Abstract
Mice infected with myopathic coxsackievirus B1 Tucson (CVB1T) develop chronic inflammatory myopathy (CIM) consisting of hind limb weakness and inflammation. Amyopathic virus variants are infectious but attenuated for CIM. In this report, viral clones, chimeras, and sequencing were used to identify viral determinants of CIM. Chimeras identified several regions involved in CIM and localized a weakness determinant to nucleotides 2493 to 3200 of VP1. Sequencing of multiple clones and viruses identified five candidate determinants that were strictly conserved in myopathic viruses with one located in the 5′ untranslated region (UTR), three in the VP1 capsid, and one in the 3C protease. Taken together, these studies implicate Tyr-87 and/or Val-136 as candidate determinants of weakness. They also indicate that there are at least two determinants of inflammation and one additional determinant of weakness encoded by myopathic CVB1T.
The group B coxsackieviruses cause a variety of human diseases ranging from mild flu-like illnesses to life-threatening conditions such as aseptic meningitis and myocarditis (15). Coxsackievirus infections are also associated with the development of certain chronic diseases, including diabetes, dilated cardiomyopathy, inflammatory myopathy, and chronic fatigue syndrome (4, 12, 13, 17, 29). All enteroviruses, including coxsackieviruses, are structurally similar and consist of a naked icosahedral capsid enclosing a single-stranded, messenger-sense, polyadenylated RNA genome of approximately 7,400 nucleotides (nt) in length. The viral genome contains a single open reading frame flanked by a 5′ untranslated region (5′ UTR) and a 3′ UTR that are involved in viral RNA translation and genome replication. While coxsackieviruses are best known as cytopathic viruses that cause death of the host cell, viral RNA may persist in skeletal muscle, cardiac muscle, spleen, and lymph node (10, 26, 27).
In 1979, Ray et al. described a model of chronic inflammatory myopathy (CIM) caused by infection of newborn mice with coxsackievirus B1 isolated from a patient with pleurodynia (19). This virus was later referred to as the Tucson strain (CVB1T) (22). In this model, the acute viral infection resolves within 2 weeks and is followed by chronic postviral myopathy that peaks at 1 month and is manifest as inflammation and weakness of proximal hind limb skeletal muscle. Weakness is apparent clinically as a change in hind limb mobility and gait. Chronic histopathology consists of perimysial and endomysial infiltration of muscle by mononuclear cells, accompanied by ongoing muscle necrosis and regeneration (22). CIM development is driven by a T-cell-dependent immunopathic response, as nude or neonatally thymectomized mice do not develop chronic weakness or inflammation (30, 31), and the H-2 haplotype influences disease severity (25).
The importance of the infecting strain of virus in the pathogenesis of CIM is illustrated by studies of CVB1T variants (24). Infection with myopathic variants causes CIM, whereas amyopathic variants are equally infectious but attenuated for the later development of CIM. Thus, acute cytopathic infection of muscle is not sufficient by itself to induce CIM. The objective of the present study was to identify the viral genetic determinants that cause CIM. We used a reverse genetics approach, producing infectious cDNA clones of myopathic and amyopathic variants combined with construction of viral chimeras and whole-genome sequencing.
Induction of CIM by cloned virus.
Infectious cDNA clones were produced from myopathic and amyopathic CVB1T (24). SY8 is a descendant of the original Tucson strain (22). Virus MP1 was derived directly from SY8 and has a myopathic phenotype equivalent to that of its SY8 parent (24). AMP2 is an amyopathic variant produced by passage of MP1 in BGMK cells (24). To produce viral clones, viral RNA was purified with TriPure (Roche, Indianapolis, Ind.) and full-length cDNA clones were constructed by long reverse transcriptase PCR(RT-PCR) (2, 26). First-strand viral cDNA was synthesized with 200 U of reverse transcriptase (SuperScript II; Invitrogen, Carlsbad, Calif.) and 5 pmol of primer 3DTSP6-CP3 (5′-CGTGTCAAGCTTATTTAGGTGACACTATAGAT14-3′) or primer 3DT-CP4 (5′-CGAGAGCGTGTCAAGCTTACGT24) according to the manufacturer's recommendations. Primer components included a HindIII site (italicized), an SP6 promoter (underlined), and an oligo(dT) region. Viral cDNA was amplified in a 100-μl hot-start PCR containing 25 pmol of each primer and 5 U of rTth DNA polymerase XL (PE Applied Biosystems, Foster City, Calif.). The downstream primer was 3DTSP6-CP3 or 3DT-CP4. The upstream primer was 5UTR-CP2 (5′-CGAGGTTCTAGATAATACGACTCACTATAGTTAAAACAGCCTGTGGGTTG-3′), which contains a 5′ XbaI site (italicized) followed by a T7 promoter (underlined) and the first 20 bases of the CVB1N sequence (bold) (7). The amplification profile consisted of a single cycle of 94°C for 1 min, 50°C for 1 min, and 72°C for 6 min followed by 30 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 6 min, with a 6-s extension per cycle. The 7.4-kb amplicon was digested with XbaI and HindIII, gel purified, and ligated into pUC18 with T4 ligase. Plasmid DNA was transformed into Escherichia coli SURE2 cells (Stratagene, La Jolla, Calif.), and all clones were grown at a reduced temperature of 30°C to maintain the stability of the viral insert. Clones containing a 7.4-kb insert were transfected into BGMK cells with Lipofectin (Invitrogen) according to the manufacturer's protocol. When cultures exhibited 100% cytopathic effect, virus was extracted by three freeze-thaw cycles, centrifugation at 2,500 × g, and storage of the supernatant at −80°C.
