Abstract
The hepatitis C virus (HCV) causes chronic hepatitis in most infected individuals by evading host immune defenses. In this investigation, we show that HCV-infected cells may go undetected in the immune system by suppressing major histocompatibility complex (MHC) class I antigen presentation to cytotoxic T lymphocytes. Cells expressing HCV subgenomic replicons have lower MHC class I cell surface expression. This is due to reduced levels of properly folded MHC class I molecules. HCV replicons induce endoplasmic reticulum (ER) stress (K. Tardif, K. Mori, and A. Siddiqui, J. Virol. 76:7453-7459, 2002), which results from a decline in protein glycosylation. Decreasing protein glycosylation can disrupt protein folding, preventing the assembly of MHC class I molecules. This results in the accumulation of unfolded MHC class I. Therefore, the persistence and pathogenesis of HCV may depend upon the ER stress-mediated interference of MHC class I assembly and cell surface expression.
Hepatitis C virus (HCV) infection often leads to chronic hepatitis, which can progress to liver cirrhosis and hepatocellular carcinoma (13). HCV consists of a 9.6-kb positive-sense single-stranded RNA genome (6, 36). Translation of the HCV genome is initiated by an internal ribosome entry site located in the 5′ untranslated region (38, 43, 44). The resulting 3,000-amino-acid polyprotein of translation is cleaved into at least three structural proteins (core, E1, and E2) and six nonstructural proteins (NS2 to NS5A/B) (6, 36). Many of the nonstructural protein products are essential for productive viral replication (23). Changes in intracellular events resulting from HCV replication are poorly understood. The development of selectable and efficiently replicating HCV subgenomic replicons in a human hepatoma cell line, Huh7, has provided a suitable model for investigating the mechanisms of viral persistence and pathogenesis in the context of HCV replication (27).
For HCV to establish persistent infections, the virus must evade host immune responses. An immune response begins with the presentation of viral antigens to cytotoxic T lymphocytes (CTLs). To present viral antigen at the cell surface to CTLs, major histocompatibility complex (MHC) class I heavy chains must be glycosylated and correctly folded in the endoplasmic reticulum (ER) to form a complex with β2-microglobulin and a short peptide derived from the viral antigen (reviewed in reference 34). Failure to glycosylate or properly fold MHC class I heavy chains results in their slow or inefficient transport to the cell surface (9, 11, 31, 34, 35). However, once they are sufficiently processed, MHC class I heavy chains are loaded with antigenic peptides generated by the proteasome, and stable class I-β2-microglobulin-peptide complexes can be transported to the cell surface through the Golgi.
Viruses have numerous mechanisms for interfering with MHC class I antigen presentation (reviewed in reference 42). Some viral proteins bind to and retain class I molecules in the ER (1, 4, 8, 21). Others target class I molecules for degradation by the proteasome or lysosome (20, 45). Viral proteins have also been shown to suppress proteasome degradation of antigens in the cytoplasm (15) or interfere with the transfer of antigen peptide fragments into the ER (2, 3, 19). The strategies employed by HCV to avoid detection and elimination by the immune system have not been investigated.
HCV nonstructural proteins are associated with the ER in a ribonucleoprotein replication complex (18, 29), and we have shown that their expression from an HCV replicon induces ER stress (16, 41). This stress results from the inhibition of protein glycosylation, which disrupts protein folding, causing the accumulation of unfolded proteins in the ER. In response to ER stress, cells activate an intracellular signaling pathway known as the unfolded protein response (reviewed in reference 30). The unfolded protein response activates the transcriptional induction of chaperones and enzymes required for protein folding. In a cellular environment where protein folding is disrupted, cells are further burdened by the continuous production of protein. Cells alleviate this added stress by attenuating protein synthesis. However, translation is enhanced in cells expressing HCV replicons (41), suggesting that these cells are not fully capable of recovering from ER stress.
In this study, we investigated the effects of HCV replication and ER stress on MHC class I cell surface expression and antigen presentation. Cells stably expressing HCV replicons have far less MHC class I cell surface expression compared to control cells. This can be attributed to lower levels of properly folded MHC class I. Lower protein glycosylation levels may contribute to the accumulation of unfolded MHC class I. HCV persistence and pathogenesis may rely upon the ER stress-mediated suppression of MHC class I cell surface expression.
MATERIALS AND METHODS
Cell culture.
Huh7 and FCA4 human hepatoma cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U of penicillin per ml, and 100 μg of streptomycin sulfate per ml. FCA4 and GS4.3 cells (kindly provided by C. Seeger of the Fox Chase Center, Philadelphia, Pa.) were also grown in G418 (500 μg/ml; Gibco-BRL). FCA4 and GS4.3 cells are HCV replicon-containing Huh7 cell lines. FCA4 cells stably express a HCV subgenomic replicon with a single adaptive mutation, a deletion of serine residue 1176 (17). The replicons expressed in FCA4 and GS4.3 cells are bicistronic constructs composed of the HCV internal ribosome entry site (nucleotides 1 to 377 of the 5′ noncoding region), the neomycin phosphotransferase (neo) gene, the encephalomyocarditis internal ribosome entry site, which mediates the translation of HCV nonstructural proteins NS3 through NS5, and the 3′ noncoding region (27). A neomycin-resistant Huh7 cell line was constructed by transfecting Huh7 cells with pRc/CMV, a plasmid encoding a neomycin resistance gene, and selecting for transfected cells with G418 (500 μg/ml) over a 3-week period. Cells were harvested for each experiment at 70% confluence under the same growth conditions unless otherwise indicated. Cells were maintained at 5% CO2 and 37°C.
