Abstract
Viruses have evolved various strategies to prevent premature apoptosis of infected host cells. Some of the viral genes mediating antiapoptotic functions have been identified by their homology to cellular genes, but others are structurally unrelated to genes of known function. In this study, we used a random, unbiased approach to identify such genes in the murine cytomegalovirus genome. From a library of random transposon insertion mutants, a mutant virus that caused premature cell death was isolated. The transposon was inserted within open reading frame m41. An independently constructed m41 deletion mutant showed the same phenotype, whereas deletion mutants lacking the adjacent genes m40 and M42 did not. Apoptosis occurred in different cell types, could be blocked by caspase inhibitors, and did not require p53. Within the murine cytomegalovirus genome, m41, m40, and m39 form a small cluster of genes of unknown function. They are homologous to r41, r40, and r39 of rat cytomegalovirus, but lack sequence homology to UL41, UL40, and UL37 exon 1 (UL37x1) which are located at the corresponding positions of the human cytomegalovirus genome. Unlike UL37x1 of human cytomegalovirus, which encodes a mitochondrion-localized inhibitor of apoptosis that is essential for virus replication, m41 encodes a protein that localizes to the Golgi apparatus. The murine cytomegalovirus m41 product is the first example of a Golgi-localized protein that prevents premature apoptosis and thus extends the life span of infected cells.
The programmed cell death (apoptosis) of a virus-infected cell is a strategy of the host organism to prevent virus replication and spread. Apoptosis can occur as a direct result of virus infection or can be triggered by immune effector cells that recognize infected cells. As a counterstrategy, viruses have developed mechanisms to inhibit apoptosis and extend the life span of the infected cell (31, 44).
Herpesviruses possess large double-stranded DNA genomes. They encode up to 200 genes, many of which modulate intrinsic functions of the host cell (38). Cytomegaloviruses (CMVs), prototypes of the β subgroup of the Herpesviridae, are the largest of the herpesviruses. With protracted replication cycles of ≈24 h for murine cytomegalovirus (MCMV) and 48 to 72 h for human cytomegalovirus (HCMV), these viruses are particularly vulnerable to elimination by apoptosis. Therefore, it is not surprising that CMVs have acquired a number of genes that prevent premature apoptosis of the infected host cell.
The immediate-early genes IE1 and IE2 of HCMV were the first CMV genes for which an antiapoptotic function was demonstrated. The IE1 and IE2 proteins both inhibit the induction of apoptosis by tumor necrosis factor alpha or by an E1B 19-kDa protein-deficient adenovirus (50). More recent work demonstrated that inhibition of apoptosis by IE1 and IE2 involves activation of the phosphatidylinositide 3′-OH kinase pathway and the cellular kinase Akt (49).
Two additional HCMV genes expressed at immediate-early times after infection have been shown to inhibit apoptosis at different levels. The product of open reading frame (ORF) UL37 exon 1 (pUL37x1) encodes a mitochondrion-localized protein. It forms a complex with the adenine nucleotide translocator and suppresses apoptosis by blocking permeabilization of the mitochondrial outer membrane (17). The UL37x1 protein also disrupts mitochondrial networks (28). It shows no significant amino acid homology to Bcl-2 and does not bind BAX or the mitochondrial voltage-dependent anion channel VDAC, suggesting that pUL37x1 represents a separate class of cell death inhibitors (16). The product of UL36 binds to the prodomain of caspase 8 and prevents its activation (40). It lacks sequence similarity to viral or cellular FLICE-inhibitory proteins and to other known antiapoptotic proteins.
The antiapoptotic function of these HCMV genes was studied in cells expressing the isolated genes but not in the context of viral infection. However, it has been shown that UL36 is dispensable for virus growth in cell culture (32) and that this gene is mutated in the most commonly used laboratory strains, AD169 and Towne (32), owing to a missense mutation or partial deletion that renders the UL36 protein nonfunctional (40). In fact, the HCMV laboratory strains contain a number of known (9, 13, 30, 40) and likely many more unknown mutations compared to fresh clinical isolates. Unlike the clinical HCMV strains, which infect various cell types in human patients and in cell culture (35), the laboratory strains replicate almost exclusively in cultured human fibroblasts, precluding analysis of cell type-specific functions with virus mutants.
The laboratory strains of murine cytomegalovirus (MCMV) appear to be less degenerate than the HCMV laboratory strains, as MCMV laboratory strains can replicate in various cell types in vitro and cause disease in experimentally infected mice. MCMV has been used to study the genetic basis for endothelial cell tropism. A library of random transposon insertion mutants of the MCMV genome was screened for mutants that had lost the ability to replicate in endothelial cells in culture. A gene, M45, which is required for virus growth and spread in endothelial cells by preventing rapid onset of apoptosis in infected endothelial cells was identified (6). The mechanism of action of the M45 protein has not yet been determined. M45 shows significant sequence homology to the ICP10 protein of herpes simplex virus type 2, including the N-terminal protein kinase domain of ICP10. Two laboratories have recently demonstrated that ICP10 can inhibit apoptosis and that the protein kinase domain is required for this activity. However, the mechanism by which apoptosis is suppressed remains controversial (25, 33, 34).
In a different approach, systematic analysis of an entire viral gene family revealed that another MCMV gene had antiapoptotic activity, which became apparent only in a specific cell type. Inactivation of gene M36, the genetic and functional homolog of the HCMV gene UL36, led to a virus mutant with a severe growth defect in macrophages but not in fibroblasts (29).
In the present study, we identified and analyzed a previously uncharacterized antiapoptotic gene of MCMV. We show that the product of ORF m41 is a Golgi-resident protein that is required to prevent premature apoptosis of infected cells and thus extends their life span.
MATERIALS AND METHODS
Plasmids and retroviral vectors.
ORFs m41 and UL37x1 were amplified by PCR with the primers listed in Table 1, introducing an influenza virus hemagglutinin (HA) epitope tag at the 5′ or the 3′ end of the coding sequence. PCR products were digested with BamHI and EcoRI or XhoI and EcoRI, respectively, and cloned into pcDNA3 (Invitrogen) to generate pcDNA-m41HA, pcDNA-HAm41, and pcDNA-UL37x1HA. The integrity of the cloned m41 and UL37x1 sequences was verified by sequence analysis.
TABLE 1.