To test for induction of CIM, specific-pathogen-free ICR (CD-1) mice (Harlan, Indianapolis, Ind.) were injected intraperitoneally with 50 PFU of virus in 50 μl of phosphate-buffered saline within 48 h after birth. Negative controls were injected with phosphate-buffered saline. CIM was evaluated at 1 month postinfection. For histopathology, hamstring muscle was flash-frozen in isopentane cooled in liquid nitrogen and sectioned completely (sections were 8 μm thick), with every 10th section being mounted, stained with hematoxylin and eosin, and graded as shown in Fig. 1. The level of inflammation was fairly consistent throughout the entire piece of muscle. The incidences and levels of severity of weakness and inflammation were comparable between mice infected with cloned myopathic virus MP1.23 or MP1.24 and mice infected with the respective parent virus MP1 or SY8 (Table 1). Similarly, the amyopathic cloned viruses AMP2.17 and AMP2.22 caused markedly reduced levels of weakness and inflammation that were comparable to those of their parent virus, AMP2. Pairwise comparisons indicated that all myopathic parental viruses and clones differed from AMP2 and its clones in mean weakness grade (P ≤ 0.0004) and mean inflammation grade (P ≤ 0.003). Weakness and inflammation in amyopathic virus-infected mice did not differ from those in the uninfected control (P > 0.14). These results are consistent with the original characterization of the viruses MP1 and AMP2, which revealed only minor differences in virulence (24), and indicate that the myopathic phenotype of the parental virus is preserved in the cloned viruses.
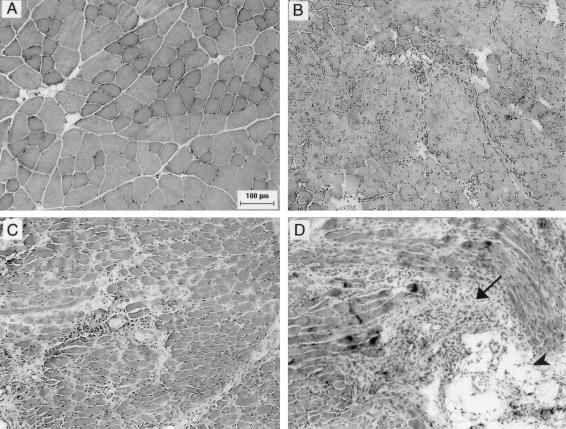
FIG. 1.
Inflammatory histopathology grades 0 to 3 in hamstring muscle at 1 month postinfection. Representative examples of the different grades of inflammation in hamstring muscle are shown (original magnification, ×150). (A) Grade 0, uninfected control; (B) grade 1, MP1.24-infected mouse showing small numbers of interstitial inflammatory cells; (C) grade 2, MP1.24-infected mouse with more widespread infiltration and occasional medium sized infiltrates; (D) grade 3, MP1.24-infected mouse with widespread inflammation and many medium- to large-sized infiltrates (arrow). This sample also shows fatty replacement (arrowhead). At the experimental end point, mice were euthanized by cervical dislocation under ketamine plus xylazine anesthesia according to the Public Health Service policy on Humane Care and Use of Laboratory Animals and the research protocol approved by the Institutional Animal Care and Use Committee at the University of Minnesota.
TABLE 1.