Antibodies.
W6/32 is a monoclonal antibody that recognizes correctly folded HLA-A, -B, and -C molecules associated with β2-microglobulin (5). The monoclonal antibody CD13 was a gift from K. V. Holmes (University of Colorado Health Sciences Center, Denver, Colo.). Fluorescein isothiocyanate (FITC)-coupled anti-mouse immunoglobulin was used as a secondary antibody in fluorescence-activated cell sorting experiments. The polyclonal antibody HC70 recognizes unfolded free MHC class I heavy chains (40).
Flow cytometry.
Cells were detached from culture dishes with phosphate-buffered saline (PBS) supplemented with 0.5 mM EDTA. The detached cells were washed two times with PBS. The cells were finally resuspended in PBS containing 2% FBS at a concentration of 106 cells/ml. MHC class I cell surface staining was detected with the W6/32 antibody. FITC-conjugated anti-mouse immunoglobulin was used as the secondary antibody. The primary and secondary antibodies were incubated with the cells for 30 min at 4°C. The cells were washed three times with PBS supplemented with 2% FBS after each incubation period. Flow cytometry was performed at the University of Colorado Health Sciences Center Flow Cytometry Core Facility in a Coulter XL with System II software (Beckman Coulter, Hialeah, Fla.). Cytotoxicity assays were performed as described previously (24). Huh7, FCA4, and GS4.3 cells were incubated with Na251CrO4 for 1 h at 37°C in 5% CO2. After these cells were washed three times with R-10 medium, the hepatoma cell lines were incubated with HCV-specific CTL clones at 37°C for 4 h in three replicate wells. Cellular release of 51CrO4 into the supernatant was measured with a Top Count Mircroplate scintillation counter. The HCV CTL clones were restricted by HLA A11. HLA typing of Huh7, FCA4, and GS4.3 cells was performed by ClinImmune Labs.
Pulse-chase experiments.
Cells were incubated for 30 min in methionine- and cysteine-free DMEM supplemented with 2 mM glutamine, antibiotics, and 10% FBS. Cells were then labeled for the indicated times with 200 μCi of Trans 35S label (1,175 Ci/mmol; ICN) per milliliter of the above medium. To chase 35S-labeled proteins, cells were incubated in complete DMEM for the indicated times. Cells were lysed in a commonly used nondenaturing Nonidet P-40 lysis buffer (50 mM Tris-HCl [pH 7.4], 0.5% NP-40, 5 mM MgCl2) (20, 40). This lysis buffer was used to extract MHC class I from the ER through the cell surface. The lysates were centrifuged to pellet nuclei and debris. Lysates used for immunoprecipitations were normalized to equal trichloroacetic acid-precipitable 35S-labeled protein. Immunoprecipitations were carried out with specific antiserum and protein G-Sepharose (Amersham Pharmacia Biotech) at 4°C. The immunoprecipitates were washed five times with 1 ml each of NP-40 wash buffer (50 mM Tris-HCl [pH 7.4], 0.5% NP-40, 5 mM EDTA, 150 mM NaCl) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Brefeldin A (10 μg/ml; Sigma), MG132 (25 μM; Sigma), and lactacystin (20 μM; Sigma) were added to the pulse and chase media at the indicated concentrations. Endoglycosidase Hf (New England Biolabs) was added to immunoprecipitates according to the manufacturer's instructions.
Cell permeabilization and fractionation by squeeze-out centrifugation.
Cells were pulse-labeled as described above. They were resuspended in PB (25 mM HEPES [pH 7.3], 115 mM potassium acetate, 5 mM sodium acetate, 2.5 mM MgCl2, 0.5 mM EGTA) containing 0.025% digitonin and protease inhibitors (1 mM phenylmethylsulfonyl fluoride, aprotinin [10 μg/ml], and leupeptin [10 μg/ml]) (40). Soluble cytoplasmic proteins were squeezed out of permeabilized cells by centrifugation at 14,000 rpm at 4°C for 10 min. Two samples were taken for each permeabilization reaction. The supernatant and pellet of one sample were remixed to represent the total starting material. The pellet fraction was resuspended in PB containing digitonin and protease inhibitors. SDS, dithiothreitol, and NP-40 were added to the lysates to final concentrations of 0.1%, 0.2 mM, and 0.5%, respectively. After a clarifying spin, unfolded MHC I heavy chain was immunoprecipitated with HC70 antibody and protein G-Sepharose. The immunoprecipitates were washed five times in PB containing digitonin and protease inhibitors and analyzed by SDS-PAGE.
Plasmids.
MHC class I transcriptional activity was measured with wild-type, −205/+2 ATG, and mutant, −335BII102/+2 ATG, HLA-A11 luciferase reporters, gifts from C. Gazin (Massachusetts General Hospital, Boston) (25). The construct −335BII102/+2 ATG contains a mutation disrupting the NF-Y binding site (−105 to −83) (25). The NF-Y binding site is an essential element for MHC class I transcription (28, 39). This mutant luciferase reporter was used as a negative control.