Oligonucleotide primers used in this study
| Primer or oligonucleotide | Sequence |
|---|---|
| Primersa | |
| m41-HA | 5′-aaa gga tcc acc atg gga gac gat gat cgt c-3′ |
| 5′-aaa gaa ttc aAG CGT AGT CTG GGA CGT CGT ATG GGT Atc tgt caa tga tca cga cga-3′ | |
| HA-m41 | 5′-aaa gga tcc acc atg TAC CCA TAC GAC GTC CCA GAC TAC GCT atg gga gac gat gat cgt c-3′ |
| 5′-aaa gaa ttc tca tct gtc aat gat cac gac-3′ | |
| UL37x1-HA | 5′-aaa gaa ttc cac cat gtc tcc agt cta cgt gaa t-3′ |
| 5′-aaa ctc gag tca AGC GTA GTC TGG GAC GTC GTA TGG GTA ctg gtg aga ctg ctg ggg | |
| Oligonucleotidesb | |
| MCMVΔm41 | 5′-AAT AGT CAT CCG ATG ATC GTG TCG CCG CCC GAC CGC CCT CCT CCC CCA ATt gtt gac aat taa tca tcg gca t-3′ |
| 5′-CGC CGT TTC CTC ACA TTC CGT TGT CGT GCG CAG GTT CCT CCG AAC CTT TGt cag tcc tgc tcc tcg gcc a-3′ | |
| MCMVΔm40 | 5′-ACC ATC CCC TCG ACT CGC GTC TTC TCT GTC GAT CTG GGT GTC GGG GAG GGt cag tcc tgc tcc tcg gcc a-3′ |
| 5′-TAG CTA TCA CTT TCG AAA CGC ATA AAC GAC GCT ATG TCC CAC TCA TCT CAt gtt gac aat taa tca tcg gca t-3′ | |
| MCMVΔm42 | 5′-ATA GAG GAC GTG GTT ATG CTA CTT TTG TGA ACG GAC GCG GTT GGG AGG ACg aat tca gtc ctg ctc ctc ctc ggc ca-3′ |
| 5′-AGA ACG AGT ATG ACC AAG AGG GAC ACA CAG ACT CCG ACC GTG ATG GCG GTt gtt gac aat taa tca tcg gca t-3′ | |
| MCMVΔM45 | 5′-GCT AGA GAA GTT CTA CGT CGA CGT CGG GCC CCT CGT CGA GTT CGC GTG ACg aat tca gtc ctg ctc ctc ggc ca-3′ |
| 5′-TCG TGC AGG CGA TGG GGC TCG ATC TTG ACG GAG CGC ACG CAC TCA TCG AGt gtt gac aat taa tca tcg gca t-3′ | |
| AD169ΔUL37x1 | 5′-TTT CAA GAC GAC GTG AGA CCC ACA CGC GGG TTT CAC TTC TTT CTT TAA Tta ccc ata cga cgt ccc ag-3′ |
| 5′-TTT CTT AAC CAA GGC GGG AGA GGA TCT TCA AGG CGT TTT CGC TGG ATC Cag gac gac gac gac aag taa-3′ | |
| MCMV-m41HA | 5′-TCC TCG CAG TGA TCT GCA TCG TCA TCC TGA TCG TCG TGA TCA TTG ACA GAt acc cat acg acg tcc cag-3′ |
| 5′-AGT CAT CCG ATG ATC GTG TCG CCG CCC GAC CGC CCT CCT CCC CCA ATT CAa gga cga cga cga caa gta a-3′ | |
| AD169-UL37x1HA | 5′-GGC AAC GAG CTC GGA TGC TGC AGC ACA ACG GCC CCC AGC AGT CTC ACC AGt acc cat acg acg tcc cag-3′ |
| 5′-CCT TCT TAT ACT ATC CCG GAG TCT GTG GTT TTT TTG TTT ACC CCT GCT TAa gga cga cga cga caa gta a-3′ |
The HA tag is in capital letters, and restriction sites are in italics.
Homologous sequences are in capital letters, and introduced EcoRI sites are in italics.
Epitope-tagged genes were excised from pcDNA3 with the same enzymes as above and cloned into pRetroEBNA (23) to obtain pRetro-m41HA, pRetro-HAm41, and pRetro-UL37x1HA. Amphotropic retroviral vectors were generated by use of the Phoenix packaging cell line as described previously (23).
Mutagenesis of CMV genomes.
All mutant CMV genomes were generated in Escherichia coli based on full-length bacterial artificial chromosome (BAC) clones of the HCMV AD169 genome (22), the MCMV genome (pSM3fr) (47), and the MCMV-GFP genome. MCMV-GFP (kindly provided by M. Messerle) contains the enhanced green fluorescent protein (EGFP) gene from pEGFP-C1 (Clontech) inserted into the MCMV ie2 locus. The ie2 gene has been shown to be dispensable for virus growth in vitro and in vivo (8, 27).
A library of MCMV transposon insertion mutants has been published previously (6). The transposon insertion mutants were generated by a single-step transposon mutagenesis protocol (5, 7) and transferred to fibroblasts by bacterial invasion to obtain a library of mutant viruses (6). The transposon contains a kan gene for selection in E. coli and the EGFP gene driven by an HCMV immediate-early promoter from pEGFP-C1. The location of the transposon insertion was determined by direct sequencing as described previously (7).
Gene deletion mutants were constructed by homologous recombination in E. coli with linear, PCR-generated fragments. Briefly, a zeocin resistance gene (zeo) was amplified by PCR from pEM7/Zeo (Invitrogen). The primers contained an additional 50 nucleotides for homologous recombination. See Table 1 for a complete list of the primers used in this study. Recombination of BACs with linear DNA fragments was carried out in E. coli strain DY380 as described previously in detail elsewhere (26). DY380 expresses the recombination genes exo, bet, and gam of bacteriophage λ in a temperature-dependent fashion.
Tagging of viral genes was also performed by homologous recombination with linear fragments, essentially as described above. A kan gene flanked by FRT sites was excised with EcoRI from plasmid pSLFRTKn (1) and inserted into pcDNA-m41HA. The resulting plasmid, pcDNA-m41HA-FK, served as the template for PCR amplification of the influenza virus HA epitope sequence and the FRT-flanked kan gene. This cassette was inserted at the 3′ end of the gene to be tagged. The kan gene was subsequently removed with FLP recombinase expressed in E. coli strain DH10B from plasmid pCP20 (11). For a detailed description of the gene tagging strategy, see the report by Uzzau et al. (45).
For molecular analysis, BACs were digested with different restriction enzymes and separated on large 0.6% agarose gels. Southern blot hybridizations were performed with a nonradioactive digoxigenin labeling and detection kit (Roche). Mutations introduced were verified by PCR or sequencing.
Cells and viruses.
MCMV was grown and titered on NIH 3T3 cells by standard procedures. Virus titers were determined according to the median tissue culture infectious dose (TCID50) method (37). NIH 3T3, M2-10B4, and SVEC4-10 cells were obtained from the American Type Culture Collection. The murine 10.1 cell line and life-extended human foreskin fibroblasts (HFFs) have been described previously (4, 19).