Induction of CIM by cloned viruses and chimeras of CVB1T
| Virus | Chimera construction boundariesa
|
Weaknessb
|
Inflammationc
|
|||||
|---|---|---|---|---|---|---|---|---|
| nt 1-747 (UTR-706) | nt 748-2492 | nt 2493-3200 (VP1 Tyr-87) (VP1 Val-136) | nt 3201-7391 (VP1 Thr-276) (3C Lys-53) | % (no. positive/total) | Graded | % (no. positive/total) | Grade | |
| SY8 | 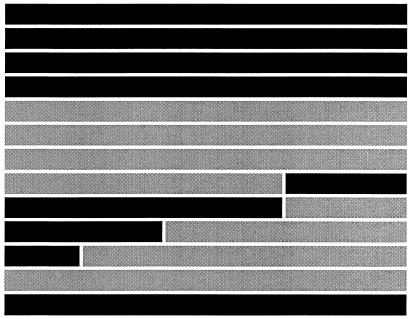 |
62 (8/13) | 1.8 ± 1.0 | 57 (8/13) | 1.3 ± 0.7 | |||
| MP1.24 | 72 (21/29) | 2.1 ± 0.6 | 69 (20/29) | 1.4 ± 0.4 | ||||
| MP1 | 75 (9/12) | 2.2 ± 1.0 | 77 (9/12) | 1.1 ± 0.5 | ||||
| MP1.23 | 78 (21/27) | 2.8 ± 0.8 | 74 (20/27) | 1.4 ± 0.4 | ||||
| AMP2 | 9 (1/11) | 0.1 ± 0.2 | 18 (2/11) | 0.3 ± 0.4 | ||||
| AMP2.17 | 7 (2/29) | 0.1 ± 0.1 | 21 (6/29) | 0.2 ± 0.2 | ||||
| AMP2.22 | 25 (4/16) | 0.3 ± 0.3 | 25 (4/16) | 0.4 ± 0.4 | ||||
| AMP-3200 | 25 (11/44) | 0.4 ± 0.2 | 27 (10/37) | 0.4 ± 0.2 | ||||
| MP-3200 | 53 (31/58) | 1.0 ± 0.3 | 35 (15/43) | 0.6 ± 0.3 | ||||
| MP-2492 | 10 (2/20) | 0.2 ± 0.3 | 44 (8/18) | 0.7 ± 0.5 | ||||
| MP-747 | 10 (2/21) | 0.1 ± 0.2 | 33 (7/21) | 0.4 ± 0.3 | ||||
| AMP2.17 sham | 12 (1/8) | 0.1 ± 0.2 | 12 (1/8) | 0.1 ± 0.2 | ||||
| MP1.24 sham | 58 (11/19) | 1.8 ± 0.9 | 67 (12/18) | 1.3 ± 0.5 | ||||
| None | 6 (2/36) | 0.1 ± 0.1 | 0 (0/36) | 0 | ||||
The sizes of the viral genomic regions are not shown to scale. Nucleotide numbers are given for each region, with the location(s) of the determinants mapped subsequently by sequencing shown in parentheses. The MP genome is shown in black, and the AMP genome is shown in gray. Cloned viruses MP1.24, MP1.23, AMP2.17, and AMP2.22 are listed beneath their respective parental viruses SY8, MP1, and AMP2. All manipulations of recombinant DNA and infectious virus were performed according to the National Institutes of Health Guidelines for Research Involving Recombinant DNA Molecules and the Institutional Biosafety Committee at the University of Minnesota.
Weakness was determined on a scale of 0 to 3, with grade 0 indicating no weakness and grade 3 representing the most severe weakness (27). The final weakness grade is the sum of the grades for the left and right legs of each mouse.
The level of inflammation in the right hamstring was graded on a scale of 0 to 3 as described for Fig. 1.
Values for the grade of weakness and inflammation were obtained at 1 month postinfection and are presented as means ± 2 standard errors of the mean. Comparison of means was performed by analysis of variance with a Student-Newman-Keuls test used for post hoc pairwise comparisons when P was ≤0.05. Some muscle samples could not be scored because of technical difficulties in freezing and cutting. Thus, in some groups, the number of samples scored for inflammation was reduced compared to the number evaluated for weakness.
Use of viral chimeras to localize myopathic determinants.