Cell transfections and luciferase assays.
Cells at approximately 70% confluence were transfected by using Lipofectin reagent (Gibco-BRL). Huh7 and FCA4 cells were transfected with 2 μg of the indicated luciferase reporter plasmid. Luciferase activity was determined by standard procedures. The transfection efficiency of Huh7 and FCA4 cells was normalized with a luciferase reporter plasmid.
Metabolic labeling.
The rate of glycoprotein and protein synthesis in Huh7 and FCA4 cells was followed by labeling cells with d-[1-14C]glucosamine (53 mCi/mmol; ICN) and Trans 35S label (1,175 Ci/mmol; ICN). To follow the rate of glycoprotein synthesis, cells were labeled in 1 ml of complete medium containing [14C]glucosamine (1 μCi/ml). Protein synthesis was monitored in cells labeled with Trans 35S label (20 μCi/ml) in 1 ml of methionine- and cysteine-free DMEM supplemented with 2 mM glutamine, antibiotics, and 10% FBS. At the indicated times, the trichloroacetic acid-precipitable radioactivity in 30 μg of protein from cell lysates was determined by liquid scintillation counting. This assay was performed three times, producing comparable results.
RESULTS
HCV replicons interfere with MHC class I cell surface expression.
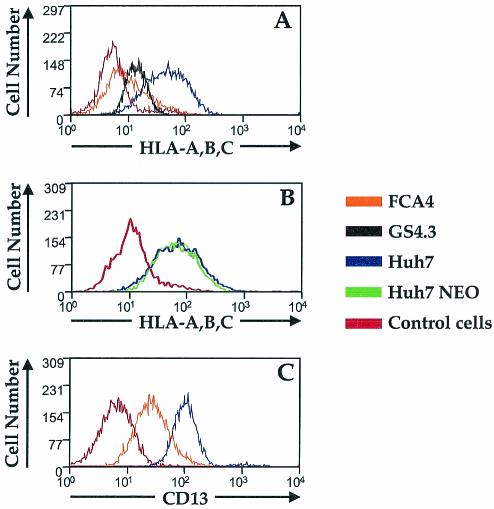
Viruses can escape eradication from CTLs by reducing the amounts of MHC class I molecules on the surface of infected cells. In FCA4 cells expressing HCV subgenomic replicons, fluorescence-activated cell sorting analysis revealed significantly lower cell surface labeling with the conformation-specific class I antibody W6/32, an antibody which only recognizes properly folded MHC class I-β2-microglobulin complexes (Fig. 1A). FCA4 cells displayed 85% less W6/32-specific staining than Huh7 control cells. Another HCV replicon-expressing cell line, GS4.3, also showed a significant reduction in class I cell surface expression (Fig. 1A), suggesting that the lower surface expression is not a unique characteristic of FCA4 cells. The interference with MHC class I expression influences the ability of CTLs to target both FCA4 and GS4.3 cells. HCV-specific CTL lines were unable to recognize and lyse either FCA4 or GS4.3 cells (data not shown). These results indicate that the expression of HCV replicons leads to reduced cell surface expression of MHC class I.
FIG. 1.
HCV replicons affect MHC class I cell surface expression. (A) Huh7 (blue), FCA4 (orange), and GS4.3 (black) cells were analyzed by flow cytometry after being stained with W6/32 monoclonal antibody against folded HLA-A, -B, -C, and FITC-conjugated anti-mouse antibody. (B) Neomycin-resistant Huh7 cells (green) and parental Huh7 cells (blue) were analyzed by flow cytometry after being stained with W6/32 antibody and FITC-conjugated anti-mouse antibody. (C) Huh7 (blue) and FCA4 (orange) cells were stained with CD13 monoclonal antibody and FITC-conjugated anti-mouse antibody before being analyzed by flow cytometry. The red histograms in panels A, B, and C are negative controls, where cells were incubated only with FITC-conjugated anti-mouse antibody.
Unlike Huh7 cells, both FCA4 and GS4.3 cells are under neomycin (G418) selection pressure. To determine if this could account for the differences in class I cell surface expression, we analyzed the cell surface expression of class I in Huh7 cells stably expressing a neomycin resistance gene. G418-resistant Huh7 cells exhibited the same degree of MHC class I expression at the cell surface as the parental Huh7 cells (Fig. 1B).
The reduction in MHC class I cell surface expression in HCV replicon-expressing cells does not appear to be specific to class I molecules. Like MHC class I, CD13 is cotranslationally glycosylated in the ER before it is transported through the Golgi to the cell surface (reviewed in reference 37). CD13 surface expression in FCA4 cells was also reduced 65% (Fig. 1C). These results imply that HCV replicons interfere with the general expression of glycoproteins at the cell surface.
Lower levels of properly folded MHC class I molecules in cells expressing HCV replicons are not the result of reduced class I heavy chain expression.