To reconstitute infectious virus from mutant MCMV genomes, BAC DNA was transfected into NIH 3T3 cells with Superfect transfection reagent (Qiagen) as described previously (7). For HCMV, BAC DNA was transfected into HFFs by electroporation as previously described (2). AD169ΔUL37x1 virus was obtained by transfection of BAC DNA into HFFs transduced with Retro-UL37x1HA and propagated on the same cells.
Cell death assays.
Cell viability was determined by a neutral red inclusion assay essentially as described previously (24). To analyze nuclear DNA fragmentation, cells were grown on coverslips, fixed with 2% paraformaldehyde, and stained with a terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay kit (Roche) according to the manufacturer's recommendations. Nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI). For detection of phosphatidylserine on the outer leaflet of the cell membrane as an early sign of apoptosis, cells on coverslips were incubated with labeled annexin V (Roche) and propidium iodide following the recommended protocol. The percentage of apoptotic cells was determined by counting a total of at least 400 cells in four or more random visual fields. The caspase inhibitors Z-VAD-FMK and Boc-D-FMK (Calbiochem) were added to the cells 30 min prior to infection at a final concentration of 100 μM. All assays were performed in duplicate or triplicate.
Immunofluorescence.
Cells were grown on coverslips, fixed with 4% paraformaldehyde, and stained with primary and secondary antibodies according to standard procedures. HA-tagged proteins were detected with a monoclonal rat antibody, 3F10 (Roche). A mouse monoclonal antibody against protein disulfate isomerase (PDI) was purchased from StressGen. An antibody recognizing the Golgi membrane protein p115 (48) was kindly provided by M. G. Waters. Secondary antibodies anti-rat immunoglobulin G-Alexa 488 and anti-mouse immunoglobulin G-Alexa 568 and the mitochondrion-specific dye Mitotracker Red CMXRos were obtained from Molecular Probes.
Immunoprecipitation and Western blotting.
Metabolic labeling with [35S]methionine and [35S]cysteine and immunoprecipitation experiments were performed essentially as described previously elsewhere (1). The 3F10 anti-HA antibody and protein G-Sepharose were used for precipitation. Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gels were dried and exposed to Kodak X-Omat AR films. For Western blotting, proteins were transferred to nitrocellulose membranes and probed with the monoclonal mouse anti-HA antibody 16B12 (Covance Research Products). Signals where detected by enhanced chemiluminescence.
RESULTS
MCMV mutant causes apoptosis of infected cells.
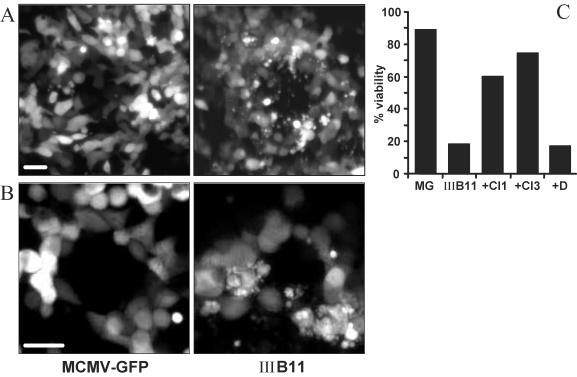
In a previous study, a library of mutant MCMV genomes was established by random transposon mutagenesis of a full-length MCMV BAC. The uncharacterized library of mutant viral genomes was subsequently converted into a library of mutant viruses by direct transfer of the BACs from E. coli to fibroblasts (6). Each mutant virus carries a transposon inserted at a random position within the MCMV genome. The transposon contains the EGFP gene driven by an HCMV immediate-early promoter. Therefore, cells infected with the virus mutants can be identified by their green fluorescence. In this library, we found a virus mutant, IIIB11, that caused an interesting phenotype in infected NIH 3T3 fibroblasts: membrane blebbing and release of vesicles reminiscent of apoptotic bodies (Fig. 1A). These phenomena were seen in only a few cells infected with the control virus, MCMV-GFP, which contains the EGFP gene inserted at an innocuous position. A similar phenotype was observed upon infection of SVEC4-10 endothelial cells (Fig. 1B) and M2-10B4 bone marrow stromal cells (not shown), suggesting that the phenotype is not cell type specific. Premature cell death was even more apparent when cells were infected at a high multiplicity of infection. At 36 h after infection, the viability of cells infected with IIIB11 was markedly lower than that of cells infected with the MCMV-GFP control virus (Fig. 1C). This effect was inhibited when the infection was done in the presence of broad-spectrum caspase inhibitors, although inhibition of cell death was not complete (Fig. 1C).
FIG. 1.
Apoptotic cell death induced by an MCMV transposon insertion mutant. (A) NIH 3T3 and (B) SVEC4-10 cells were infected at a multiplicity of infection of 0.01 TCID50/cell with mutant IIIB11 and with the control virus, MCMV-GFP. The fluorescent images show virus plaques with morphological signs of apoptosis (membrane blebbing, disintegration of cells with formation of apoptotic bodies) in individual IIIB11-infected cells, most prominently seen around the center of the plaque. Bar, 20 μm. (C) The viability of NIH 3T3 cells infected with IIIB11 was markedly reduced compared to that of cells infected with MCMV-GFP (MG) 36 h after infection at a multiplicity of 10 TCID50/cell. Cell death could be largely inhibited by treating cells with caspase inhibitor I or III (CI1 or CI3, i.e., Z-VAD-FMK or Boc-D-FMK, respectively) but not by addition of dimethyl sulfoxide (D), the solvent used for the caspase inhibitors.
Identification of ORF m41.
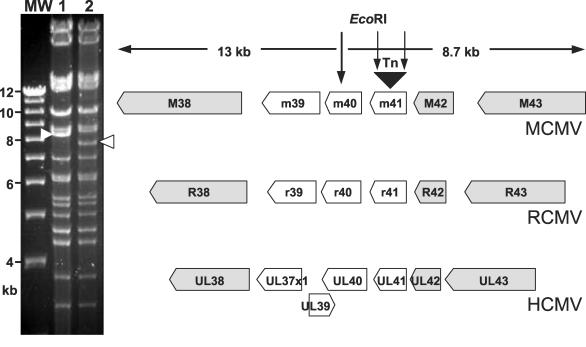
The corresponding BAC clone was used to determine the transposon insertion site in mutant IIIB11. The transposon was found to be inserted at nucleotide position 54022 of the MCMV genome, within a short ORF of 414 nucleotides (Fig. 2). This ORF, m41, is located in a group of ORFs that are conserved between MCMV and rat CMV, but not between the rodent CMVs and HCMV (10, 30, 36, 46) (Fig. 2). However, HCMV contains a gene encoding a mitochondrion-localized inhibitor of apoptosis, UL37x1, at a similar position within its genome (17).