The MP-3200 chimeras were prepared by exchanging the XbaI/AvrII fragment (viral nt 1 to 3200) of AMP2.17 or AMP2.22 with the homologous region of MP1.23 or MP1.24. These four chimeras behaved similarly in vivo, and the data were pooled for the final analysis (Table 1). Likewise, the four reciprocal chimeras were grouped together as AMP-3200. The MP-2492 chimera was derived from the MP-3200 chimera of MP1.23 and AMP2.17 by exchanging the SfiI/AvrII fragment (nt 2493 to 3200) with that of AMP2.17. Similarly, MP-747 was prepared by replacing the BssHII/AvrII fragment (nt 747 to 3200). Sham chimeras were constructed by replacing the XbaI/AvrII fragment of AMP-3200 or MP-3200 to regenerate intact MP1.24 or AMP2.17, respectively. All chimera junctions were verified by sequencing. The MP-3200 and AMP-3200 chimeras caused significantly reduced levels of weakness and inflammation compared to levels caused by MP1.24 and MP1.23 (P < 0.0001), indicating that myopathic viral determinants were located both upstream and downstream of the nt 3200-nt 3201 junction (Table 1). Levels of inflammation were similar between MP-3200- and AMP-3200-infected mice (P = 0.42), but MP-3200 caused more weakness than AMP-3200 (P = 0.045). Inflammation caused by MP-3200 or AMP-3200 was higher than in uninfected controls (P = 0.0037 or P = 0.041, respectively) but did not differ from that caused by AMP2.17 or AMP2.22. The MP-2492 chimera caused a significant reduction in the incidence and severity of weakness compared to the incidence and severity of weakness caused by MP-3200 (P = 0.011), indicating that there is a viral determinant of weakness located in the region from nt 2493 to 3200. The use of viruses that are closely related makes it less likely that this interpretation is hampered by the masking of disease determinants, which has been described in reports of Theiler's virus chimeras, as reviewed by Jakob and Roos (8). Taken as a whole, the chimeras suggest that weakness and inflammation are caused by multiple viral determinants, with at least one weakness determinant being located in the region of VP1 from nt 2493 to 3200, a determinant of inflammation being located upstream of nt 3201, and additional determinants of weakness and/or inflammation being located downstream of nt 3200.
Viral clones and chimeras were also evaluated for their ability to cause mortality, replicate in skeletal muscle, and induce antiviral immunoglobulin G (IgG) antibodies (Table 2). Mice infected with myopathic virus showed a slightly lower rate of survival than that of mice infected with amyopathic virus. Mice infected with MP1 or its clone MP1.23 showed the lowest survival rates, which differed from those of mice infected with AMP2 and its clones, all the viral chimeras, and the uninfected control (P ≤ 0.038). Survival of mice infected with MP1.24 differed only from that of mice infected with AMP2.17, AMP2, or MP-2492 or the uninfected control mice (P ≤ 0.036), while the survival of mice infected with SY8 or the MP1.24 sham control did not differ from the survival of mice infected with any other viruses or chimeras (P ≥ 0.098). Although myopathic viruses achieved slightly higher titers in muscle during acute infection, none of the virus titers differed significantly from each other. The chimeras and parental viruses also showed comparable levels of replication as determined with one-step growth curves of viruses in BGMK cells (data not shown). Similar results were obtained in our original studies of the parental CVB1T variants, where a more detailed analysis of growth kinetics indicated that the parental AMP2 virus is not defective for replication in vivo or in vitro (24).
TABLE 2.
Comparison of levels of virus virulence and replication and the host antiviral antibody responses in mice infected with CVB1T clones and chimeras
| Virus | % Survivala | Log10 PFU/g of muscleb | Antivirus IgG (U/ml)c |
|---|---|---|---|
| SY8 | 80 ± 9 | 7.94 ± 0.57 | 10,854 ± 2,254 |
| MP1 | 59 ± 26 | 8.69 ± 0.05 | 11,320 ± 3,510 |
| MP1.23 | 63 ± 22 | 7.84 ± 0.49 | 8,973 ± 2,154 |
| MP1.24 | 69 ± 8 | 8.24 ± 0.54 | 9,499 ± 1,746 |
| AMP2 | 93 ± 10 | 7.45 ± 2.04 | 5,445 ± 2,100 |
| AMP2.17 | 89 ± 11 | 6.19 ± 1.44 | 4,930 ± 1,962 |
| AMP2.22 | 87 ± 12 | 6.81 ± 0.94 | 3,060 ± 962 |
| AMP-3200 | 83 ± 16 | 6.67 ± 1.20 | 3,120 ± 800 |
| MP-3200 | 82 ± 10 | 6.62 ± 1.19 | 5,682 ± 1,162 |
| MP-2492 | 95 ± 5 | NT | 6,421 ± 1,862 |
| MP-747 | 96 ± 9 | NT | 5,302 ± 1,606 |
| AMP2.17 sham | 96 ± 7 | NT | 6,133 ± 3,640 |
| MP1.24 sham | 78 ± 24 | NT | 9,423 ± 2,172 |
| None | 95 ± 5 | ND | 0 |
% Survival is the mean percentage of mice that survived per litter ± 2 standard error of the means (at least two litters of mice were infected).
