with 1 ml of TRIzol. Extraction of total RNA was carried out according to the TRIzol protocol. To facilitate precipitation of embryonic RNA, 5 μg of glycogen was added as a carrier. RNA was dissolved in 50 μl (embryos) or 200 to 500 μl (deciduae and tissues) of H2O. Ten microliters of embryonic RNA (20% of the total amount) or 5 μg of decidua and tissue RNA was treated for 30 min at 37°C with DNase I (Invitrogen) in a volume of 50 μl according to the Invitrogen protocol. To test for the complete removal of any contaminating DNA, a PCR using 2 μl of DNase-digested RNA, 2.5 mM MgCl2, 200 μM deoxynucleoside triphosphates (dNTPs), 200 nM (each) primer complementary to intracisternal A-particle (IAP) sequences (Table 1), and 0.1 U of Taq polymerase (Invitrogen) per μl was carried out in 1× polymerase buffer. Reaction conditions were 95°C for 5 min followed by 35 cycles of the amplification step (95°C for 45 s, 60°C for 45 s, and 72°C for 1 min). PCRs were loaded onto a 2% agarose gel and visually checked for the presence of IAP products. Forty-five microliters of DNase-digested RNAs with a negative IAP PCR were reverse transcribed with random hexamer primers for 1 h at 42°C using Superscript II (Invitrogen) in a final volume of 70 μl according to the instructions of the manufacturer. Reverse transcription (RT) reactions were subsequently diluted fivefold in H2O and stored at −20°C.
TABLE 1.
Oligonucleotides used for real-time PCR
| Elements or target gene | Primer (sequence) | MgCI2 (mM) | Primer concentration (each) (nM) | Amplicon length (bp) |
|---|---|---|---|---|
| ETnII-α | ETnI/II_3400s2 [CCAGC(C/T)(A/C)TTCTAACTCAATC], MusD_4094as (GCAGGGAGTAATCTATGTAAG) | 2 | 0.15 | 204 |
| ETnII-β | ETnI/II_3400s2 [CCAGC(C/T)(A/C)TTCTAACTCAATC], ETnII_3636as [CATT(T/C)(G/A)TTAGT(C/T)AGGGGGTATTAAGTGAC] | 2 | 0.2 | 130 |
| ETnII-γ | MusD_677s (GAGTTGTTTCAGGCCAGAGGAGTAAGG) ETnII_1125as (TACCATTGTCAAACACATTAATCATGAACC) | 2.5 | 0.2 | 168 |
| MusD (absolute quantification) | MusD_922s [GTGGTATCTCAGGA(G/A)GAGTGCO], MusD_1058as (GGGCAGCTCCTCTATCTGAGTG) | 1 | 0.15 | 136 |
| MusD (relative quantification) | MusD_1731s (GATTGGTGGAAGTTTAGCTAGCAT), MusD_1878as (TAGCATTCTCATAAGCCAATTGCAT) | 2 | 0.2 | 148 |
| ETnI | ETnI_457s (GTTAAACCCGAGCGCTGGTTC), ETnI/II_665as (GCTATAAGGCCCAGAGAGAAATTTC) | 1.5 | 0.2 | 203 |
| IAP | IAP_4185s (AAGCAGCAATCACCCACTTTGG), IAP_4277as [CAATCATTAGATG(T/C)GGCTGCCAAG] | 1 | 0.2 | 93 |
| GAPDH (RT-PCR) | GAPDH_139s (AACGACCCCTTCATTGAC), GAPDH_331as (CTCCACGACATACTCAGCAC) | 2.5 | 0.2 | 193 |
| Pthlh | PTHrP_ex3_s (GAACACCCGCGTTTGAAGAG), PTHrP_ex3_as (GCTGTGGCTCCCATAGCAA) | 2.5 | 0.2 | 90 |
Cloning and sequencing of ETnII and MusD transcripts.
To clone expressed ETnII and MusD sequences, we used 7.5-day embryonic RNA from two different litters, reverse transcribed each of them in two separate reactions as described above, and then amplified a 1.05-kb ETnII-β fragment (nucleotides [nt] 2550 to 3600) with the forward primer 5′-CATTATTGTAAAGTCATTTTTCTCATCC-3′ and the reverse primer 5′-GTGACCAATTGCTCTTTAATTAAC-3′ or a 1.2-kb MusD fragment (nt 680 to 1880) with the forward primer 5′-GAGTTGTTTCAGGCCAGAGGAGTAAGG-3′ and the reverse primer 5′-TAGCATTCTCATAAGCCAATTGCAT-3′. PCR was carried out with a 5-μl RT reaction, 2.5 mM MgSO4, 200 μM dNTPs, 200 nM (each) primers, and 0.1 U of HiFi Platinum Taq polymerase (Invitrogen) per μl in 1× high-fidelity polymerase buffer. Three PCRs were performed in parallel with different annealing temperatures (58, 60, and 62°C). Reaction conditions were 95°C for 5 min followed by 28 cycles of the amplification step (95°C for 45 s, 58 to 62°C for 45 s, and 72°C for 1 min). A mixture of all PCR products was cloned into the pGEM-T vector (Promega, Madison, Wis.) according to the manufacturer's protocol, and 10 to 12 clones were sequenced per fragment and strain.
Database mining and phylogenetic analysis.
Subfragments of ETnI (nt 240 to 400 of M16478), ETnII (nt 700 to 3500 and 5300 to 6700 of AC074208), and MusD (nt 1 to 320 and 800 to 3800 of BK001485) were used as query sequences for searching the February 2002 release of the C57BL/6J sequence supplied by the Mouse Genome Sequencing Consortium (MGSC version 3), which is based on the ARACHNE assembly of a whole-genome shotgun approach with 7.7-fold sequence coverage but also has incorporated finished clone information. The databases were searched using the Ensemble SSAHA search server (http://www.ensembl.org/Mus_musculus/). The five resulting “hit lists” were transferred to the MS Office program Excel, color coded, combined, and then sorted according to chromosomal location. Full-length elements were easily recognizable by a distinct color pattern. Partial elements and solitary LTRs were identified by aligning the sequence surrounding the LTR with ETn or MusD elements. Hits on sequence reads with unknown chromosomal locations were not analyzed further. They were mostly short contigs that contained only partial ETn or MusD sequences. We also searched the nonredundant high-throughput database (htgs) on the National Center for Biotechnology Information mouse server using BlastN (http://www.ncbi.nlm.nih.gov/genome/seq/MmBlast.html) and found two additional ETnII elements: one of them was partially represented by a short MGSC clone and the other one was not contained in the MGSC assembly.
For phylogenetic analysis, MusD sequences were aligned using the CLUSTALW algorithm (41). After sequence alignment optimization and sequence divergence calculations, programs NEIGHBOR and DRAWGRAM of PHYLIP (Phylogeny Interference Package, version 3.5c; J. Felsenstein, Department of Genetics, University of Washington, Seattle) were used to calculate the branch length and to draw the neighbor-joining dendrogram, respectively.
Quantitative real-time PCR.
Amplification was carried out with the SYBRGreen amplification kit from Perkin-Elmer (Wellesley, Mass.) in a total volume of 25 μl. Every reaction was performed in triplicate in 96-well plates using the ICycler (Bio-Rad, Hercules, Calif.). The template (5 ng of DNA or 0.5 μl of an RT reaction in a volume of 5 μl) was mixed with 1× SYBRGreen buffer, 1 to 2.5 mM MgCl2, 200 μM dNTPs (containing dUTP), 0.016 U of AmpliTaq Gold polymerase per μl, and 150 to 200 nM (each) primer. Primer sequences are given in Table 1. Reaction conditions were 95°C for 10 min followed by 40 cycles of the amplification step (95°C for 30 s, 62°C for 45 s, and 72°C for 30 s). The well factors were determined by preparing an external well factor plate with 1× well factor solution.