FIG. 2.
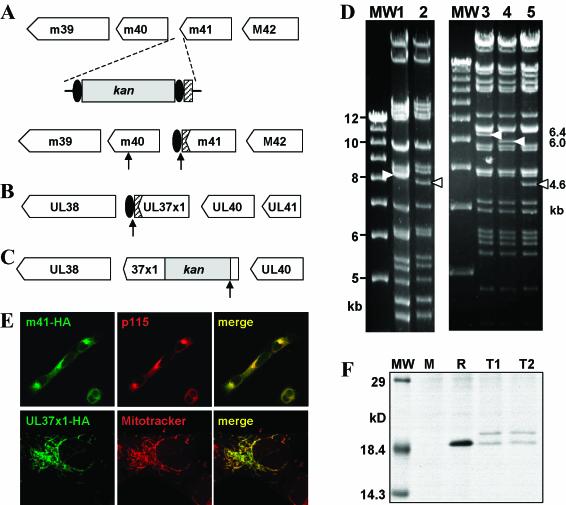
Location of the transposon insertion in mutant IIIB11. The transposon insertion within ORF m41 of the MCMV genome results in a change of the EcoRI restriction pattern (white arrowheads) in IIIB11 BAC (lane 2) compared to the MCMV wild-type BAC (lane 1), because the transposon (Tn) contains EcoRI sites within its inverted repeats. The exact location of the transposon insertion was determined by sequencing. ORFs m39, m40, and m41 show no sequence homology to the HCMV ORFs at the corresponding positions but are homologous to r39 to r41, respectively, of rat CMV (RCMV). Genes conserved among the three CMVs are shown in gray. Lane MW, molecular size markers.
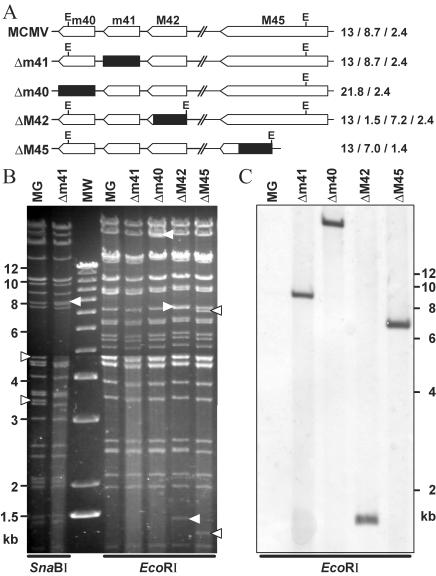
The phenotype observed in mutant IIIB11 could result from disruption of ORF m41, an effect on the expression of one of the adjacent genes m40 and M42, or from an additional adventitious mutation elsewhere in the viral genome. To discriminate among these possibilities, we constructed targeted deletion mutants MCMVΔm40, MCMVΔm41, MCMVΔM42, and MCMVΔM45 (Fig. 3). The ΔM45 mutant was included as a positive control virus, as viruses carrying mutations in this gene have previously been shown to induce apoptosis rapidly in infected endothelial cells (6). The mutant MCMV genomes were constructed by homologous recombination in E. coli with the BAC technology. The individual ORFs were deleted and replaced with a zeocin resistance gene. For the ΔM42 and ΔM45 mutants, short sequences at the 3′ end of the ORFs were left intact to avoid deletion of potential promoter sequences of downstream genes. In these two mutants, additional EcoRI restriction sites were introduced to facilitate detection of the mutations by restriction digestion. Figure 3 shows a schematic representation of the mutations and the mutant genomes in an ethidium bromide-stained gel and in a Southern blot analysis.
FIG. 3.
Construction of MCMV deletion mutants. (A) ORFs m41, m40, M42, and M45 were deleted from the MCMV-GFP BAC (MG) and replaced with a zeo gene (black box). This resulted in loss or gain of an EcoRI restriction site (E) in mutants MCMVΔm40, ΔM42, and ΔM45 but not in Δm41. The calculated sizes of the EcoRI restriction fragments in the m40 to M45 region are given on the right. (B) Restriction patterns visualized by ethidium bromide staining. Bands with sizes different from that of the parental MCMV-GFP BAC are indicated by arrowheads. An SnaBI restriction site within m41 is lost in MCMVΔm41, resulting in fusion of two SnaBI fragments of 4.8 and 3.3 kb into a new fragment of 8.1 kb (arrowheads). (C) Southern blot of EcoRI-digested BACs, hybridized with a probe specific for the zeo gene sequence. Lane MW, molecular size markers.
The deletion mutants shown in Fig. 3 were derived from MCMV-GFP and can thus be visualized in the same way as IIIB11. Cells infected with MCMVΔm41 displayed the same phenotype as shown for IIIB11 in Fig. 1, whereas cells infected with MCMVΔm40 or MCMVΔM42 did not (data not shown). To compare the importance of m41 and M45 for inhibition of virus-induced apoptosis, we used different cell death assays. Annexin V detects phosphatidylserine on the outer leaflet of the cell membrane, an early sign of apoptosis. At later stages of apoptosis and during nonapoptotic cell death, cell membrane integrity becomes compromised, allowing annexin V to enter the cell and bind to phosphatidylserine molecules at the inner leaflet of the cell membrane. Costaining with propidium iodide is used to control for this. The TUNEL assay, by contrast, detects nuclear DNA fragmentation, which occurs at late stages of apoptosis. The neutral red inclusion assay measures cell viability and is a very reliable test to quantitate cell survival and death. However, this assay cannot discriminate between apoptotic and nonapoptotic cell death.
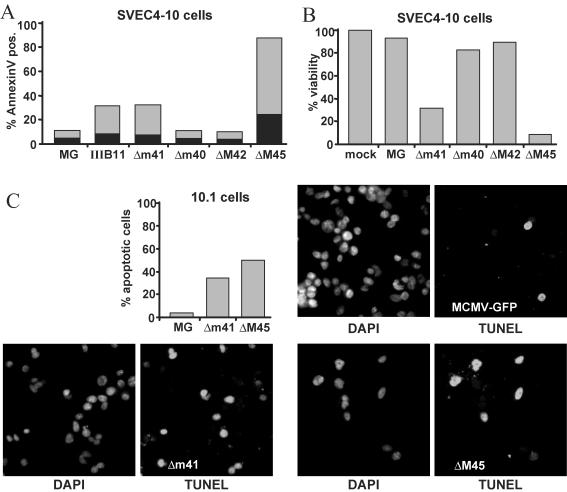
SVEC4-10 endothelial cells were infected at a high multiplicity of infection with the deletion mutants. At 24 h after infection, an annexin V assay was used to detect early signs of apoptosis in infected cells (Fig. 4A). At a later time point, cell viability was measured by neutral red inclusion (Fig. 4B). Both the m41 and M45 mutants induced apoptotic cell death, but the effect of the ΔM45 mutant appeared to be stronger than the effect of the m41 mutants. Similar results were obtained when nuclear DNA fragmentation as a late sign of apoptosis was measured with a TUNEL assay (not shown). Mutants Δm40 and ΔM42 did not differ significantly from the MCMV-GFP parental virus, demonstrating that the phenotype of IIIB11 and MCMVΔm41 is caused by inactivation of m41 rather than an effect on neighboring genes. As determined by the TUNEL assay, MCMVΔm41 and MCMVΔM45 also induced premature apoptosis in 10.1 cells, a spontaneously immortalized, p53-deficient cell line unrelated to SVEC4-10 cells (Fig. 4C). This indicated that p53 is not required for viral induction of apoptosis.