Acute titers of virus were determined at 7 days after infection in proximal hind limb muscle (hamstring and quadriceps) (24). Values are means ± 2 standard errors of the mean of results for at least three mice. NT, not tested; ND, not detected.
The level of antivirus antibody (IgG) was determined by ELISA as described previously (25) and is expressed as units per milliliter based on a laboratory standard assigned a value of 5,000 U/ml. All samples were tested in triplicate, and the number of mice tested per group was the same as the number used to determine weakness in Table 1.
Production of neutralizing antibodies to myopathic CVB1T correlates with the severity of chronic myopathy in this model (9). In the present study, we found that the host antibody response to virus was consistently higher in mice infected with the intact myopathic viruses MP1, SY8, MP1.24, and MP1.23 and the MP1.24 sham chimera than in mice infected with the remaining amyopathic viruses or chimeras (P ≤ 0.0001 to P = 0.042). This finding indicates that differences in the immunogenicities of viral determinants are linked to the pathogenic phenotype of the virus, resulting in either a qualitative or quantitative difference in the immune response to infection. This difference is also apparent in the production of autoantibodies to muscle and nuclear antigens, which are found in mice infected with myopathic, but not amyopathic, virus (23).
Candidate determinants identified by sequencing.
Two myopathic (MP1.23 and MP1.24) and two amyopathic (AMP2.17 and AMP2.22) cDNA clones were completely sequenced. Automated sequencing was performed by PCR using Big Dye Terminator cycle sequencing protocols (PE Applied Biosystems). Sequence was first obtained by gene walking for pMP1.24 (Amplicon Express, Pullman, Wash.). These same primers were then used to sequence clones MP1.23, AMP2.17, and AMP2.22 on an ABI 377 sequencer through the Advanced Genetic Analysis Center at the University of Minnesota. Both strands were sequenced for all clones, with coverage at two to three times per base. Contig assembly and alignments were performed with SeqMan and MegAlign, respectively (DNAStar, Madison, Wis.), with manual editing (16). The nucleotide sequences of MP1.23, MP1.24, and AMP2.17 indicated that the CVB1T genome was 7391 nt in length. The AMP2.22 clone contained three rather than two terminal guanine residues and thus was 7392 nt in length.
CVB1N is the only other type B1 coxsackievirus that has been completely sequenced (7). Sequence information obtained from MP1.24 was compared to that from CVB1N (GenBank accession number M16560) (7), CVB2 Ohio (AF081485), CVB3 (M88483), CVB4 Benschoten (X05690), CVB5 Faulkner (AF114383), and CVB6 Schmitt (AF114384). In the 5′ UTR, pMP1.24 showed the highest percentage of identity with CVB6 (85.3%), followed by CVB5 and CVB3 (84.5%), CVB1 (83.9%), CVB2 (83.3%), and CVB4 (80.6%). The base composition for MP1.24 showed a higher adenine content (28.3% A, 24.6% G, 23.5% C, and 23.7% U), similar to that of CVB1N. Comparative alignments were used to predict the translational start site at nt 743 and cleavage sites within the polyprotein. Seven AUG codons were located upstream of the translational start site. Two of these, at nt 381 and 393, did not occur in CVB1N, but all seven were conserved in CVB2, CVB3, CVB5, and CVB6. The predicted polyprotein of CVB1T consists of 2,182 residues and aligns with no deletions or additions in the coding region relative to that of CVB1N. The percent identity of the capsid region for these two CVB1 strains was 94.6%. In contrast, the capsid region of CVB1T showed at most 82.9% identity with CVB types 2 through 6. The amino acids flanking all predicted cleavage sites of CVB1T were identical to those in CVB1N, CVB3, CVB4, and CVB5. Thus, CVB1T appears to be a typical member of the coxsackievirus group with a high degree of similarity to CVB1N.