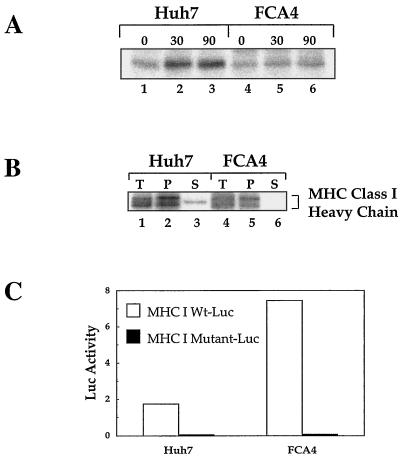
To determine whether HCV replicons affect MHC class I maturation, correctly folded MHC class I molecules were immunoprecipitated from 35S-labeled FCA4 cell lysates with W6/32 antibody. In cells expressing HCV replicons, the amount of W6/32-reactive class I heavy chain was significantly lower in the course of the 90-min chase period compared to the amount in Huh7 cells (Fig. 2A). However, MHC class I molecules slightly increased in FCA4 cells at later chase times (Fig. 2A, lanes 4 to 6), suggesting a mild stabilization of MHC class I.
FIG. 2.
Lower levels of folded MHC class I protein in FCA4 cells expressing HCV replicons are not due to the downregulation of class I expression. (A) HCV replicons have smaller amounts of folded MHC class I. Huh7 and FCA4 cells were 35S-pulse-labeled for 1 h and chased for the indicated times. The cells were washed, lysed, and immunoprecipitated with W6/32, an antibody that recognizes folded MHC class I. (B) FCA4 cells and Huh7 cells have similar amounts of class I heavy chain protein expression. After cells were 35S-pulse-labeled for 1 h, MHC class I heavy chains were immunoprecipitated from Huh7 and FCA4 cell lysates under denaturing conditions with HC70, an antibody that recognizes unfolded MHC class I heavy chains. Fractionation of permeabilized Huh7 and FCA4 cells was performed by squeeze-out centrifugation. Total starting material (T), pellet (P), and supernatant (S) fractions are labeled. (C) FCA4 cells with HCV replicons activate MHC I transcription. Huh7 and FCA4 cells were transfected with MHC I wild-type (Wt, open bars) and mutant (solid bars) luciferase reporters. The mutant MHC I reporter has a disrupted NF-Y binding site.
A number of viruses suppress MHC class I assembly and antigen presentation by downregulating the expression of class I genes (14). To analyze the protein levels of MHC class I heavy chains, heavy chains were immunoprecipitated from 35S-labeled Huh7 and FCA4 cell lysates under denaturing conditions with antibody specific for unfolded MHC class I heavy chains (HC70). The Huh7 and FCA4 cell lines had similar amounts of class I heavy chains (Fig. 2B, compare lanes 1 and 4). By performing a fractionation experiment in which the ER and other membrane-bound components were separated from the cytosol, we were able to show that the majority of MHC class I heavy chain found in Huh7 and FCA4 cells were localized to the ER and other membranes (Fig. 2B, lanes 2 and 5). Only small amounts of class I heavy chain were found in the cytoplasmic fraction of Huh7 cells (Fig. 2B, lane 3), and no heavy chain was detected in the cytoplasm of FCA4 cells (Fig. 2B, lane 6). This indicates that MHC class I heavy chains are inserted into the ER during translation. Further examination of class I expression showed that FCA4 cells expressing HCV subgenomic replicons stimulated the transcription of an MHC class I heavy chain promoter about fourfold over that in Huh7 cells (Fig. 2C). Therefore, smaller amounts of folded MHC I complexes in FCA4 cells are not the result of either suppressed class I heavy chain protein synthesis or mRNA expression.
HCV replicons inhibit the production of glycosylated proteins.
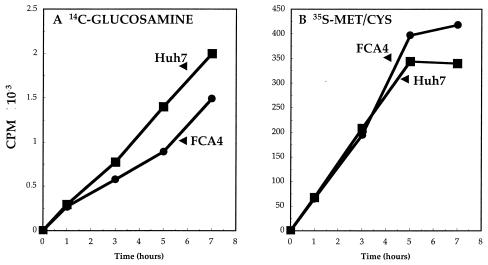
The glycosylation of MHC class I heavy chains is necessary for efficient class I folding and assembly (9). To determine the effect of HCV replicons on glycoprotein synthesis, we followed the time course of [14C]glucosamine incorporation into FCA4 and Huh7 cells (Fig. 3A). It is evident that glucosamine labeling was lower by approximately 30% in cells expressing HCV replicons. The reduction in glucosamine incorporation in FCA4 cells cannot be attributed to lower protein synthesis. [35S]methionine/cysteine labeling showed that translation occurred to a greater extent in FCA4 cells, especially at later time points (Fig. 3B). Reduced protein glycosylation causes ER stress in HCV replicon-containing cells (41).
FIG. 3.
Lower glucosamine incorporation into macromolecules observed in cells expressing HCV replicons. Huh7 and FCA4 cells were grown in 1 ml of medium containing [14C]glucosamine (1 μCi/ml) (A) or [35S]methionine and [35S]cysteine (20 μCi/ml) (B). At the indicated times, the trichloroacetic acid-precipitable radioactivity was determined in Huh7 (▪) and FCA4 (•) cells.