For standardization of DNA PCRs, amplification was performed in parallel (again in triplicates) on a 90-bp fragment of the single-copy mouse gene (pthlh) for parathyroid hormone-related protein (NM_008970) (7, 26). The relative copy numbers of the retroviral gene and of the pthlh gene were calculated by use of separate standard curves (serial dilution [0.1 to 10 ng] of SELH or LM DNA on the same plate), and standardization was carried out by division of the relative number of retroviral genes by the relative number of pthlh amplicons. To standardize RT-PCRs, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA was amplified in parallel. The expression levels of retroviral and GAPDH transcripts were estimated by use of standard curves (serial dilution of a mixture of RT reactions), and the retroviral transcript level was then normalized by division by the expression level of GAPDH.
To compare MusD expression with the expression of ETnII-α and ETnII-β, plasmids containing the retroviral amplicons were constructed and serially diluted in order to obtain standard curves. The use of quantifiable standard plasmids was made necessary by unequal primer efficiencies, precluding a direct comparison of the relative quantification results. The obtained absolute number of retroviral transcripts was then normalized by dividing by the expression level of GAPDH, which itself was obtained by using a standard curve derived from serial dilutions of a mixture of RT reactions. To construct the standard plasmids, SELH DNA was amplified with the primers MusD_823s (5′-CACTCAGGCAGATAATTTTACC-3′) and MusD_1192as (5′-TCATTAGTCAGCCTCTGAAGCTT-3′) for MusD and the sense primer ETnI/II_3375s (5′-TGATTTTTCTC(T/C)TTTCTTCAGTGTGACCAG-3′) together with the antisense primer MusD_4094as (5′-GCAGGGAGTAATCTATGTAAG-3′) or ETnII_3636as (5′-CATT(T/C)(G/A)TTAGT(C/T)AGGGGGTATTAAGTGAC-3′) for ETnII-α and ETnII-β, respectively. The amplicons (MusD, 135 bp; ETnII-α, 237 bp; ETnII-β, 156 bp) were then cloned into BS/SK+ (Promega), and the constructs were verified by sequencing and then photometrically quantified.
Nucleotide sequence accession numbers.
The sequences for the six young MusD elements were deposited in GenBank under accession numbers BK001485 to BK001490.
RESULTS AND DISCUSSION
ETn copy number in C57BL/6J mice.
To evaluate the populations of ETnI, ETnII, and MusD elements in mice, we searched the C57BL/6J (B6) draft sequence as described in Materials and Methods. In total, we identified 201 full-length or partial ETnI elements, 39 ETnII elements, and 93 MusD elements (Table 2) randomly distributed over all chromosomes. In 17 cases, insufficient sequence data were available to distinguish between ETnII and MusD and in two cases a distinction between ETnI and ETnII was not possible. At least two of the MusD elements (BK001485 and BK001486, located on chromosomes 2 and 16, respectively) are fully coding competent: they contain intact open reading frames for gag, pro, and pol. However, no env-related sequences were detectable within any of the 93 MusD elements. Interestingly, a MusD element located at position 157645000 of chromosome 1 contains a 1.4-kb insertion upstream of its 3′ LTR that has the structure of a classical processed pseudogene, with 99% identity to the cDNA sequence of the predicted casein kinase II-α gene (Csnk2a1-rs4) located on chromosome 2. Finally, we found about 40 MusD-related sequences whose LTRs could not be identified by aligning the 3′ and 5′ ends of the element or showed no homology to typical MusD LTRs. It is likely that these sequences were created by recombinatory events, resulting in MusD-related elements with variant LTRs. Indeed, in a separate study, we have identified several new families of mouse retroviral elements belonging to the same betaretroviral superfamily as MusD (G. J. Baillie et al., unpublished data).
TABLE 2.
Number of ETn elements and solitary LTRs in C57BL/6J mice
| Element or gene | No. of full-length or partial elements (total no./with insertion)a | No. of solitary LTRs (total no./with insertion) |
|---|---|---|
| ETnI | 201/30 | 500/250 |
| ETnII | 39/2 | |
| MusD | 93/3 | 156/30 |
Full-length, contains 5′ and 3′ LTR and internal sequences; partial, contains at least one LTR and some internal sequence; insertion, 13 or 14 bp at position 91 of the LTR.
Our results are more accurate but are in accord with Southern blot analyses in which the number of MusD and ETnII elements was roughly estimated to be near 100 (24). However, the list of elements in B6 is still not complete since the database searched covers only 96% of the euchromatic DNA. In some cases, elements could not be positively identified due to missing sequence data, and several hits in unsorted, fragmentary clones were not further analyzed. Also, and even more importantly, in some cases the assembly of the mouse draft sequences might be erroneous due to the repetitive nature of the elements, which makes an accurate assembly difficult by whole-genome shotgun methods.
Overview of LTR sequences.
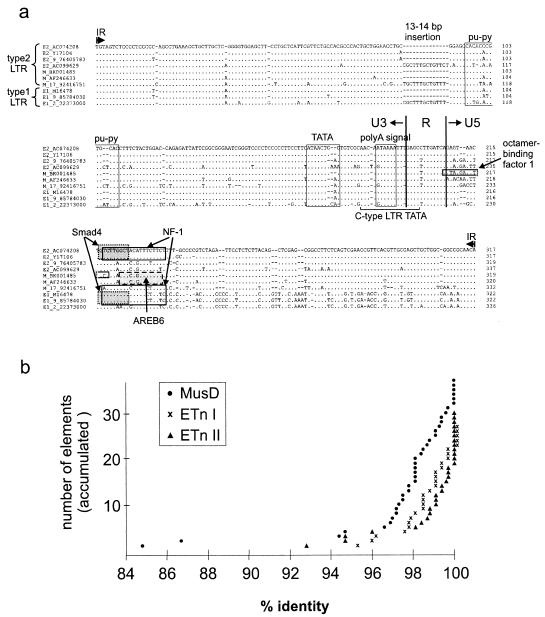
LTRs of most ETnII and MusD elements are highly similar (Fig. 1a). They display 92 to 100% identity within each group, but also between groups. There are indeed some ETnII elements that can only be distinguished from MusD by comparing internal sequences. We collectively named ETnII and MusD LTRs “type 2 LTRs,” in contrast to the “type 1 LTRs” of ETnI elements. ETnI LTRs are only about 80 to 85% identical to the type 2 LTRs due to sequence differences in the last 100 bp of the LTR, but they are more than 92% identical to each other. One sequence feature found in some LTRs is a 13- to 14-bp length difference in the U3 region at position 91 of the LTR. This 13- to 14-bp sequence is found in all LTR groups but is more frequent in ETnI (Table 2). Leong et al. (21) first described this length difference in an ETnI element, a type 1 and a type 2 solitary LTR (sLTR), and they hypothesized that the LTRs may have been created by homologous recombination between two ETn elements of different families. Due to the higher number of ETnI LTRs containing the 13- to 14-bp segment, we suggest that this variation originally occurred in an ETnI LTR and then spread to type 2 LTRs through recombination. Brûlet et al. (2) noted that the LTRs contain a series of alternating purines and pyrimidines (pu-py) with the potential to switch to the Z form. Z-DNA structures have been associated with gene rearrangements (25, 42). Since the purine-pyrimidine stretch is located very close to the insertion site at positions 95 to 103 of the LTR, it is possible that it might have facilitated recombination between LTRs.