Comparison of the four CVB1T clones that were completely sequenced revealed one mutation in the 5′ UTR and four coding changes that differed consistently between myopathic and amyopathic clones (Table 3). Three of the coding changes occurred in capsid protein VP1, with the Thr-to-Ala-276 mutation located only three residues upstream of the VP1/2A cleavage site. The fourth coding mutation occurred in protease 3C. Sequencing of additional virus clones and of amplicons produced by RT-PCR of parental virus RNA indicated that these five primary nucleotide differences were strictly associated with the pathogenic phenotype of the virus. Six additional conserved differences were located within the coding region but were translationally silent, and the possibility remains that one or more of these changes may be involved through effects on RNA secondary structure (5).
TABLE 3.
Nucleotide sequence and coding differences between myopathic and amyopathic viruses
| nt (amino acid or mutation)b | Viral gene | Nucleotide differencesa:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cloned virus (parental virus)
|
Parental virus
|
||||||||||
| MP1.23 (MP1) | MP1.24 (SY8) | MP1.25 (SY8) | MP1.26 (SY8) | AMP2.17 (AMP2) | AMP2.22 (AMP2) | AMP2.15 (AMP2) | SY8 | MP1 | AMP2 | ||
| 485 | UTR | G | G | G | — | G | A | G | — | — | — |
| 706 | UTR | C | C | C | C | T | T | T | C | C | T |
| 996 | VP2 | T | T | T | T | T | T | C | T | T | T |
| 1174 (Ser) | VP2 | G | A | A | A | A | A | G | A | A | A |
| 1426 (Gln) | VP2 | G | G | G | G | A | A | A | G | G | A |
| 1591 (Asn) | VP2 | C | C | C | C | T | T | T | C | C | T |
| 1610 (Leu) | VP2 | C | T | — | — | T | T | T | T | C | T |
| 2332 (Ile) | VP3 | T | C | — | — | T | T | — | — | — | — |
| 2712 (Tyr→Phe-87) | VP1 | A | A | A | A | T | T | T | A | A | T |
| 2815 (Thr) | VP1 | G | G | G | G | T | T | T | G | G | T |
| 2859 (Val→Ala-136) | VP1 | T | T | T | T | C | C | C | T | T | C |
| 3157 (Val) | VP1 | T | A | A | A | A | A | A | A | A | — |
| 3278 (Thr→Ala-276) | VP1 | A | A | A | A | G | G | G | A | A | G |
| 3415 (Ser) | 2A | C | C | C | C | T | T | T | C | C | T |
| 3652 (Gly) | 2A | A | G | A | A | A | A | A | — | — | — |
| 3784 (Gly) | 2B | T | T | T | T | C | C | C | T | T | C |
| 3791 (Asp→Asn) | 2B | G | A | A | A | A | A | A | A | A | A |
| 4080 (Lys→Arg) | 2C | A | A | A | A | G | A | A | A | A | A |
| 5510 (Lys→Glu-53) | 3C | A | A | A | A | G | G | G | A | A | G |
| 5617 (Gly) | 3C | C | C | C | C | A | A | A | C | C | A |
| 5930 (Lys→Glu) | 3D | A | G | A | A | A | A | A | A | A | A |
| 6178 (Thr) | 3D | C | T | T | — | T | T | T | T | T | T |
| 6681 (Arg→His) | 3D | G | A | G | G | A | A | A | A | G | A |
| 6694 (Asn) | 3D | T | C | T | T | C | C | C | — | — | — |
| 6907 (Tyr) | 3D | C | C | C | C | T | C | C | — | — | — |
| 7075 (Pro) | 3D | C | T | C | C | C | C | C | — | — | — |
Viral cDNA clones MP1.23, MP1.24, AMP2.17, and AMP2.22 were completely sequenced, and any position which differed between any of these four clones is shown. The remaining clones and parental viruses were sequenced only in the areas of interest. A dash indicates that the position was not sequenced. All positions with nucleotide differences in the UTR or coding regions that were completely conserved among the MP and AMP viruses are shown in bold.
All amino acid changes are shown relative to the sequence of MP1.23. Predicted coding changes are as indicated in parentheses. The five primary nucleotide differences that occurred in the UTR or resulted in a coding change are in italics, and the amino acid number within the viral protein is given.