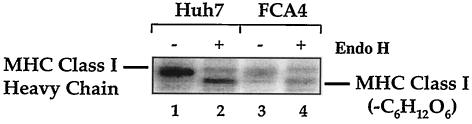
The MHC class I heavy chain is converted to a higher-molecular-mass form after its glycosylation in the ER and early Golgi with high-mannose glycans. After the glycosylated class I heavy chain forms a stable complex with β2-microglobulin and peptide, the class I molecules are transported to the medial Golgi, where they are further modified with complex glycans, including sialic acid (32). Endoglycosidase H cleaves glycans found on proteins in the ER and early Golgi but is resistant to complex glycans found on glycoproteins in the medial Golgi. To characterize the oligosaccharide modifications found on MHC class I heavy chains in Huh7 and FCA4 cells, we pulse-labeled cells in an experiment identical to that shown in lanes 1 and 4 of Fig. 2A and treated the immunoprecipitated samples with endoglycosidase H (Fig. 4). When W6/32 immunoprecipitates from Huh7 and FCA4 cells were treated with endoglycosidase H, a single glycan was removed from the higher-molecular-mass species of class I heavy chains (Fig. 4, compare lanes 2 and 4). After an hour of pulse-labeling, most MHC class I heavy chains were glycosylated in Huh7 and FCA4 cells. However, more MHC class I heavy chain molecules were resistant to endoglycosidase H in Huh7 cells than in FCA4 cells (Fig. 4, compare lanes 2 and 4), suggesting that fewer MHC class I heavy chains are transported to the Golgi in FCA4 cells.
FIG. 4.
Analysis of the glycan modifications on MHC class I heavy chains indicates that less folded class I can be found in the Golgi of cells expressing HCV replicons. Huh7 and FCA4 cells were 35S-radiolabeled for 1 h and immunoprecipitated with W6/32. The immunoprecipitated samples were digested with endoglycosidase H (Endo H) or not and subjected to SDS-PAGE. The endoglycosidase H-sensitive forms of MHC class I heavy chains are labeled as MHC class I (−C6H12O6).
Reduced levels of folded MHC class I in HCV replicon-expressing cells are not influenced by inhibiting ER-to-Golgi transport or proteasome activity.
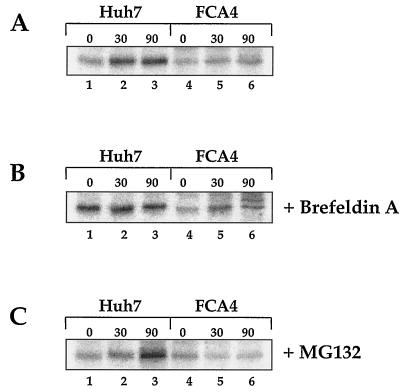
MHC class I transverses the Golgi en route to the cell surface. Brefeldin A inhibits ER-to-Golgi traffic. Treating FCA4 cells expressing HCV replicons with brefeldin A had little effect on the amount of folded MHC class I (Fig. 5). There was a slight increase in folded MHC class I in FCA4 cells after a 30-min chase period (Fig. 5B, compare lanes 4 and 5), but the 90-min time point (lane 6) appeared to have as much MHC class I as the sample at the starting point of the chase (lane 4). The higher-molecular-weight band in Fig. 5B (lane 6) is likely a protein in FCA4 cells that coimmunoprecipitates with MHC class I. Since brefeldin A redistributes Golgi-resident proteins from the Golgi to the ER, this band may be from a protein typically localized in the Golgi. More importantly, each time point in the chase for FCA4 cells shows significantly lower levels of W6/32-reactive MHC class I molecules than Huh7 cells treated with brefeldin A (Fig. 5B). These results imply that the decreased production of folded MHC class I in FCA4 cells is the result of events preceding ER-to-Golgi transport.
FIG. 5.
Lower levels of folded MHC class I found in HCV replicons expressing cells are not influenced by inhibiting ER-to-Golgi transport or proteasome activity. Huh7 and FCA4 cells were 35S-pulse-labeled for 1 h and chased for the indicated times (A) in the absence of inhibitor, (B) in the presence of 10 μg of brefeldin A per ml, or (C) in the presence of 25 μM MG132.
The generation of antigenic peptides by the proteasome is essential to MHC class I assembly. When cells were exposed to the proteasome inhibitor MG132, folded MHC class I levels seen in FCA4 cells were unchanged (Fig. 5). In Huh7 cells, the amounts of folded class I appeared to increase during the 90-min chase period. While inhibiting antigenic peptide synthesis limited the assembly of MHC class I in Huh7 cells, the reduction of antigenic peptide did not constrain the amounts of folded class I in FCA4 cells. Consequently, the reduced amount of properly folded class I in HCV replicon-expressing cells cannot be linked to the proteasome.
HCV replicons induce the accumulation of unfolded MHC class I molecules.