FIG. 1.
Similarity of ETn and MusD LTRs. (a) Alignment of LTR sequences. 5′-LTR sequences of representative elements of ETnII (E2_AC074208, E2_Y17106, E2_9_76405783, and E2_AC099629), MusD (M_BK001485, M_AF246633, and M_17_92416751), and ETnI (E1_M16478, E1_9_85784030, and E1_2_22373000) are shown. ETnII and MusD LTRs are designated type 2 sequences, and ETnI LTRs are designated type 1 sequences. AC074208 is an ETnII element that has integrated into the wiz gene of B6 mice (1) and is located on chromosome 17 at position 31588150. E2_Y17106 was found as a new insertion in SELH mice (9). M_BK001485 is located on chromosome 2 of B6 mice and contains full-length open reading frames for gag, pro, and pol. This element is a member of the young MusD subfamily discussed in the text. M_AF246633 corresponds to the MusD2 sequence identified by Mager and Freeman (24). E1_M16478 represents the first ETnI element, which was found by Brûlet et al. (2) in BALB/c mice. A structural comparison of the full-length elements is shown in Fig. 2 for all four ETnII elements, M_BK001485, E1_M16478, and E1_9_85784030. Points indicate identity with the E2_AC074208 sequence, whereas dashes indicate gaps in the sequence. The inverted repeats (IR) and the locations of the purine-pyrimidine stretch (pu-py), putative TATA box, and poly(A) signal are marked as suggested by Brûlet et al. (2). Note that a MatInspector search (32) identified nt 178 to 195 of E2_AC074208 as a mammalian C-type LTR TATA box with a core similarity of 1.0 and a matrix similarity of 0.921. The locations of a 13- or 14-bp insertion, putative transcription factor binding sites, and the borders of U3, R, and U5 are marked. (b) Comparison of 5′ and 3′ LTR sequences. The identity of 5′ and 3′ LTRs of 37 MusD, 30 ETnII, and 26 ETnI elements was determined with the Diblast program on the National Center for Biotechnology Information server. The y axis shows the accumulated number of elements.
Comparative analysis of ETn and MusD elements.
Comparative sequence analysis revealed that ETnI LTRs consistently group in one distinct cluster as expected, a second major branch contains ETnII LTRs, and a third branch contains MusD LTRs intermingled with a few ETnII LTRs (data not shown). We have not attempted an in-depth phylogenetic analysis of ETnII and MusD LTRs, as trees constructed with these LTRs have poor bootstrap support, suggesting frequent recombinations between forms. Based on internal sequences, four of the nine freshly integrated elements (those at the sites of new mutations) are ETnII sequences, three of which cluster in the ETnII-specific branch and one of which clusters in the mixed ETnII-MusD branch. The identity of the other five fresh insertions is not known due to insufficient sequence data for the internal regions. However, the LTR of an element integrated into the leptin gene of ob2JB6 mice (29) falls within the ETnII branch, suggesting that this element is most likely an ETnII. Interestingly, none of the type 2 sLTRs were found in the ETnII-only cluster, but rather all grouped with the sequences in the MusD-ETnII branch. This result suggests that MusD elements, rather than ETnII, are the antecedent of most type 2 sLTRs.
To determine the relative ages of ETnI, ETnII, and MusD elements, we compared the level of identity of 5′ and 3′ LTRs within full-length elements and found that the LTRs were 100% identical in more than one-third of the ETnII elements (12 of 30) but in less than one-fifth of MusD and ETnI elements (6 of 37 and 5 of 26, respectively). In addition, the proportion of elements with a low level of identity of 5′ and 3′ LTRs (84 to 98%) is higher for MusD than for ETnI and ETnII (Fig. 1b). The high percentage of ETnII elements with identical LTRs supports our previous Southern blot data (24) indicating that the ETnII family has arisen most recently.
The evolutionary position of ETnI is somewhat of a mystery. Analysis of the 5′ and 3′ LTR identities places them between MusD and ETnII elements in terms of age (Fig. 1b). In addition, the ratio of sLTRs to full-length or partial elements (Table 2) is higher for type 1 than for type 2 LTRs. This remains true even under the assumption that most type 2 sLTRs are derived from MusD elements (see above). A high proportion of sLTRs is often indicative of an old age, which would suggest that ETnI is older than MusD. Conversely, the simplest scenario based on structure is that the ETnI group was derived from ETnII, as the two groups differ only within a 270-bp region at the 3′ end of the LTR and immediately downstream of the 5′ LTR. In any event, regardless of their origin, ETnI elements must have undergone a wave of amplification at some point in mouse evolutionary history. These elements have a putative primer binding site which is necessary for the transposition of LTR retroelements (17). In addition, we identified with the Mfold program of Zuker (27) a putative packaging signal consisting of a stable stem-loop structure at position 450 with an ACC motif in the 7-bp loop (data not shown). A potential primer binding site and a stem-loop structure have also been described for MusD and ETnII elements, despite their different nucleotide sequences (24). It seems that ETnI elements have lost their retrotranspositional activity after amplification, since none of these elements have been found at new insertion sites. One possible reason for this apparent inactivity is that the retroviral proteins necessary for ETnI retrotransposition are no longer present or expressed in the mouse germ line.
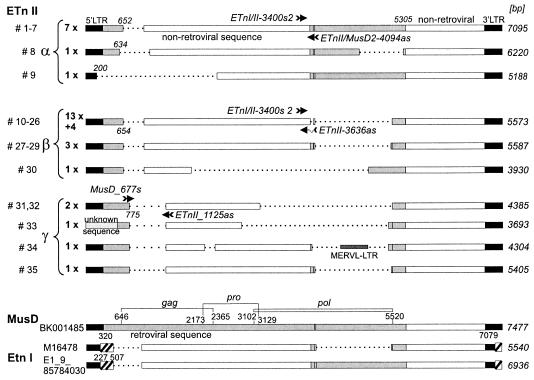
Structural analysis of ETnII elements.
Thirty-one of the 39 ETnII elements found in the B6 database contain 5′ and 3′ LTRs and intervening sequences. A compilation of these elements, as well as of four elements identified previously as new insertions in non-B6 mice, is shown in Table 3. Their structures are displayed schematically in Fig. 2, with a full-length MusD element and two ETnI elements for comparison. Most ETnII elements share high levels of similarity with MusD elements throughout the LTRs and in sections of the 5′ and 3′ internal regions, including at least the first 13 bp of the gag open reading frame and the last 166 bp of pol (24). However, most of the retroviral coding region present in MusD (gag, pro, and pol) is replaced in ETnII elements by an unknown (nonretroviral) sequence. As mentioned above, ETnII elements are almost identical to ETnI elements, except for an ETnI-specific sequence comprising the 3′ portion of the LTRs and 200 bp immediately downstream of the 5′ LTR.
TABLE 3.