The weakness determinant that mapped to the region from nt 2493 to 3200 (Table 1) includes the coding differences identified at Tyr-87 and Val-136. If CVB1T is aligned with the three-dimensional structure of CVB3 determined by Muckelbauer et al. (11), Tyr-87 is expressed on the virion surface adjacent to the BC loop, and Val-136 is located within the DE loop of VP1. The BC loop has been mapped as an important neutralizing immunogenic site (20) and is involved in receptor binding of poliovirus 2-Lansing (14). The DE loop of poliovirus type 2 contains a major attenuation determinant (3, 21) and also affects antigenicity and host range (21, 28). Furthermore, Thr-129 located within the DE loop of CVB4 is part of an immunodominant conformational B-cell epitope and a major determinant of pancreatitis and hypoglycemia (1, 6, 18). These parallels between the potential involvement of Tyr-87 and/or Val-136 in the pathogenesis of CIM with related antigenic determinants in other enteroviruses lead us to speculate that there may be shared underlying mechanisms of pathogenesis mediated by an immunopathic response to VP1. While we have not yet conclusively identified which of the candidate determinants are important, these mapping studies point to the presence of multiple viral determinants which interact with the host immune response and promote the development of chronic immunopathic muscle disease.
Nucleotide sequence accession numbers.
Sequences for all four clones starting with nt 21 to the 3′ end are available in GenBank (accession numbers AY186745 for pMP1.24, AY186746 for pMP1.23, AY186747 for pAMP2.17, and AY186748 for pAMP2.22).
Acknowledgments
This work was supported by Public Health Service grant AI36223 from the National Institute of Allergy and Infectious Diseases and a grant from the North Central Chapter of the Arthritis Foundation (P.E.T.).
We thank Stacey Orvik and Wei Li for excellent technical assistance. We also thank Maribeth Sandager and the College of Biological Sciences Imaging Center at the University of Minnesota for assistance with digital imaging.
REFERENCES
- 1.Caggana, M., P. Chan, and A. Ramsingh. 1993. Identification of a single amino acid residue in the capsid protein VP1 of coxsackievirus B4 that determines the virulent phenotype. J. Virol. 67:4797-4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 3.Equestre, M., D. Genovese, F. Cavalieri, L. Fiore, R. Santoro, and R. Perez Bercoff. 1991. Identification of a consistent pattern of mutations in neurovirulent variants derived from the sabin vaccine strain of poliovirus type 2. J. Virol. 65:2707-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galbraith, D. N., C. Nairn, and G. B. Clements. 1997. Evidence for enteroviral persistence in humans. J. Gen. Virol. 78:307-312. [DOI] [PubMed] [Google Scholar]
- 5.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halim, S., and A. I. Ramsingh. 2000. A point mutation in VP1 of coxsackievirus B4 alters antigenicity. Virology 269:86-94. [DOI] [PubMed] [Google Scholar]
- 7.Iizuka, N., S. Kuge, and A. Nomoto. 1987. Complete nucleotide sequence of the genome of coxsackievirus B1. Virology 156:64-73. [DOI] [PubMed] [Google Scholar]
- 8.Jakob, J., and R. P. Roos. 1996. Molecular determinants of Theiler's murine encephalomyelitis-induced disease. J. Neurovirol. 2:70-77. [DOI] [PubMed] [Google Scholar]
- 9.Jongen, P. J., F. W. Heessen, H. J. ter Laak, J. M. Galama, and F. J. Gabreels. 1994. Coxsackie B1 virus-induced murine myositis: relationship of disease severity to virus dose and antiviral antibody response. Neuromusc. Disord. 4:17-23. [DOI] [PubMed] [Google Scholar]
- 10.Klingel, K., B. M. McManus, and R. Kandolf. 1995. Enterovirus-infected immune cells of spleen and lymph nodes in the murine model of chronic myocarditis: a role in pathogenesis? Eur. Heart J. 16:42-45. [DOI] [PubMed] [Google Scholar]
- 11.Muckelbauer, J. K., M. Kremer, I. Minor, G. Diana, F. J. Dutko, J. Groarke, D. C. Pevear, and M. G. Rossmann. 1995. The structure of coxsackievirus B3 at 3.5 Å resolution. Structure 3:653-667. [DOI] [PubMed] [Google Scholar]
- 12.Muir, P., and L. C. Archard. 1994. There is evidence for persistent enterovirus infections in chronic medical conditions in humans. Rev. Med. Virol. 4:245-250. [Google Scholar]