FIG. 4.
MCMV deletion mutations cause apoptosis. SVEC4-10 endothelial cells were infected at a multiplicity of 5 TCID50/cell with MCMV-GFP (MG) or deletion mutants. (A) Phosphatidylserine on the outer leaflet of the cell membrane was detected with annexin V at 24 h after infection. The percentage of cells staining positive with both annexin V and propidium iodide (indicating late-stage apoptosis or nonapoptotic cell death) is shown in black. (B) Cell viability was determined 32 h after infection by the neutral red inclusion assay. The viability of cells infected with the Δm41 or the ΔM45 mutant was markedly reduced, whereas cells infected with MCMVΔm40 or MCMVΔM42 were as viable as cells infected with MCMV-GFP. (C) Cells of p53-deficient fibroblast cell line 10.1 also underwent apoptosis after infection with the Δm41 or the ΔM45 mutant. Cells were stained on coverslips by the TUNEL assay and counterstained with DAPI. Fewer cells are seen in the photomicrographs of cells infected with the Δm41 or ΔM45 mutant virus because dead and disintegrated cells detached from the coverslips.
Growth properties of mutant viruses.
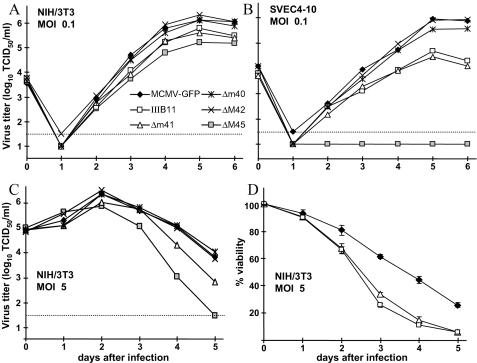
In a previous study we showed that inactivation of the M45 gene precluded growth of MCMV in endothelial cells but allowed almost normal growth in fibroblasts, bone marrow stromal cells, and hepatocytes (6). Multistep growth curves, in which cells were infected with the mutant viruses at a multiplicity of 0.1 TCID50/cell, revealed that, in comparison to the wild-type virus, the m41 mutant viruses grew to a slightly reduced level in fibroblasts but were diminished by up to 50-fold in endothelial cells (Fig. 5A and B), probably because SVEC4-10 endothelial cells are more prone to apoptosis than NIH 3T3 fibroblasts (unpublished observation). Single-step growth curves in which NIH 3T3 cells were infected at a multiplicity of 5 TCID50/cell showed a more rapid decline in viral titers for the m41 and M45 deletion mutants 3 to 5 days after infection (Fig. 5C). The decline in virus titer was probably caused by premature cell death, as seen in a time course analysis of cell viability (Fig. 5D). When SVEC4-10 cells were used instead of NIH 3T3 cells, cell viability was even more compromised (not shown), consistent with our previous observation that SVEC4-10 cells are more sensitive to virus-induced apoptosis. However, the titer drop was delayed compared to the decline in cell viability. Virus release by dying cells and stability of previously released virus particles could account for this delay. Taken together, the results suggests that the antiapoptotic function of the m41 gene product became apparent predominantly at late stages after infection and when cells were infected at a high multiplicity of infection (see also Fig. 1 and 4).
FIG. 5.
Growth properties of MCMV mutants. (A) NIH 3T3 fibroblasts and (B) SVEC4-10 endothelial cells were infected at a multiplicity of 0.1 TCID50/cell for multistep growth curves. Deletion and insertion mutagenesis of the m41 ORF (mutants MCMVΔm41 and IIIB11) had no obvious effect on virus growth in NIH 3T3 fibroblasts and resulted in only moderately reduced titers in SVEC4-10 endothelial cells. By contrast, the ΔM45 mutant failed to grow on SVEC4-10 cells, consistent with previous results (6). (C) After infection at a multiplicity of 5 TCID50/cell, the titers of the Δm41 and the ΔM45 mutants declined faster than the titers of the control viruses. All titers represent mean values of two or three parallel experiments. Dotted line, detection limit. (D) A time course analysis showed a faster decline of cell viability for the m41 mutant viruses (Δm41 and IIIB11) compared to MCMV-GFP. Values were determined in triplicate with standard deviations (error bars).
Subcellular localization of the m41 proteins.
According to the published MCMV sequence (36), ORF m41 encodes a polypeptide of 138 amino acids. It contains a hydrophobic stretch close to the C terminus and is predicted to be a type II transmembrane protein of 14.6 kDa.
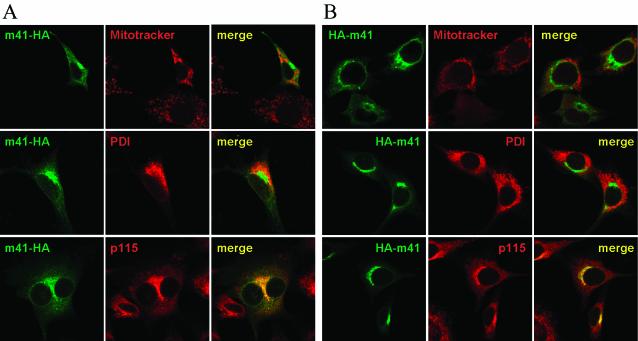
To determine the subcellular distribution of the protein encoded by the m41 ORF, the m41 sequence was cloned by PCR and fused to an HA epitope tag sequence at either the 5′ or the 3′ end. In addition, the HCMV UL37x1 gene was cloned and HA tagged at the 3′ end. Proteins HA-m41, m41-HA, and UL37x1-HA were expressed in NIH 3T3 cells by using retroviral expression vectors and visualized by immunofluorescence with an antibody directed against the HA epitope. Figure 6 shows that the m41 protein colocalized with a marker for the Golgi apparatus but not with markers for the endoplasmic reticulum or mitochondria. This localization was observed for both the N- and C-terminally tagged m41 proteins. By contrast, UL37x1-HA localized to the mitochondria (not shown), consistent with previous reports and its function as a mitochondrion-localized inhibitor of apoptosis (12, 17).