Compilation of ETnII elements
| Element no.a | % Identity of 5′ and 3′ LTR (nt identity) | Chromosomal localizationb (accession no.)c |
|---|---|---|
| ETnII-α | ||
| 1 | 100 (317/317) | Chr17_31588176-31595292 (AC074208) |
| 2 | 100 (319/319) | Chr14_41452531-41459612 |
| 3 | 100 (318/318) | Chr11_19377210-19384289 (AL604026) |
| 4 | 98 (316/320) | Chr13_22581335-22574256 (AC606486) |
| 5 | NDd | ChrX_18799835-18806000 (3′ LTR not sequenced) |
| 6 | 98 (316/321) | Chr6 98473758-98466674 |
| 7 | 94 (301/318) | (AC079497) (13-bp deletion in 5′ LTR) (found in MGSCv3 on unsorted contig) |
| 8 | 100 (318/318) | (AC090879) (deletion in pol) (no corresponding contig in MGSCv3 found) |
| 9 | 100 (200/200) | Chr6_132803728-132808915 (internal deletion from positions 199 to 2136) |
| ETnII-β | ||
| 10 | 99 (317/320) | Chr12_63172077-63177635 |
| 11 | 100 (317/317) | Chr7_126261947-126267492 |
| 12 | 99 (316/318) | Chr11_9893680-9888129 (AL663103) |
| 13 | 99 (318/319) | Chr1_45386498-45380976 (has a tandem repeat which is only partially sequenced) |
| 14 | 98 (317/321) | Chr15_56243714-56249236 |
| 15 | 99 (315/316) | Chr13_8665719-8660180 |
| 16 | 100 (317/317) | Chr13_111323164-111328713 (AC079540) |
| 17 | 99 (316/319) | Chr6_89443125-89437558 |
| 18 | 98 (314/318) | Chr1_89529865-89524356 |
| 19 | 100 (317/317) | Chr3_35292118-35297660 |
| 20 | 100 (180/180) | Chr5_88845450-88840003 (sequence gap upstream of 3′ LTR; second half of 3′ LTR is divergent, possibly recombination or insufficient assembly) |
| 21 | ND | Chr14_103573278-103568229 (600 bp at 3′ end not sequenced) |
| 22 | 99 (315/318) | Chr16_25289489-25294967 |
| 23 | 100 (317/317) | (Y17106) (SELH mice, not present in C57BL/6) (reference 9) |
| 24 | 100 (317/317) | (Y17107) (SELH mice, not present in C57BL/6) (reference 9) |
| 25 | ND | dea/Hk1 (A/J, not present in C57BL/6) (reference 31) |
| 26 | ND | (M55665, X15598) (BALB/c, not present in C57BL/6) (references 5 and 36) |
| 27 | 99 (318/319) | Chr3_136259507-136253896 |
| 28 | 99 (317/319) | Chr9_76405783-76400628 |
| 29 | 99 (230/231) | Chr10_24792982-24798774 (3′ LTR only partially sequenced) |
| 30 | 100 (317/317) | Chr6_6765778-6769709 |
| ETnII-γ | ||
| 31 | 96 (313/326) | Chr17_56640925-56645328 (AC079510) |
| 32 | 98 (314/320) | Chr4_142074493-142078754 (AL626773) |
| 33 | ND | Chr10_130717075-130720772 (first 500 nucleotides replaced by an unknown, nonrepetitive sequence) |
| 34 | 92 (310/334) | Chr9_35951778-35946201 (additional 12 bp at position 91 of the LTRs; MERVL-LTR insertion at position 3000; 17-bp deletion at position 231 of 5′ LTR) |
| 35 | 94 (319/337) | Chr15_44010973-44015322 (AC099629; additional 14 bp at position 91 of the LTRs) |
Element structures are shown in Fig. 1.
Obtained by screening of the MGSCv3 database released on February 2002 (http://www.ensembl.org/Mus_musculus/).
If present in the NCBI nr or htgs database (October 2002).
ND, no or insufficient data available.
FIG. 2.
Schematic representation of ETnII elements. Thirty-five ETnII elements (for reference, see Table 3) were sorted into three different groups (α, β, and γ) according to structural similarities. Representatives of each group are shown, with their lengths (in base pairs) given on the right. Black bars, LTRs; gray bars, retroviral genes (gag, pro, pol); white bars, nonretroviral sequences of unknown origin; hatched bars, sequences specific to ETnI. To better illustrate the position of homologous regions, length differences are indicated by dotted lines. Some nucleotide positions are shown, with the numbers referring to the respective element. The identities and the overall number of elements in the B6 draft sequence showing the same structure are indicated on the left of the respective exemplary element. ETnII group α encompasses 9 elements, group β comprises 21 elements, and group γ comprises 5 elements. Four of the 17 group β elements which share an identical structure were isolated from non-B6 mice (indicated by “13x + 4”), and all other elements were from B6. In element 33, the 5′ LTR was replaced by an unknown sequence, and element 34 contains a MERVL sLTR. The locations of group α-, β-, and γ-specific primer pairs used for expression analysis are indicated by arrows.
We grouped the ETnII elements into three subtypes. The nine elements in subtype α contain the 3′ half of the MusD pol gene. All other elements contain only rudimentary parts of this sequence. Subtype β consists of 21 elements in which the transition from gag to the unknown nonretroviral sequence takes place around nt 650. All four non-B6 elements (23 to 26 in Table 3) are members of this group. In the five elements of subtype γ, the transition from gag to the unknown sequence occurs 120 bp further downstream. This grouping scheme was useful for choosing PCR primers for subsequent analysis. Since ETnII elements are structurally heterogeneous and additionally share many similarities with MusD and ETnI, it was not possible to choose a single primer pair to analyze all ETnII elements. By grouping ETnII elements into three subtypes we were able to choose three different primer pairs to allow detection of the majority of the ETnII elements or transcripts. As for the MusD and ETnI elements shown at the bottom of Fig. 2, it is important to note that they are representatives of many elements present in B6 mice, some of which are partially deleted. For example, many MusD elements have large deletions in the gag gene, and most ETnI elements contain only a short pol-related segment.
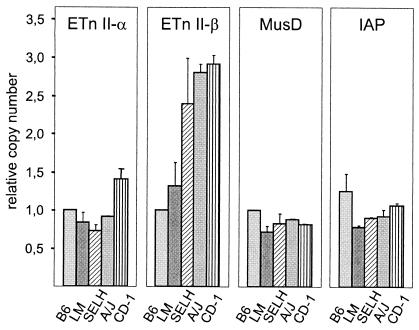
Comparison of ETn and MusD copy number between B6 and non-B6 mice.
To determine if there are detectable differences in copy number in different mouse strains, quantitative real-time PCR was performed with genomic DNA from the B6, LM, SELH, A/J, and CD-1 strains. A/J and SELH mice were chosen because six ETn-related insertion mutations have been reported for these strains (four in A/J and two in SELH) (1). SELH mice are of additional interest because this strain has a high frequency of exencephaly at birth and has an unusually high mutation rate (9, 14). For comparison, we determined the relative number of retrovirus-like IAP elements, which have also caused several new mouse mutations (for reviews, see references 18 and 19). No significant difference was found in the relative copy numbers of IAP, ETnII-α, or MusD elements, except for a slightly elevated number of ETnII-α copies in the outbred strain CD-1 (Fig. 3). However, SELH, A/J, and CD-1 mice contained significantly more ETnII-β elements than B6 or LM mice. Given the number of full-length ETnII-β elements in B6 mice (17 elements [Table 3, no. 10 to 22 and 27 to 30]), we estimate the copy number in SELH, A/J and CD-1 mice to be higher than 40. Notably, the three ETnII elements found in A/J and SELH mutations, for which sufficient sequence information is available, belong to the ETnII-β subtype. No ETn-induced mutations have been described for CD-1 mice, but since CD-1 is an outbred strain, mutations are likely not as frequently analyzed as in the common inbred strains. Interestingly, it has been reported that tumor incidence in CD-1 mice is extraordinarily high: up to 30% of mice aged 18 months or older develop lung and liver tumors and up to 15% develop lymphomas (M. L. A. Giknis and C. B. Clifford, technical report, Charles River Laboratory, Wilmington, Mass., http://www.criver.com/techdocs/tech_pdf/CRLtoxdata2000.pdf). It is tempting to speculate that ETnII insertions might contribute to tumorigenesis in this strain.