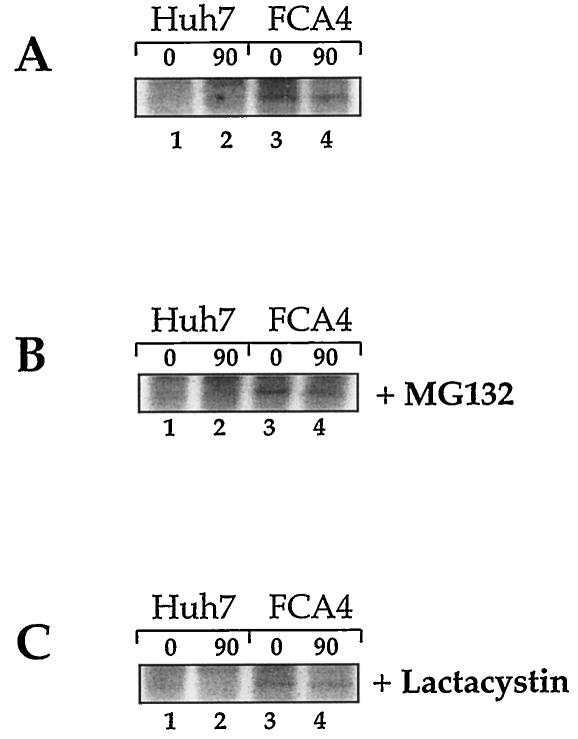
Lower levels of intracellular protein glycosylation induce ER stress, and this leads to an accumulation of unfolded proteins in the ER (reviewed in reference 30). Antibody specific to unfolded MHC class I heavy chains (HC70) was used to investigate whether unfolded MHC class I accumulates in HCV replicon-expressing cells. After recovering unfolded MHC class I heavy chain from 35S-labeled FCA4 cell lysates under nondenaturing conditions by immunoprecipitation, it was clear that FCA4 cells had elevated levels of unfolded class I heavy chains (Fig. 6A). Unfolded protein can be transported out of the ER, where they are degraded by the proteasome (reviewed in reference 7). The unfolded heavy chains isolated from FCA4 cells were stable over the course of a 90-min chase (Fig. 6A, lanes 3 and 4). Treating cells with either of the proteasome inhibitors, MG132 or lactacystin, did not increase the amounts of unfolded heavy chain found in FCA4 cells (Fig. 6). This suggests that unfolded MHC class I is not targeted for proteasome degradation in cells expressing HCV replicons.
FIG. 6.
FCA4 cells expressing HCV replicons have elevated levels of unfolded MHC class I and the levels of MHC class I are not affected by the proteasome inhibitors MG132 and lactacystin. Huh7 and FCA4 cells were 35S-pulse-labeled for 1 h and chased for the indicated times (A) in the absence of inhibitor, (B) in the presence of 25 μM MG132, or (C) in the presence of 20 μM lactacystin. The cells were washed, lysed, and immunoprecipitated with HC70 under nondenaturing conditions.
DISCUSSION
In acute HCV infections, CTLs mount a specific response against virus-infected cells resulting in immediate viral clearance (reviewed in reference 22). In contrast to acute hepatitis, chronic hepatitis elicits a weak CTL response (reviewed in reference 10). The role of CTLs in chronic infections is not clearly understood. HCV causes chronic hepatitis in most infected individuals. For HCV to persist in infected cells, the virus must evade immune detection and subsequent cytolysis. The MHC class I cell surface presentation of viral antigens to CTLs is required for an immune response. Many persistent viruses interfere with MHC class I antigen presentation to escape the immune system.
In the present study, we investigated the strategy used by HCV to elude host immune defenses. Cells expressing HCV replicons have reduced cell surface expression of MHC class I molecules (Fig. 1). This is a direct result of smaller amounts of correctly folded MHC class I (Fig. 2A). Reduced protein glycosylation in HCV replicon-expressing cells (Fig. 3) may suppress MHC class I assembly, because heavy chain glycosylation contributes to class I processing (9). Furthermore, endoglycosidase H digestion of class I molecules shows that HCV replicon-expressing cells also have smaller amounts of class I molecules that proceed as far as the medial Golgi (Fig. 4). Lower protein glycosylation also induces ER stress, and this may trigger the accumulation of unfolded MHC class I in the ER (Fig. 6).
A number of viral proteins have been shown to downregulate MHC class I antigen presentation at every conceivable level. Among all of these viral proteins, only the adenovirus E3/19K protein has been shown to induce ER stress (33). Adenovirus E3 associates with MHC class I and physically retains class I in the ER. However, unlike adenovirus E3, HCV nonstructural proteins do not associate with MHC class I heavy chains (data not shown), suggesting that HCV replicons induce physiological changes in cells to decrease the surface expression of MHC class I. HCV replicons do not cause MHC class I to completely disappear from the cell surface. Elimination of class I from the cell surface would put the virus at risk because natural killer cells can recognize cells deficient in MHC cell surface expression.
When HCV replicons initiate ER stress, cells respond by activating the expression of chaperones and enzymes involved in protein folding in the ER. Although HCV replicon-expressing cells have lower levels of folded MHC class I, the levels do increase slightly after a 90-min chase period compared to those in Huh7 cells (Fig. 2A). This observation is probably a consequence of the upregulation of protein-folding machinery in response to ER stress. The smaller amounts of folded MHC class I molecules in FCA4 cells cannot be due to decreased class I expression, as heavy-chain protein synthesis and mRNA expression are not downregulated in these cells (Fig. 2B and 2C).
The inhibition of protein glycosylation induces a number of physiological changes in cells, including ER stress. In cells under ER stress, protein folding is disturbed, and this can cause MHC class I heavy chains and other unfolded proteins to accumulate in the ER (Fig. 6). The alterations found in MHC class I protein processing in HCV replicon-expressing cells do not appear to be specifically directed against MHC class I. While the cell surface expression of MHC class I is suppressed in cells expressing HCV replicons, another glycoprotein, CD13, also has reduced expression at the cell surface (Fig. 1). HCV replicons do not specifically target MHC class I to interfere with viral antigen presentation to CTLs. In fact, the ER stress induced by HCV replicons causes alterations in intracellular events that apparently disrupts overall glycoprotein folding and transport to the surface of the cell. Although ER stress induced by HCV replicons may cause the reduction in MHC class I cell surface expression, the precise mechanism is currently unknown and under investigation.