FIG. 6 and 7.
Figure 6 (top panels) shows subcellular distribution of the m41 protein. ORF m41 was tagged with an HA epitope sequence at the 3′ or the 5′ end and expressed in NIH 3T3 cells with a retroviral vector (panels A and B, respectively). The m41 protein colocalized with a marker for the Golgi (p115) but not with markers for the endoplasmic reticulum (PDI) or mitochondria (Mitotracker). Colocalizations are shown in yellow in the merged pictures.
Similar protein distributions were seen when m41 and UL37x1 were expressed in NIH 3T3 cells by transient transfection with pcDNA3 plasmid expression vectors. However, the extremely high expression levels obtained with these vectors sometimes resulted in cell toxicity and partly aberrant subcellular distribution patterns, which were interpreted as overexpression artifacts. Therefore, retroviral expression vectors were preferred for colocalization studies.
To test the hypothesis that the m41 gene product requires interactions with other viral proteins to acquire its proper structure or to be transported to a specific compartment, we analyzed its intracellular localization during MCMV infection. As an m41-specific antibody was not available, we created a mutant MCMV BAC in which the m41 sequence was tagged in situ (Fig. 7A). For comparison, we also created recombinant HCMV AD169 BACs, in which the UL37x1 gene was tagged or inactivated (Fig. 7B and C). As the UL36-38 region contains spliced genes (41), we wanted to minimize the sequence alterations in this region. Therefore, only a short sequence encoding amino acids 5 through 34 was deleted and replaced with a prokaryotic kan gene for inactivation of UL37x1. This short sequence has previously been shown to be essential for both mitochondrial localization and antiapoptotic activity of the UL37x1 protein (20).
FIG. 7.
Figure 7 (bottom panels) shows construction of epitope-tagged CMV mutants. (A) An HA epitope tag sequence (hatched box) was fused to the 3′ end of the MCMV m41 ORF by homologous recombination. A kan gene flanked by FRT sites (black ovals) was inserted for selection of recombinant BACs in E. coli. The kan gene was subsequently removed with FLP recombinase, leaving a single FRT site behind. (B) In an analogous fashion, an AD169-UL37x1 HA-tagged mutant was constructed. (C) In mutant AD169ΔUL37x1, a domain essential for UL37x1 protein function was deleted and replaced with a kan gene. EcoRI restriction sites are indicated by arrows. (D) EcoRI restriction patterns of the MCMV wild-type BAC (lane 1), MCMV-m41HA (lane 2), the AD169 wild-type BAC (lane 3), AD169ΔUL37x1 (lane 4), and AD169-UL37x1HA (lane 5). Changes in the restriction pattern are indicated by arrowheads. Lane MW, molecular size markers. (E) The m41 protein expressed in NIH 3T3 cells by a tagged MCMV mutant colocalizes with a marker for the Golgi apparatus (p115), whereas the HA-tagged pUL37x1 expressed in HFF cells by AD169-UL37x1HA colocalizes with a mitochondrial marker (Mitotracker). (F) NIH 3T3 cells were infected with wild-type MCMV (M) or MCMV-m41HA (clones T1 and T2) or transduced with Retro-m41HA (R). After metabolic labeling with [35S]methionine and [35S]cysteine, HA-tagged proteins were immunoprecipitated and separated by SDS-PAGE. Two m41 protein products were expressed by MCMV-m41HA but only one by Retro-m41HA. Similar results were obtained in Western blot experiments (not shown).
Recombinant viruses were obtained by transfecting the MCMV-m41HA and AD169-UL37x1HA BACs (Fig. 7D) into fibroblasts. In infected cells, the tagged proteins were detected by Western blotting (not shown) and by immunofluorescence microscopy (Fig. 7E). Both proteins, UL37x1 and m41, exhibited the same localization within CMV-infected cells as was observed for cells that received the individual protein in the absence of CMV infection (Fig. 6).
Transfection of human fibroblasts with the AD169-ΔUL37x1 BAC did not yield recombinant virus. However, when fibroblasts transduced with a retroviral vector expressing UL37x1 were used, AD169-ΔUL37x1 virus could be grown. The mutant virus grew slowly on complementing cells, with obvious signs of apoptosis in infected cells, suggesting that the fibroblasts expressed UL37x1 at levels insufficient for full complementation of the defect (not shown). Recombinant virus generated on the complementing cells could not be propagated in noncomplementing fibroblasts. Although the same results were obtained with two separate AD169-ΔUL37x1 BAC clones, a second-site mutation that contributes to the observed phenotype cannot be entirely excluded. However, results recently presented by others also indicated that UL37x1 is essential for AD169 replication in human fibroblasts (G. Hahn, S. T. Eichhorst, B. Korn, P. H. Krammer, and R. Greaves, presentation at the 26th International Herpesvirus Workshop, abstr. 7.06, 2001). Our findings are in accordance with these results.
ORF m41 encodes at least two distinct protein products.
As we analyzed the protein products of the m41 ORF by immunoprecipitation and in Western blot experiments with the tagged MCMV-m41HA virus, we identified two distinct protein products with apparent molecular masses of approximately 19 and 21 kDa (Fig. 7F). When the HA-tagged ORF was expressed by a retroviral or a plasmid expression vector, only the smaller product of 19 kDa was detected (Fig. 7F). Moreover, the apparently larger product was also absent in cells transduced with Retro-m41HA and superinfected with MCMV (not shown), ruling out the possibility that the larger product resulted from a posttranslational modification by MCMV proteins.
DISCUSSION
In this study, we provide genetic evidence that the m41 ORF is required to prevent premature apoptosis of MCMV-infected cells. Apoptosis of cells infected with m41 mutant viruses was shown by morphology and by apoptosis-specific cell death assays (annexin V, TUNEL, and caspase inhibitors). Systematic deletion mutagenesis of the region surrounding the m41 ORF confirmed that the neighboring ORFs, m40 and M42, did not encode the function. This function of an m41 gene product could not be predicted by sequence analysis, as m41 shows no apparent homology with any protein of known function. This demonstrates that random unbiased mutagenesis and screening procedures are valuable tools with which to identify viral gene functions.