FIG. 3.
Analysis of retroviral copy number by quantitative real-time PCR. The relative copy number of elements in the strains C57BL/6J (B6), LM/Bc, SELH/Bc, A/J, and CD-1 was determined by using primers specific for ETnII-α, ETnII-β, MusD, and IAP elements (see Materials and Methods). Note that due to different primer efficiencies, graphs can only be compared within a group.
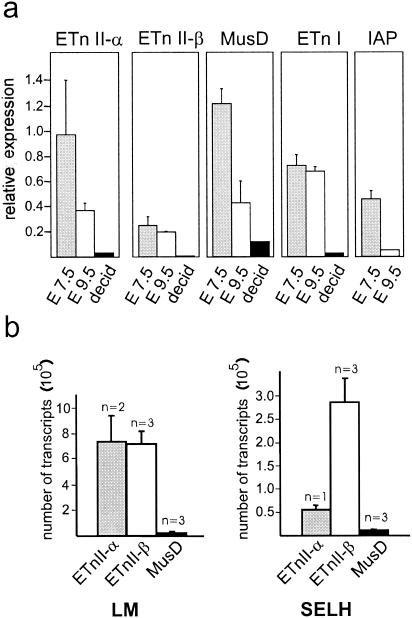
Expression of retroviral elements declines with embryonic age.
Brûlet et al. (2, 3) showed that ETn elements are highly transcribed in embryonic carcinoma cells and during early mouse embryogenesis, with the expression peaking between days 3.5 and 7.5. However, these results were obtained with hybridization probes that did not differentiate between ETnI, ETnII, and MusD. To investigate the expression of these elements during mouse embryogenesis, we isolated 7.5-day-old embryos of SELH and LM mice and analyzed their ETn expression by quantitative real-time PCR. Older embryos (9.5 days old) and maternal decidual tissue were also analyzed. SELH mice were chosen because two new ETnII insertions have been identified in this strain (9). LM mice are a healthy inbred strain used in comparison studies of the exencephaly-prone SELH strain (15, 16). Our results indicate that all elements (ETnII-α, ETnII-β, MusD, and ETnI) are expressed in the ES cell line R1 from 129 mice (data not shown) and at embryonic stage E7.5 of LM and SELH mice (shown in Fig. 4a for the LM strain). Transcripts were also found at E9.5, although at a significantly reduced number. In maternal tissue (decidua), transcript levels from all elements were low or almost undetectable.
FIG. 4.
Expression of retroviral elements in embryos and decidua. (a) Relative RNA levels of different elements in 7.5- and 9.5-day-old embryos and in surrounding tissue (decidua) are shown. RNA quantification was performed by real-time PCR. For this representative experiment, expression was determined in LM/Bc mice (E7.5, three samples; E9.5, two samples; decidua, one sample). Similar results were obtained for SELH/Bc mice (at least two samples each for E7.5, E9.5, and decidua; data not shown). Note that values are only comparable within a boxed group. (b) Comparison of ETnII and MusD expression in 7.5-day-old embryos of LM and SELH mice. The relative numbers of ETnII-α, ETnII-β, and MusD transcripts were determined by real-time PCR. Serial dilutions of plasmids containing the retroviral amplicons were used to construct standard curves (see Materials and Methods). n, number of mice tested.
Since the expression of all elements declined with embryonic age, we hypothesize that a change in the methylation status of embryonic DNA might be responsible for the reduction of expression with time. A role of CG methylation in silencing transcription has been shown for IAP elements in several studies (6, 11, 12, 23, 43). It is known that DNA is demethylated upon fertilization of the egg but is remethylated during early embryogenesis (for review, see reference 33). For IAP, de novo methylation commences soon after implantation (28, 34). We therefore analyzed the expression of IAP elements in E7.5 and E9.5 embryos (Fig. 4a). As with ETns, IAP transcription is reduced at the later stage, supporting the suggestion that the observed differences are due to a global regulatory effect and not due to a specific change in transcriptional regulation of ETn elements. Attempts to demonstrate differences in methylation of ETns with methods based on bisulfite-treated DNA and PCR amplification have so far proven to be unsuccessful due to the highly repetitive character of the elements.
ETnII expression exceeds MusD expression in 7.5-day-old embryos.
To compare the relative expression of ETnII and MusD elements, we performed real-time PCR with group-specific primers. The high similarity of these elements limits the possibilities of choosing PCR primers with equal amplification efficiencies. When primer efficiencies are different, results from relative quantification of one element group cannot be readily compared with the results from another group. We therefore used reference plasmids containing cloned target sequences of the various groups in our PCR experiments to make standard curves for comparisons between groups. Our results indicate that ETnII-α and ETnII-β transcript levels in 7.5-day-old LM embryos are up to 30-fold higher than those for MusD (Fig. 4b, left panel). Similar results were obtained for SELH embryos, although only one sample was examined for ETnII-α (Fig. 4b, right panel). This large difference between ETnII and MusD expression levels was surprising given that the LTRs of the two groups are very similar. In addition, it is estimated that LM mice contain at least twice as many MusD as ETnII elements, since in B6 mice MusD elements are 2.5-fold more abundant than ETnII elements (Table 2) and LM and B6 have similar copy numbers (Fig. 3). Thus, regardless of the near identity of their LTRs and despite a higher copy number, expression of MusD is significantly lower than that of ETnII. Screening of the (mainly B6) expressed sequence tag database with group-specific sequences also indicated that ETnII transcripts are more abundant than MusD transcripts (26 versus 7 hits) (1).
There are several mechanisms that could be responsible for the higher expression of ETnII elements than of MusD elements. First, slight differences in the LTR could confer a higher ETnII promoter activity. With the Web-based program MatInspector (32) we analyzed 17 MusD and 29 ETnII LTRs for potential transcription factor binding sites. We found only one possibly significant difference at nt 212 to 230, where an overlapping Smad4 and NF-1 site was predicted for two-thirds of the analyzed ETnII LTRs but for none of the MusD LTRs (Fig. 1a). Smad4 is involved in the transforming growth factor beta pathway and can positively or negatively influence transcription by dimerization with other Smads. However, it was recently found that Smad4 can also bind to AP-1 and facilitate transcriptional activation of the mouse gonadotropin-releasing hormone receptor gene (30). It is intriguing that an AP-1 factor which is known to act as a positive activator of the mouse mammary tumor virus promoter (20) may enhance the transcription of ETnII in conjunction with Smad4 by binding to the Smad/AP-1 binding site. Most MusD LTRs (14 of 17) contain a potential binding site for the zinc finger homeodomain transcription factor AREB6 instead of the Smad/AP-1 sequence. AREB6 can act as a repressor by interacting with proteins, including transcription factors in specific tissues and in different developmental stages (13), and might contribute to the low transcription levels of MusD elements. Another potential explanation for the higher ETnII transcript levels is that the non-LTR internal sequences of ETnII or MusD elements contain cis-acting enhancer or repressor motifs. Experiments are currently under way to test the functional significance of LTR and non-LTR sequence differences in controlling transcription of these elements.