- 13.Muir, P., F. Nicholson, S. J. Illavia, T. S. McNeil, J. F. Ajetunmobi, H. Dunn, W. G. Starkey, K. N. Reetoo, N. R. Cary, J. Parameshwar, and J. E. Banatvala. 1996. Serological and molecular evidence of enterovirus infection in patients with end-stage dilated cardiomyopathy. Heart 76:243-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray, M. G., J. Bradley, X.-F. Yang, E. Wimmer, E. G. Moss, and V. R. Racaniello. 1988. Poliovirus host range is determined by a short amino acid sequence in neutralization antigenic site I. Science 241:213-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pallansch, M. A. 1997. Coxsackievirus B epidemiology and public health concerns. Curr. Top. Microbiol. Immunol. 223:13-30. [DOI] [PubMed] [Google Scholar]
- 16.Parker, L. T., H. Zakeri, Q. Deng, S. Spurgeon, P.-Y. Kwok, and D. A. Nickerson. 1996. AmpliTaq DNA polymerase, FS dye-terminator sequencing: analysis of peak height patterns. BioTechniques 21:694-699. [DOI] [PubMed] [Google Scholar]
- 17.Ramsingh, A. I., N. Chapman, and S. Tracy. 1997. Coxsackieviruses and diabetes. Bioessays 19:793-800. [DOI] [PubMed] [Google Scholar]
- 18.Ramsingh, A. I., W. T. Lee, D. N. Collins, and L. E. Armstrong. 1997. Differential recruitment of B and T cells in coxsackievirus B4-induced pancreatitis is influenced by a capsid protein. J. Virol. 71:8690-8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ray, C. G., L. L. Minnich, and P. C. Johnson. 1979. Selective polymyositis induced by coxsackievirus B1 in mice. J. Infect. Dis. 140: 239-243. [DOI] [PubMed] [Google Scholar]
- 20.Reimann, B. Y., R. Zell, and R. Kandolf. 1991. Mapping of a neutralizing antigenic site of coxsackievirus B4 by construction of an antigen chimera. J. Virol. 65:3475-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren, R., E. G. Moss, and V. R. Racaniello. 1991. Identification of two determinants that attenuate vaccine-related type 2 poliovirus. J. Virol. 65:1377-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strongwater, S. L., Z. K. Dorovini, R. D. Ball, and T. J. Schnitzer. 1984. A murine model of polymyositis induced by coxsackievirus B1 (Tucson strain). Arthritis Rheum. 27:433-442. [DOI] [PubMed] [Google Scholar]
- 23.Tam, P. E., D. R. Fontana, and R. P. Messner. Coxsackievirus B1-induced chronic inflammatory myopathy: differences in induction of autoantibodies to muscle and nuclear antigens by cloned myopathic and amyopathic viruses. J. Lab. Clin. Med., in press. [DOI] [PubMed]
- 24.Tam, P. E., and R. P. Messner. 1997. Coxsackievirus-induced chronic inflammatory myopathy: virus variants distinguish between acute cytopathic effects and pathogenesis of chronic disease. Virology 233:199-209. [DOI] [PubMed] [Google Scholar]
- 25.Tam, P. E., and R. P. Messner. 1996. Genetic determinants of susceptibility to coxsackievirus B1-induced chronic inflammatory myopathy: effect of host background and major histocompatibility complex genes. J. Lab. Clin. Med. 128:279-289. [DOI] [PubMed] [Google Scholar]
- 26.Tam, P. E., A. M. Schmidt, S. R. Ytterberg, and R. P. Messner. 1994. Duration of virus persistence and its relationship to inflammation during the chronic phase of coxsackievirus B1-induced murine polymyositis. J. Lab. Clin. Med. 123:346-356. [PubMed] [Google Scholar]
- 27.Tam, P. E., A. M. Schmidt, S. R. Ytterberg, and R. P. Messner. 1991. Viral persistence during the developmental phase of coxsackievirus B1-induced murine polymyositis. J. Virol. 65:6654-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiegers, K., H. Uhlig, and R. Dernick. 1989. N-AgIB of poliovirus type 1: a discontinuous epitope formed by two loops of VP1 comprising residues 96-104 and 141-152. Virology 170:583-586. [DOI] [PubMed] [Google Scholar]
- 29.Yousef, G. E., D. A. Isenberg, and J. F. Mowbray. 1990. Detection of enterovirus specific RNA sequences in muscle biopsy specimens from patients with adult onset myositis. Ann. Rheum. Dis. 49: 310-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ytterberg, S. R., M. L. Mahowald, and R. P. Messner. 1987. Coxsackievirus B 1-induced polymyositis. Lack of disease expression in nu/nu mice. J. Clin. Investig. 80:499-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ytterberg, S. R., M. L. Mahowald, and R. P. Messner. 1988. T cells are required for coxsackievirus B1 induced murine polymyositis. J. Rheumatol. 15:475-478. [PubMed] [Google Scholar]