The m41 ORF encodes at least two distinct protein products. Only one of them could be detected by heterologous expression of the m41 ORF with retroviral or plasmid vectors. The second gene product, which accumulated to a similar level, was only seen when m41 expression was analyzed during virus infection. In the absence of a specific antibody, this was achieved by tagging the coding sequence within the viral genome. Such an in situ tagging procedure, which has previously been used to tag bacterial and yeast genes (39, 45), should be very useful for herpesvirus genetics as well. Although we have not yet been able to determine the origin of the second m41 gene product, preliminary results suggest that it derives from a spliced transcript (W. Brune, unpublished results). A more detailed analysis of transcription in this region will be necessary to identify all spliced and unspliced genes. At present it is not known whether the antiapoptotic function is mediated by the smaller or the larger m41 gene product or both.
Even though MCMV m41 and HCMV UL37x1 show no sequence homology, their similar positions within the two viral genomes suggested a potential functional homology. Both proteins are involved in inhibition of apoptosis induced by virus infection. However, the antiapoptotic effect of UL37x1 appears to be stronger, as UL37x1 has been shown to be essential for replication of the HCMV AD169 strain. By contrast, expression of m41 improves the virus yield but is not essential for virus replication in cell culture. On the other hand, one has to take into consideration that an AD169 strain with a mutant UL37x1 is in fact mutant for two antiapoptotic genes, as UL36 has previously been shown to be mutant in this strain (40). It is possible that UL36 and UL37x1 have additive effects and that only one of the genes is required to allow survival of the infected cell and virus replication. This question can only be resolved by use of an HCMV strain that contains a wild-type UL36 gene or by repairing the UL36 mutation in AD169.
The different subcellular distributions of the UL37x1 and m41 proteins suggest quite divergent mechanisms of action. The UL37x1 protein localizes to mitochondria, where it fulfills a Bcl-2-like function even though it has no structural homology to Bcl-2 (16, 17). By contrast, m41 encodes a protein product that localizes to the Golgi apparatus and is thus not likely to be a functional homolog of UL37x1.
How could a protein found in the secretory pathway interfere with induction of apoptosis? It is conceivable that m41 interacts with death receptors or their ligands to downmodulate their presentation on the cell surface. Such a function has been demonstrated for the RID protein complex (also known as E3-10.4K/14.5K) of adenovirus type 5 (14, 18, 42, 43). However, the RID complex does not accumulate in the Golgi as m41 does, but binds to death receptors on the cell surface and targets them to lysosomes for degradation. To our knowledge, m41 represents the first example of a Golgi-resident inhibitor of apoptosis. Interestingly, an endoplasmic reticulum-resident protein, M-T4, that is required to prevent premature apoptosis of infected lymphocytes (3, 21) was recently identified in myxomavirus, a poxvirus with numerous antiapoptotic genes (15). Although the mechanisms by which the m41 and M-T4 proteins interfere with apoptosis are still unknown, these examples highlight the use of multiple strategies by large DNA viruses like the herpesviruses and poxviruses to ensure their survival in different host cells. Identifying the mechanism of m41 action will enhance our understanding of cellular defenses against virus infection.
Acknowledgments
We thank J. Goodhouse for excellent help with confocal microscopy and S. Erhard for technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (Emmy Noether Fellowships to W.B. and M.N. and SFB 479) and by grant CA85786 from the National Cancer Institute.
REFERENCES
- 1.Atalay, R., A. Zimmermann, M. Wagner, E. Borst, C. Benz, M. Messerle, and H. Hengel. 2002. Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcγ receptor homologs. J. Virol. 76:8596-8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barry, M., S. Hnatiuk, K. Mossman, S. F. Lee, L. Boshkov, and G. McFadden. 1997. The myxoma virus M-T4 gene encodes a novel RDEL-containing protein that is retained within the endoplasmic reticulum and is important for the productive infection of lymphocytes. Virology 239:360-377. [DOI] [PubMed] [Google Scholar]
- 4.Bresnahan, W. A., G. E. Hultman, and T. Shenk. 2000. Replication of wild-type and mutant human cytomegalovirus in life-extended human diploid fibroblasts. J. Virol. 74:10816-10818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brune, W. 2002. Random transposon mutagenesis of large DNA molecules in Escherichia coli. Methods Mol. Biol. 182:165-171. [DOI] [PubMed] [Google Scholar]
- 6.Brune, W., C. Ménard, J. Heesemann, and U. H. Koszinowski. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291:303-305. [DOI] [PubMed] [Google Scholar]
- 7.Brune, W., C. Ménard, U. Hobom, S. Odenbreit, M. Messerle, and U. H. Koszinowski. 1999. Rapid identification of essential and nonessential herpesvirus genes by direct transposon mutagenesis. Nat. Biotechnol. 17:360-364. [DOI] [PubMed] [Google Scholar]
- 8.Cardin, R. D., G. B. Abenes, C. A. Stoddart, and E. S. Mocarski. 1995. Murine cytomegalovirus IE2, an activator of gene expression, is dispensable for growth and latency in mice. Virology 209:236-241. [DOI] [PubMed] [Google Scholar]
- 9.Cha, T. A., E. Tom, G. W. Kemble, G. M. Duke, E. S. Mocarski, and R. R. Spaete. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 11.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 12.Colberg-Poley, A. M., M. B. Patel, D. P. Erezo, and J. E. Slater. 2000. Human cytomegalovirus UL37 immediate-early regulatory proteins traffic through the secretory apparatus and to mitochondria. J. Gen. Virol. 81:1779-1789. [DOI] [PubMed] [Google Scholar]
- 13.Dargan, D. J., F. E. Jamieson, J. MacLean, A. Dolan, C. Addison, and D. J. McGeoch. 1997. The published DNA sequence of human cytomegalovirus strain AD169 lacks 929 base pairs affecting genes UL42 and UL43. J. Virol. 71:9833-9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elsing, A., and H. G. Burgert. 1998. The adenovirus E3/10.4K-14.5K proteins down-modulate the apoptosis receptor Fas/Apo-1 by inducing its internalization. Proc. Natl. Acad. Sci. USA 95:10072-10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett, H., and G. McFadden. 2002. Poxviruses and apoptosis: a time to die. Curr. Opin. Microbiol. 5:395-402. [DOI] [PubMed] [Google Scholar]
- 16.Goldmacher, V. S. 2002. vMIA, a viral inhibitor of apoptosis targeting mitochondria. Biochimie 84:177-185. [DOI] [PubMed] [Google Scholar]