It is also possible that the high ETnII expression level is due to transcription from a limited number of elements rather than from being associated with a low level of expression from the whole population of ETnII sequences. To address this possibility, we cloned and sequenced cDNAs generated from 7.5-day-old embryonal RNA from LM and SELH mice. To minimize the risk of clonal expansion of single transcripts during the PCRs, we used two different templates, repeated the reverse transcription and PCRs several times under various conditions, and combined the obtained PCR products before cloning. We focused on ETnII-β elements because they represent the largest subgroup of the ETnII family and also comprise the ETnII elements found in new mutations. As was to be expected from an alignment of ETnII-β elements derived from C57BL/6J, the obtained nucleotide sequences of the cDNAs were highly similar (98 to 99%). Only a few diagnostic positions were available (data not shown). In addition, almost every sequence had one or two unique nucleotide differences that could be explained by PCR errors. When unique nucleotides were ignored, 7 of 12 LM and 5 of 11 SELH clones were 100% identical with the corresponding sequence of an ETnII element located on chromosome 13 of B6 mice (Table 3, no. 16). Interestingly, the element Y17106 found in a SELH mutation shares the same sequence (99% identity over the length of the entire element). Our results suggest that many of the ETnII-β sequences expressed in LM and SELH mice as well as the ETnII element Y17106 stem from the element on chromosome 13 or from one that is nearly identical to it. It is possible that this element is in a genomic context that allows it to be highly transcribed. For example, compared to the overall population of elements, this ETnII sequence may be unmethylated or in an open chromatin configuration.
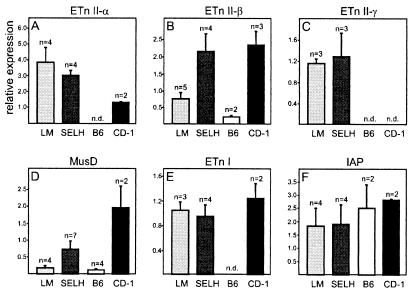
Expression of ETnII-β and MusD varies among strains.
To compare the expression of these elements among mouse strains, we analyzed transcripts of 7.5-day-old embryos from LM, SELH, B6, and CD-1 mice by real-time PCR. There is no significant difference in the expression of ETnI, ETnII-γ, or IAP elements among the strains analyzed (Fig. 5C, E, and F). For ETnII-α (Fig. 5A), CD-1 mice showed a lower level of transcripts despite a slightly elevated number of elements (compare to Fig. 3). The most interesting results were obtained for ETnII-β and MusD. Expression of these two groups in SELH and CD-1 mice was much higher than in LM and B6 mice (Fig. 5B and D). The difference in ETnII expression between SELH and LM mice varied between three- and fivefold in several repeated experiments. One reason for the increased expression could be the twofold-higher copy number of ETnII-β elements in SELH and CD-1 mice (Fig. 3), although this may not directly account for the total extent of the increase. It is possible that, in addition to the higher copy number, the expression of ETnII-β is increased in general in these mouse strains. As discussed above, analysis of ETnII-β transcripts in SELH mice did not uncover a significant difference in the composition of expressed sequences when compared to LM mice. If a few specific elements are preferentially highly active in SELH mice, then one would expect a more biased pattern of sequences, indicating that most, if not all, transcripts were derived from one or a few elements. Instead, it seems that similar elements are predominantly expressed in SELH and LM mice but that other elements are also transcribed.
FIG. 5.
Comparison of retroviral expression between 7.5-day-old embryos of different mouse strains. Relative quantification of ETnI, ETnII, MusD, and IAP expression in LM, SELH, B6, and CD-1 mice by real-time PCR is shown. n, number of mice tested; n.d., not determined. Note that values are only comparable within a boxed group.
Preferential expression of a “young” MusD subgroup.
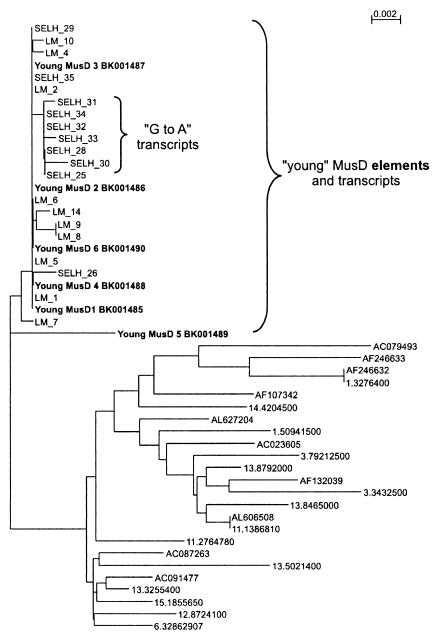
Interestingly, despite similar copy numbers among strains, the MusD group is expressed at a significantly higher level in SELH and CD-1 mice than in LM and B6 mice (Fig. 5D). A higher MusD expression level could result either from a strain-specific effect on the general level of transcription or, alternatively, from a few MusD elements which are variably present in different strains or which are present but variably regulated. To try to distinguish between these possibilities, we analyzed the composition of MusD transcripts in the same way as described for ETnII-β transcripts (see above). All cloned MusD cDNAs from SELH and LM mice (10 each) were almost identical except for a few unique nucleotide exchanges (data not shown). We compared them with 30 randomly chosen MusD elements derived from the B6 database (Fig. 6) and found that the cloned transcripts are derived from a MusD subgroup which is evolutionarily young (the 5′ and 3′ LTRs are 100% identical) and which differs from other MusD elements at about 50 nt positions distributed evenly over the entire element. Thus far, we have found six elements in the B6 database that belong to this subtype (Fig. 6), most of which contain open reading frames for gag, pro, and pol. These results indicate that the youngest and most coding-competent subgroup of MusD is preferentially expressed in LM and SELH mice. The reason for the selective transcription of these elements could be due to position effects or, more likely, different transcriptional regulation. This subgroup contains a putative octamer factor binding site at nt 203 to 217, which could contribute to transcriptional activation (see Fig. 1a, BK001485), and a possible nuclear factor-1 binding site at nt 644 to 662.
FIG. 6.
Dendrogram of transcribed MusD sequences and elements from the B6 genome. Phylogeny for 1.2-kb MusD fragments (nt 680 to 1880) was deduced from SELH and LM transcripts (10 each) and from 30 randomly chosen MusD DNA sequences of B6 mice. Six young MusD elements found in the B6 genome were submitted to GenBank as distinct entries, and their names and accession numbers are printed in bold. Names of the remaining MusD sequences refer to their chromosomal location or to the GenBank accession number from which each element was identified. The branch in the young subgroup containing the seven SELH transcripts with a G-to-A transition at position 1103 is indicated. Distances were calculated using the Kimura two-parameter model with a transition/transversion ratio of 2. The length of the bar corresponds to a nucleotide sequence divergence of 0.2%.
