Abstract
The adenovirus E1B 55-kDa protein impairs the p53 pathway and enhances transformation, although the underlying mechanisms remain to be defined. We found that Daxx binds to the E1B 55-kDa protein in a yeast two-hybrid screen. The two proteins interact through their C termini. Mutation of three potential phosphorylation sites (S489/490 and T494 to alanine) within the E1B 55-kDa protein did not affect its interaction with Daxx, although such mutations were previously shown to inhibit E1B's ability to repress p53-dependent transcription and to enhance transformation. In addition to their coimmunoprecipitation in 293 extracts, purified Daxx interacted with the E1B 55-kDa protein in vitro, indicating their direct interaction. In 293 cells, Daxx colocalized with the E1B 55-kDa protein within discrete nuclear dots, where p53 was also found. Such structures were distinct from PML (promyelocytic leukemia protein) bodies, and it appeared that Daxx was displaced from PML bodies. Thus, the Daxx concentration was diminished in dots with a prominent presence of PML and vice versa. Indeed, PML overexpression led to dramatic redistribution of Daxx from p53-E1B 55-kDa protein complexes to PML bodies. Additionally, expression of the E1B 55-kDa protein in Saos2 osteosarcoma cells reduced the number of PML bodies. Our data suggest that E1B and PML compete for available Daxx in the cell. Surprisingly, Daxx significantly augmented p53-mediated transcription and the E1B 55-kDa protein eliminated this effect. Thus, it is likely that the E1B 55-kDa protein sequesters Daxx and p53 in specific nuclear locations, where p53 cannot activate transcription. One consequence of the Daxx-E1B interaction might be an alteration of normal interactions of Daxx, PML, and p53, which may contribute to cell transformation.
Cancer arises from a cell that undergoes a number of specific changes. Transformation of primary human cells requires at least four genetic events: inactivation of both the p53 and pRb pathways, activation of mitogenic oncogenes such as ras, and telomere maintenance (11). These genetic changes may also underlie cell transformation induced by DNA tumor viruses. In fact, it is well known that several viral oncogenes involved in virus-induced cell transformation inactivate both the p53 and pRb pathways. These viral oncogenes include the simian virus 40 (SV40) large T antigen, the human papillomavirus (HPV) 16 E6 and E7 proteins, and the adenovirus (Ad) E1A and E1B proteins (1). Recent in vitro cell transformation experiments used the SV40 large T antigen and the HPV E6 and E7 oncogenes to inactivate the p53 and pRb pathways (11, 25). Interestingly, in combination with ras and the gene for the catalytic subunit of telomerase, the SV40 large T antigen effectively transformed human primary cells (11), but HPV E6 and E7 failed to do so (25), suggesting that inactivation of cellular pathways in addition to pRb and p53 may be required in malignant transformation.
The mechanisms underlying Ad-induced transformation have been a subject of extensive investigation. The E1A proteins activate the cell cycle of quiescent, nondividing cells and induce partial proliferation and immortalization (33, 35). This may stem from E1A's ability to inhibit both pRb (7) and p53 functions (17, 37). In addition, E1A binds to cellular transcriptional coactivators and acetylases p300, CREB-binding protein (CBP) (1), and PCAF (32), which may alter the expression of genes involved in proliferation and differentiation (7). However, E1A expression causes apoptosis (3, 21), thus undermining complete cell transformation. On the other hand, the E1B 19- and 55-kDa proteins can inhibit apoptosis (3, 22, 39). The E1B 19-kDa protein is a functional homologue of the apoptosis inhibitor Bc12 (34). The E1B 55-kDa protein is a potent inhibitor of p53's transactivation function (38, 43) and thus can effectively repress p53-dependent cellular processes such as cell cycle arrest and apoptosis (22, 38). Together, these Ad E1 proteins can fully transform cells (1, 33, 40).