The HCV glycoproteins E1 and E2 associate to form a heterodimeric complex (12). This complex has been proposed to form part of the HCV virion envelope. Although glycoprotein folding and cell surface expression are disrupted, they are not completely eliminated in cells expressing HCV replicons. This is important for new virus production because the formation of the glycosylated viral envelope and viral particles is essential to the propagation of HCV. Furthermore, Liberman et al. (26) showed that the E1 and E2 envelope proteins are glycosylated even under conditions in which the E2 protein induces ER stress. Future experiments with the full-length replicon, containing both structural and nonstructural HCV proteins, will determine the glycosylation status of E1 and E2 in the face of HCV replicon-induced ER stress.
In this study, we demonstrated that the translation and replication activities of HCV subgenomic replicons disrupt MHC class I cell surface expression. This is accomplished in HCV replicon-expressing cells by inhibiting protein glycosylation and inducing ER stress. This suppresses MHC class I assembly, interfering with class I expression at the cell surface. HCV may require ER stress to suppress MHC class I antigen presentation and remain undetected by host immune defenses.
Acknowledgments
We are grateful to Georg Lauer and Bruce Walker for performing cytotoxicity assays on Huh7, FCA4, and GS4.3 cells. We thank R. Tirabassi for the W6/32 and HC70 antibodies, C. Seeger for the FCA4 and GS4.3 cells, and C. Gazin for the wild-type and mutant MHC class I luciferase reporters.
K.D.T. was supported by a postdoctoral training grant from NIH (T3-DK07038). This work was supported by an NIH grant (DK061566) awarded to A.S.
REFERENCES
- 1.Ahn, K., A. Angulo, P. Ghazal, P. A. Peterson, Y. Yang, and K. Fruh. 1996. human cytomegalovirus inhibits antigen presentation by a sequential multistep process. Proc. Natl. Acad. Sci. USA 93:10990-10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, K., T. H. Meyer, S. Uebel, P. Sempe, H. Djaballah, Y. Yang, P. A. Peterson, K. Fruh, and R. Tampe. 1996. Molecular mechanism and species specificity of TAP inhibition by herpes simplex virus ICP47. EMBO J. 15:3247-3255. [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn, K., A. Gruhler, B. Galocha, T. R. Jones, E. J. Wiertz, H. L. Ploegh, P. A. Peterson, Y. Yang, and K. Fruh. 1997. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity 6:613-621. [DOI] [PubMed] [Google Scholar]
- 4.Andersson, M., S. Paabo, T. Nilsson, and P. A Peterson. 1985. Impaired intracellular transport of class I MHC antigens as a possible means for adenoviruses to evade immune surveillance. Cell 43:215-222. [DOI] [PubMed] [Google Scholar]
- 5.Barnstable, C. J., W. F. Bodmer, G. Brown, G. Galfre, C. Milstein, A. F. Williams, and A. Ziegler. 1978. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens: new tools for genetic analysis. Cell 14:9-20. [DOI] [PubMed] [Google Scholar]
- 6.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 7.Bonifacino, J. S., and A. M. Weissman. 1998. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell Dev. Biol. 14:19-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgert, H.-G., and S. Kvist. 1985. An adenovirus type 2 glycoprotein blocks cell surface expression of human histocompatibility class I antigens. Cell 41:987-997. [DOI] [PubMed] [Google Scholar]
- 9.Carreno, B. M., K. L. Schreiber, D. J. McKean, I. Stroynowski, and T. H. Hansen. 1995. Aglycosylated and phosphatidylinositol-anchored MHC class I molecules are associated with calnexin. J. Immunol. 154:5173-5180. [PubMed] [Google Scholar]
- 10.Cerny, A., and F. V. Chisari. 1999. Pathogenesis of chronic hepatitis C: immunological features of hepatic injury and viral persistence. Hepatology 30:595-601. [DOI] [PubMed] [Google Scholar]
- 11.Degen, E., M. F. Cohen-Doyle, and D. B. Williams. 1992. Efficient dissociation of the p88 chaperone from major histocompatibility complex class I molecules requires both beta 2-microglobulin and peptide. J. Exp. Med. 175:1653-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Biscegli, A. M. 1997. Hepatitis C and hepatocellular carcinoma. Hepatology 26:34S-38S. [DOI] [PubMed] [Google Scholar]
- 14.Fearon, D. T., and R. M. Locksley. 1996. The instructive role of innate immunity in the acquired immune response. Science 272:50-53. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert, M. J., S. R. Riddell, B. Plachter, and P. D. Greenberg. 1996. Cytomegalovirus selectively blocks antigen processing and presentation of its immediate-early gene product. Nature 383:720-722. [DOI] [PubMed] [Google Scholar]
- 16.Gong, G., G. Waris, R. Tanveer, and A. Siddiqui. 2001. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-κB. Proc. Natl. Acad. Sci. USA 98:9599-9604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replication. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hijikata, M., H. Mizushima, Y. Tanji, Y. Komoda, Y. Hirowatari, T. Akagi, N. Kato, K. Kimura, and K. Shimotohno. 1993. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc. Natl. Acad. Sci. USA 90:10773-10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill, A., P. Jugovic, I. York, G. Russ, J. Bennink, J. Yewdell, H. Ploegh, and D. Johnson. 1995. Herpes simplex virus turns off the TAP to evade host immunity. Nature 375:411-415. [DOI] [PubMed] [Google Scholar]