Seven of the 10 sequenced SELH transcripts but none of the LM transcripts contained a unique nucleotide difference (A instead of G) at position 1103 (of BK001485). A thorough screening of the B6 database for this sequence variant was negative. To rule out a PCR artifact, we verified the existence of these unique SELH transcripts (G-to-A transcripts) by high-stringency hybridization of RT-PCR products from 7.5-day-old embryos of LM and SELH mice with specific oligonucleotides. We obtained a strong signal with SELH, but not with LM, products (data not shown). In addition, we performed real-time PCR with primers that contained the discriminating nucleotide at the most 3′ end and with cDNA of LM, SELH, B6, and CD-1 mice. Only cDNAs of SELH and CD-1 mice gave PCR products, indicating the presence of the G-to-A transcripts. Genomic real-time PCR with DNA of the same mouse strains confirmed that B6 indeed does not contain G-to-A elements, since no PCR products were detected (data not shown). In contrast, DNA of SELH, CD-1, and also LM mice was amplifiable. Apparently, despite its presence in LM mice, the element is not transcribed or is transcribed at a very low level in this strain. Our results suggest that the high MusD expression level in SELH (and possibly CD-1) mice is due to an overexpression of one or a few MusD elements with the diagnostic feature of an A instead of a G at position 1103.This element appears to be lacking in B6 mice, and although present, it is not detectably transcribed in LM embryos. Importantly, like all other MusD transcripts found in SELH and LM mice, the overexpressed SELH transcripts belong to the young subgroup of MusD that contains elements with complete open reading frames (Fig. 6). The overexpression of functional Gag, Pro, and Pol could facilitate a high rate of retrotransposition and thus lead to an increased incidence of tumors as seen in SELH and CD-1 mice. Experiments are currently under way to determine the chromosomal localization of the overexpressed MusD element(s) in SELH embryos and to determine the functionality of MusD-encoded retroviral proteins.
Conclusions.
In this study, we have characterized the genomic structures and relationships between ETn and MusD elements and have investigated their embryonic expression and copy number in various mouse strains. While ETnI, ETnII, and MusD groups can be clearly separated, the variety of structures and the fact that some ETnII LTRs are identical to MusD LTRs indicate that frequent recombinations between elements of different groups occur, most likely via copackaging and reverse transcription. Within the population of ETnII elements, we were able to distinguish three subgroups. The ETnII-β subgroup is most numerous in the B6 draft genome and also appears to be the most active class. Six new ETn insertions have been found in SELH and A/J mice (8, 9, 22, 31, 37, 38). In all cases where a positive identification was possible, the inserted element was an ETnII-β element. We also found that the copy number and expression level of ETnII-β elements were more variable than for other ETn groups. In addition, in both LM and SELH mice, a preference towards the transcription of elements that are related in sequence to an ETnII-β element on chromosome 13 of B6 was found. Furthermore, we have detected sequences related to ETnI, ETnII-α, and ETnII-γ, but not ETnII-β, in Mus spretus (data not shown), suggesting that the β structural subtype appeared most recently in mouse evolutionary history.
The MusD elements as a group are older than ETn elements, but we have identified a young, coding-competent MusD subtype which is preferentially expressed in the strains examined. SELH mice contain at least one element belonging to this group which harbors a discriminating nucleotide at position 1103 and which contributes the majority of transcripts, suggesting that this element is overexpressed in SELH embryos. Clonal expression of a limited number of retroviral elements has also been described for IAP elements, since in the livers of aged mice, >50% of the IAP transcripts originate from a unique IAP element (4). The cause for overexpression of the MusD element is not known, but such an element was not detected in B6, indicating that it may be a variant present in only some strains. Interestingly, we found an element(s) with the nt 1103 variation in CD-1 mice, which display an even higher MusD expression than SELH mice, and in A/J mice (data not shown), in which several ETn mutations have been found. It is tempting to speculate that overexpression of this element, having the potential to produce functional retroviral proteins, could facilitate an increased level of retrotransposition of compatible RNAs. Such RNAs are most likely to be of ETnII-β origin, having primer binding sites and putative packaging signals indistinguishable from those of MusD and being much more abundant than MusD or other ETn groups, at least in E7.5 embryos. The low expression levels of MusD and ETnII-β in B6 mice provide a plausible explanation for why only one ETn-related mutation has been reported for this strain (29), even though it is one of the most commonly used strains.
It is interesting that we could detect no vestiges of an env-like gene in any of the MusD elements in the B6 database. The sequence that occupies the region upstream of the 3′ LTR is of unknown origin and we failed to find related sequences that were not associated with other MusD segments. This family could be derived from a fully intact retrovirus which lost its env gene and then amplified in the genome or it could be a true retrotransposon that never possessed an env gene. The former scenario appears more likely, however, as we have identified a wide variety of endogenous betaretroviruses that are related to the MusD family, some of which appear fully intact and contain env genes (Baillie et al., unpublished data). The ETn elements derived from MusD have lost all coding potential but are apparently undergoing retrotransposition more frequently than their parental elements. We are currently investigating potential LTR functional differences or other characteristics that may explain the ongoing activity of this intriguing family of mobile elements.
Acknowledgments
We thank Pamela Hoodless for her assistance in dissecting mouse embryos and for providing us with ES cells and tissue samples of CD-1 mice. We also thank Fumio Takei for providing us with tissue samples of C57BL/6J mice.
This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) to D.L.M. and a CIHR grant to D.M.J. and M.J.H. Core support was provided by the B. C. Cancer Agency. C.B. and G.J.B. were supported by fellowships from the German Research Association (DFG) and the New Zealand Foundation for Research Science and Technology, respectively.
REFERENCES
- 1.Baust, C., G. J. Baillie, and D. L. Mager. 2002. Insertional polymorphisms of ETn retrotransposons include a disruption of the wiz gene in C57BL/6 mice. Mamm. Genome 13:423-428. [DOI] [PubMed] [Google Scholar]
- 2.Brûlet, P., M. Kaghad, X. Yi-Sheng, O. Croissant, and F. Jacob. 1983. Early differential tissue expression of transposon-like repetitive DNA sequences of the mouse. Proc. Natl. Acad. Sci. USA 80:5641-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brûlet, P., H. Condamine, and F. Jacob. 1985. Spatial distribution of transcripts of the long repeated ETn sequence during early mouse embryogenesis. Proc. Natl. Acad. Sci. USA 82:2054-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dupressoir, A., A. Puech, and T. Heidmann. 1995. IAP retrotransposons in the mouse liver as reporters of ageing. Biochim. Biophys. Acta 1264:397-402. [DOI] [PubMed] [Google Scholar]