- 17.Goldmacher, V. S., L. M. Bartle, A. Skaletskaya, C. A. Dionne, N. L. Kedersha, C. A. Vater, J. Han, R. J. Lutz, S. Watanabe, E. D. McFarland, E. D. Kieff, E. S. Mocarski, and T. Chittenden. 1999. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc. Natl. Acad. Sci. USA 96:12536-12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gooding, L. R., T. S. Ranheim, A. E. Tollefson, L. Aquino, P. Duerksen-Hughes, T. M. Horton, and W. S. Wold. 1991. The 10,400- and 14,500-dalton proteins encoded by region E3 of adenovirus function together to protect many but not all mouse cell lines against lysis by tumor necrosis factor. J. Virol. 65:4114-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harvey, D. M., and A. J. Levine. 1991. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev. 5:2375-2385. [DOI] [PubMed] [Google Scholar]
- 20.Hayajneh, W. A., A. M. Colberg-Poley, A. Skaletskaya, L. M. Bartle, M. M. Lesperance, D. G. Contopoulos-Ioannidis, N. L. Kedersha, and V. S. Goldmacher. 2001. The sequence and antiapoptotic functional domains of the human cytomegalovirus UL37 exon 1 immediate early protein are conserved in multiple primary strains. Virology 279:233-240. [DOI] [PubMed] [Google Scholar]
- 21.Hnatiuk, S., M. Barry, W. Zeng, L. Liu, A. Lucas, D. Percy, and G. McFadden. 1999. Role of the C-terminal RDEL motif of the myxoma virus M-T4 protein in terms of apoptosis regulation and viral pathogenesis. Virology 263:290-306. [DOI] [PubMed] [Google Scholar]
- 22.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinsella, T. M., and G. P. Nolan. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405-1413. [DOI] [PubMed] [Google Scholar]
- 24.Kovacs, A., M. L. Weber, L. J. Burns, H. S. Jacob, and G. M. Vercellotti. 1996. Cytoplasmic sequestration of p53 in cytomegalovirus-infected human endothelial cells. Am. J. Pathol. 149:1531-1539. [PMC free article] [PubMed] [Google Scholar]
- 25.Langelier, Y., S. Bergeron, S. Chabaud, J. Lippens, C. Guilbault, A. M. Sasseville, S. Denis, D. D. Mosser, and B. Massie. 2002. The R1 subunit of herpes simplex virus ribonucleotide reductase protects cells against apoptosis at, or upstream of, caspase-8 activation. J. Gen. Virol. 83:2779-2789. [DOI] [PubMed] [Google Scholar]
- 26.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 27.Manning, W. C., and E. S. Mocarski. 1988. Insertional mutagenesis of the murine cytomegalovirus genome: one prominent alpha gene (ie2) is dispensable for growth. Virology 167:477-484. [PubMed] [Google Scholar]
- 28.McCormick, A. L., V. L. Smith, D. Chow, and E. S. Mocarski. 2003. Disruption of mitochondrial networks by the human cytomegalovirus UL37 gene product viral mitochondrion-localized inhibitor of apoptosis. J. Virol. 77:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ménard, C., M. Wagner, Z. Ruszics, K. Holak, W. Brune, A. Campbell, and U. Koszinowski. 2003. Role of murine cytomegalovirus US22 gene family members for replication in macrophages. J. Virol. 77:5557-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mocarski, E. S., M. N. Prichard, C. S. Tan, and J. M. Brown. 1997. Reassessing the organization of the UL42-UL43 region of the human cytomegalovirus strain AD169 genome. Virology 239:169-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Brien, V. 1998. Viruses and apoptosis. J. Gen. Virol. 79:1833-1845. [DOI] [PubMed] [Google Scholar]
- 32.Patterson, C. E., and T. Shenk. 1999. Human cytomegalovirus UL36 protein is dispensable for viral replication in cultured cells. J. Virol. 73:7126-7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perkins, D., E. F. Pereira, and L. Aurelian. 2003. The herpes simplex virus type 2 R1 protein kinase functions as a dominant regulator of apoptosis in hippocampal neurons involving activation of the ERK survival pathway and upregulation of the antiapoptotic protein Bag-1. J. Virol. 77:1292-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perkins, D., E. F. Pereira, M. Gober, P. J. Yarowsky, and L. Aurelian. 2002. The herpes simplex virus type 2 R1 protein kinase (ICP10 PK) blocks apoptosis in hippocampal neurons, involving activation of the MEK/MAPK survival pathway. J. Virol. 76:1435-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plachter, B., C. Sinzger, and G. Jahn. 1996. Cell types involved in replication and distribution of human cytomegalovirus. Adv. Virus Res. 46:195-261. [DOI] [PubMed] [Google Scholar]
- 36.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 38.Roizman, B., and A. E. Sears. 1996. Herpesviridae, p. 2221-2230. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 39.Schneider, B. L., W. Seufert, B. Steiner, Q. H. Yang, and A. B. Futcher. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11:1265-1274. [DOI] [PubMed] [Google Scholar]
- 40.Skaletskaya, A., L. M. Bartle, T. Chittenden, A. L. McCormick, E. S. Mocarski, and V. S. Goldmacher. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. USA 98:7829-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tenney, D. J., and A. M. Colberg-Poley. 1991. Expression of the human cytomegalovirus UL36-38 immediate early region during permissive infection. Virology 182:199-210. [DOI] [PubMed] [Google Scholar]
- 42.Tollefson, A. E., T. W. Hermiston, D. L. Lichtenstein, C. F. Colle, R. A. Tripp, T. Dimitrov, K. Toth, C. E. Wells, P. C. Doherty, and W. S. Wold. 1998. Forced degradation of Fas inhibits apoptosis in adenovirus-infected cells. Nature 392:726-730. [DOI] [PubMed] [Google Scholar]
- 43.Tollefson, A. E., K. Toth, K. Doronin, M. Kuppuswamy, O. A. Doronina, D. L. Lichtenstein, T. W. Hermiston, C. A. Smith, and W. S. Wold. 2001. Inhibition of TRAIL-induced apoptosis and forced internalization of TRAIL receptor 1 by adenovirus proteins. J. Virol. 75:8875-8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tschopp, J., M. Thome, K. Hofmann, and E. Meinl. 1998. The fight of viruses against apoptosis. Curr. Opin. Genet. Dev. 8:82-87. [DOI] [PubMed] [Google Scholar]
- 45.Uzzau, S., N. Figueroa-Bossi, S. Rubino, and L. Bossi. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. USA 98:15264-15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vink, C., E. Beuken, and C. A. Bruggeman. 2000. Complete DNA sequence of the rat cytomegalovirus genome. J. Virol. 74:7656-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waters, M. G., D. O. Clary, and J. E. Rothman. 1992. A novel 115-kD peripheral membrane protein is required for intercisternal transport in the Golgi stack. J. Cell Biol. 118:1015-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, Y., and J. C. Alwine. 2002. Human cytomegalovirus major immediate-early proteins and simian virus 40 large T antigen can inhibit apoptosis through activation of the phosphatidylinositide 3′-OH kinase pathway and the cellular kinase Akt. J. Virol. 76:3731-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu, H., Y. Shen, and T. Shenk. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 69:7960-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]