- 20.Hudson, A. W., P. M. Howley, and H. L. Ploegh. 2001. A human herpesvirus 7 glycoprotein, U21, diverts major histocompatibility complex class I molecules to lysosomes. J. Virol. 75:12347-12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, T. R., E. J. Wiertz, L. Sun, K. N. Fish, J. A. Nelson, and H. L. Ploegh. 1996. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc. Natl. Acad. Sci. USA 93:11327-11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klenerman, P., M. Lucas, E. Barnes, and G. Harcourt. 2002. Immunity to hepatitis C virus: stunned but not defeated. Microbes Infect. 4:57-65. [DOI] [PubMed] [Google Scholar]
- 23.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauer, G. M., K. Ouchi, R. T. Chung, T. N. Nguyen, C. L. Day, D. R. Purkis, M. Reiser, A. Y. Kim, M. Lucas, P. Klenerman, and B. D. Walker. 2002. Comprehensive analysis of CD8+-T-cell responses against hepatitis C virus reveals multiple unpredicted specificities. J. Virol. 76:6104-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lefebvre, S., P. Moreau, J. Dausset, E. D. Carosella, and P. Paul. 1999. Downregulation of HLA class I gene transcription in choriocarcinoma cells is controlled by the proximal promoter element and can be reversed by CIITA. Placenta 20:293-301. [DOI] [PubMed] [Google Scholar]
- 26.Liberman, E., Y.-L. Fong, M. J. Selby, Q.-L. Choo, L. Cousens, M. Houghton, and T. S. B. Yen. 1999. Activation of the grp78 and grp94 promoters by hepatitis C virus E2 envelope protein. J. Virol. 73:3718-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohmann, V., F. Korner, J.-O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 28.Mansky, P., W. M. Brown, J. H. Park, J. W. Choi, and S. Y. Yang. 1994. The secondary kB element, kB2, of the HLA-A class I regulatory element is an essential part of the promoter. J. Immunol. 153:5082-5090. [PubMed] [Google Scholar]
- 29.Moradpour, D., P. Kary, C. M. Rice, and H. E. Blum. 1998. Continuous human cell lines inducibly expressing hepatitis C virus structural and nonstructural proteins. Hepatology 28:192-201. [DOI] [PubMed] [Google Scholar]
- 30.Mori, K. 2000. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell 101:451-454. [DOI] [PubMed] [Google Scholar]
- 31.Noessner, E., and P. Parham. 1995. Species-specific differences in chaperone interaction of human and mouse major histocompatibility complex class I molecules. J. Exp. Med. 181:327-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Owen, M. J., A. M. Kissonerghis, and H. F. Lodish. 1980. Biosynthesis of HLA-A and HLA-B antigens in vivo. J. Biol. Chem. 255:9678-9684. [PubMed] [Google Scholar]
- 33.Pahl, H. L., M. Sester, H.-G. Burgert, and P. A. Baeuerle. 1996. Activation of transcription factor NF-κB by adenovirus E3/19K requires ER retention. J. Cell Biol. 132:511-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parham, P. 1996. Functions for MHC class I carbohydrates inside and outside the cell. Trends Biochem. Sci. 21:427-433. [DOI] [PubMed] [Google Scholar]
- 35.Rajagopalan, S., and M. B. Brenner. 1994. Calnexin retains unassembled major histocompatibility complex class I free heavy chains in the endoplasmic reticulum. J. Exp. Med. 180:407-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 37.Riemann, D., A. Kehlen, and J. Langner. 1999. CD13 - not just a marker in leukemia typing. Immunol. Today 20:83-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rijibrand, R., and S. Lemon. 1999. Internal ribosome entry site-mediated translation in hepatitis C virus replication. Curr. Top. Microbiol. Immunol. 242:86-116. [DOI] [PubMed] [Google Scholar]
- 39.Schoneich, J., J. L. Lee, P. Mansky, M. Sheffery, and S. Y. Yang. 1997. The pentanucleotide ATTGG, the “inverted CCAAT, ” is an essential element for HLA class I gene transcription. J. Immunol. 158:4788-4796. [PubMed] [Google Scholar]
- 40.Shamu, C. E., C. M. Story, T. A. Rapoport, and H. L. Ploegh. 1999. The pathway of US11-dependent degradation of MHC class I heavy chains involves a ubiquitin-conjugated intermediate. J. Cell Biol. 147:45-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tardif, K. D., K. Mori, and A. Siddiqui. 2002. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J. Virol. 76:7453-7459. [DOI] [PMC free article] [PubMed]
- 42.Tortorella, D., B. E. Gewurz, M. H. Furman, D. J. Schust, and H. L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861-926. [DOI] [PubMed] [Google Scholar]
- 43.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, C., P. Sarnow, and A. Siddiqui. 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiertz, E. J., T. R. Jones, L. Sun, M. Bogyo, H. J. Geuze, and H. L. Ploegh. 1996. The human cytomegalovirus US11 gene product dislocates MHC I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84:769-779. [DOI] [PubMed] [Google Scholar]