- 5.Elenich, L. A., and W. A. Dunnick. 1991. Sequence at insertion site of ETn retrotransposon into an immunoglobulin switch region suggests a role for switch recombinase. Nucleic Acids Res. 19:396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feenstra, A., J. Fewell, K. Lueders, and E. Kuff. 1986. In vitro methylation inhibits the promoter activity of a cloned intracisternal A-particle LTR. Nucleic Acids Res. 14:4343-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendy, G. N., A. Y. Sakaguchi, P. A. Lalley, L. Martinez, T. Yasuda, D. Banville, and D. Goltzman. 1990. Gene for parathyroid hormone-like peptide is on mouse chromosome 6. Cytogenet. Cell Genet. 53:80-82. [DOI] [PubMed] [Google Scholar]
- 8.Herrmann, B. G., S. Labeit, A. Poustka, T. R. King, and H. Lehrach. 1990. Cloning of the T gene required in mesoderm formation in the mouse. Nature 343:617-622. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann, M., M. Harris, D. Juriloff, and T. Boehm. 1998. Spontaneous mutations in SELH/Bc mice due to insertions of early transposons: molecular characterization of null alleles at the nude and albino loci. Genomics 52:107-109. [DOI] [PubMed] [Google Scholar]
- 10.Hogan, B., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 11.Hojman-Montes de Oca, F., J. Lasneret, L. Dianoux, M. Canivet, R. Ravicovitch-Ravier, and J. Peries. 1984. Regulation of intracisternal A particles in mouse teratocarcinoma cells: involvement of DNA methylation in transcriptional control. Biol. Cell 52:199-204. [DOI] [PubMed] [Google Scholar]
- 12.Hsiao, W. L., S. Gattoni-Celli, and I. B. Weinstein. 1986. Effects of 5-azacytidine on expression of endogenous retrovirus-related sequences in C3H 10T1/2 cells. J. Virol. 57:1119-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda, K., J. P. Halle, G. Stelzer, M. Meisterernst, and K. Kawakami. 1998. Involvement of negative cofactor NC2 in active repression by zinc finger-homeodomain transcription factor AREB6. Mol. Cell. Biol. 18:10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juriloff, D. M., K. B. MacDonald, and M. J. Harris. 1989. Genetic analysis of the cause of exencephaly in the SELH/Bc mouse stock. Teratology 40:395-405. [DOI] [PubMed] [Google Scholar]
- 15.Juriloff, D. M., M. J. Harris, C. Tom, and K. B. MacDonald. 1991. Normal mouse strains differ in the site of initiation of closure of the cranial neural tube. Teratology 44:225-233. [DOI] [PubMed] [Google Scholar]
- 16.Juriloff, D. M., T. M. Gunn, M. J. Harris, D. G. Mah, M. K. Wu, and S. L. Dewell. 2001. Multifactorial genetics of exencephaly in SELH/Bc mice. Teratology 64:189-200. [DOI] [PubMed] [Google Scholar]
- 17.Kaghad, M., L. Maillet, and P. Brûlet. 1985. Retroviral characteristics of the long terminal repeat of murine ETn sequences. EMBO J. 4:2911-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kazazian, H. H., Jr. 1998. Mobile elements and disease. Curr. Opin. Genet. Dev. 8:343-350. [DOI] [PubMed] [Google Scholar]
- 19.Kuff, E. L. 1990. Intracisternal A particles in mouse neoplasia. Cancer Cells 2:398-400. [PubMed] [Google Scholar]
- 20.Kusk, P., S. John, G. Fragoso, J. Michelotti, and G. L. Hager. 1996. Characterization of an NF-1/CTF family member as a functional activator of the mouse mammary tumor virus long terminal repeat 5′ enhancer. J. Biol. Chem. 271:31269-31276. [DOI] [PubMed] [Google Scholar]
- 21.Leong, W. L., M. J. Dobson, J. M. Logsdon, Jr., R. M. Abdel-Majid, L. C. Schalkwyk, D. L. Guernsey, and P. E. Neumann. 2000. ETn insertion in the mouse Adcy1 gene: transcriptional and phylogenetic analyses. Mamm. Genome 11:97-103. [DOI] [PubMed] [Google Scholar]
- 22.Letts, V. A., R. Felix, G. H. Biddlecome, J. Arikkath, C. L. Mahaffey, A. Valenzuela, F. S. Bartlett, Y. Mori, K. P. Campbell, and W. N. Frankel. 1998. The mouse stargazer gene encodes a neuronal Ca2+-channel gamma subunit. Nat. Genet. 19:340-347. [DOI] [PubMed] [Google Scholar]
- 23.Lueders, K. K., J. W. Fewell, V. E. Morozov, and E. L. Kuff. 1993. Selective expression of intracisternal A-particle genes in established mouse plasmacytomas. Mol. Cell. Biol. 13:7439-7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mager, D. L., and J. D. Freeman. 2000. Novel mouse type D endogenous proviruses and ETn elements share long terminal repeat and internal sequences. J. Virol. 74:7221-7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majewski, J., and J. Ott. 2000. GT repeats are associated with recombination on human chromosome 22. Genome Res. 10:1108-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangin, M., K. Ikeda, and A. E. Broadus. 1990. Structure of the mouse gene encoding parathyroid hormone-related peptide. Gene 95:195-202. [DOI] [PubMed] [Google Scholar]
- 27.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 28.Monk, M., M. Boubelik, and S. Lehnert. 1987. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development 99:371-382. [DOI] [PubMed] [Google Scholar]
- 29.Moon, B. C., and J. M. Friedman. 1997. The molecular basis of the obese mutation in ob2J mice. Genomics 42:152-156. [DOI] [PubMed] [Google Scholar]
- 30.Norwitz, E. R., S. Xu, J. Xu, L. B. Spiryda, J. S. Park, K. H. Jeong, E. A. McGee, and U. B. Kaiser. 2002. Direct binding of AP-1 (Fos/Jun) proteins to a SMAD binding element facilitates both gonadotropin-releasing hormone (GnRH)- and activin-mediated transcriptional activation of the mouse GnRH receptor gene. J. Biol. Chem. 277:37469-37478. [DOI] [PubMed] [Google Scholar]
- 31.Peters, L. L., P. W. Lane, S. G. Andersen, B. Gwynn, J. E. Barker, and E. Beutler. 2001. Downeast anemia (dea), a new mouse model of severe nonspherocytic hemolytic anemia caused by hexokinase [HK(1)] deficiency. Blood Cells Mol. Dis. 27:850-860. [DOI] [PubMed] [Google Scholar]
- 32.Quandt, K., K. Frech, H. Karas, E. Wingender, and T. Werner. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reik, W., W. Dean, and J. Walter. 2001. Epigenetic reprogramming in mammalian development. Science 293:1089-1093. [DOI] [PubMed] [Google Scholar]
- 34.Sanford, J. P., H. J. Clark, V. M. Chapman, and J. Rossant. 1987. Differences in DNA methylation during oogenesis and spermatogenesis and their persistence during early embryogenesis in the mouse. Genes Dev. 1:1039-1046. [DOI] [PubMed] [Google Scholar]
- 35.Shell, B., P. Szurek, and W. Dunnick. 1987. Interruption of two immunoglobulin heavy-chain switch regions in murine plasmacytoma P3.26Bu4 by insertion of retrovirus-like element ETn. Mol. Cell. Biol. 7:1364-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shell, B. E., J. T. Collins, L. A. Elenich, P. F. Szurek, and W. A. Dunnick. 1990. Two subfamilies of murine retrotransposon ETn sequences. Gene 86:269-274. [DOI] [PubMed] [Google Scholar]
- 37.Shiels, A., and S. Bassnett. 1996. Mutations in the founder of the MIP gene family underlie cataract development in the mouse. Nat. Genet. 12:212-215. [DOI] [PubMed] [Google Scholar]
- 38.Shiels, A., D. Mackay, S. Bassnett, K. Al Ghoul, and J. Kuszak. 2000. Disruption of lens fiber cell architecture in mice expressing a chimeric AQP0-LTR protein. FASEB J. 14:2207-2212. [DOI] [PubMed] [Google Scholar]
- 39.Sonigo, P., S. Wain-Hobson, L. Bougueleret, P. Tiollais, F. Jacob, and P. Brûlet. 1987. Nucleotide sequence and evolution of ETn elements. Proc. Natl. Acad. Sci. USA 84:3768-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka, I., and H. Ishihara. 2001. Enhanced expression of the early retrotransposon in C3H mouse-derived myeloid leukemia cells. Virology 280:107-114. [DOI] [PubMed] [Google Scholar]
- 41.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wahls, W. P., L. J. Wallace, and P. D. Moore. 1990. The Z-DNA motif d(TG)30 promotes reception of information during gene conversion events while stimulating homologous recombination in human cells in culture. Mol. Cell. Biol. 10:785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walsh, C. P., J. R. Chaillet, and T. H. Bestor. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20:116-117. [DOI] [PubMed] [Google Scholar]