The Ad type 2 (Ad2) and Ad5 E1B 55-kDa protein binds to the amino-terminal transcriptional transactivation domain of p53 (19, 43). This interaction is thought to impair p53's transactivation activity, the key function of p53 as a tumor suppressor (42, 43). The E1B 55-kDa protein binds to E4orf6 (27), which plays a role in targeting p53 for ubiquitin-dependent proteolysis (12, 31). Apart from modulating p53 activities and stability, we recently showed that the E1B 55-kDa protein from Ad2 and Ad12 can specifically inhibit acetylation of p53 by PCAF (20), thus preventing p53 activation, because acetylation of p53 at specific sites is important for p53's transactivation function (10, 29). The E1B 55-kDa protein is a transcriptional repressor, probably through interaction with transcription machinery (23, 24) and/or histone deacetylase and corepressor mSin3A complexes (30). While it has been shown that the E1B 55-kDa protein is involved in several processes that could potentially contribute to Ad-induced transformation, including promotion of preferential export of viral mRNAs during lytic viral growth (4, 8, 12, 41) and, in the case of Ad12 E1B, induction of chromosomal fragility at specific loci in the human genome (16, 44), only the transcriptional repression function of E1B has been demonstrated to be important for Ad-induced transformation (39, 42, 43). In particular, there is a positive correlation between E1B's transcriptional repression function and its ability to repress apoptosis and to cooperate with E1A in transformation. Teodoro et al. showed that mutations that convert several potential phosphorylation sites near E1B's C terminus abolish its ability to act as a transcription repressor and as an inhibitor of apoptosis and to transform cells with the E1A and E1B 19-kDa proteins (39). As such a mutant E1B protein can still bind to p53 (39), the interaction of E1B with other cellular proteins may be required for the transcriptional repression and transformation mediated by the E1B 55-kDa protein. Therefore, identification of new E1B-binding proteins may provide further insights into the roles of E1B in transformation.
To identify cellular proteins that may be exploited by E1B 55-kDa proteins to bring about transcriptional repression and cell transformation, we have searched human cDNA libraries by using E1B as a probe in yeast two-hybrid screens. The human Daxx protein was identified as an E1B 55 kDa-binding protein. We report here data that support a specific interaction between the E1B 55-kDa protein and Daxx and the potential impact of this interaction on the p53 and PML pathways. The implications of our findings for mechanisms by which the E1B 55-kDa oncoprotein modifies the functions of important cell growth regulators such as p53 and PML are discussed.
MATERIALS AND METHODS
Plasmids and antibodies.
Vectors pGAD-C(x) and pGBDU-C(x), used for yeast two-hybrid assays, were as previously described (14). Various DNA fragments encoding the Ad2 and Ad12 E1B 55-kDa proteins were cloned into pGAD-C(x) and pGDBU-C(x) as reported previously (20). The full-length Daxx cDNA clone was purchased from Invitrogen. Daxx fragments covering different regions of the cDNA were cloned into pGAD-C(x) and pGBDU-C(x) for two-hybrid assays. The green fluorescent protein (GFP)-Daxx fusion was constructed by inserting full-length Daxx cDNA into pEGFP-C2 (Clontech).
Antibodies against Daxx (M-112), PML (PG-M3), and p53 (DO-1 and anti-p53-393FL) were purchased from Santa Cruz Biotechnology. Mouse anti-Flag monoclonal (M2) and rabbit polyclonal antibodies were purchased from Sigma. Rabbit polyclonal anti-DNMT1 (DNA methyltransferase 1) antibody was purchased from New England Biolabs. Rabbit anti-Ad12 E1B 55-kDa protein polyclonal antibody was described previously (16). Hybridoma cells (2A6) producing mouse monoclonal anti-Ad5 E1B 55-kDa protein antibody were provided by Arnold Levine. Monoclonal antifibrillarin antibody was obtained from John Aris.
Yeast two-hybrid screens.
The Ad2 E1B 55-kDa protein amino acid (aa) 155 to 495 segment was fused with the Gal4 DNA-binding domain (BD) in plasmid pGBDU-C1 and used as bait (we were unable to clone the full-length open reading frame of the Ad2 E1B 55-kDa protein into this and other high-copy-number plasmids, as explained previously [16]). The HeLa cDNA library consists of cDNA fragments cloned into pGADGH (a Clontech Matchmaker library). The bait and the cDNA library were introduced into yeast strain PJ69-4A (14). The transformed yeast cells were replica plated on synthetic dropout (SD) medium lacking histidine but containing 5 mM 3-aminotriazole and SD medium lacking adenine. Positive clones were selected in accordance with the manufacturer's (Clontech) protocol. Each recovered positive plasmid was reintroduced into yeast along with Ad2 E1B in pGBDU-C1 or empty pGBDU-C1. Only the clones that supported yeast growth in SD medium in the presence of Ad2 E1B in pGBDU-C1 were considered truly positive. About 106 yeast transformants were screened in this way.
Immunofluorescence microscopy.
Antibody staining of transfected cells was done in accordance with the previously published protocol (16). Briefly, 24 h after transfection, cells grown on glass coverslips were fixed with 4% paraformaldehyde for 20 min at room temperature and permeabilized with 0.2% Triton X in phosphate-buffered saline (PBS). The slides were then incubated with blocking buffer (2% fetal bovine serum, 0.1% sodium azide, 0.1% Tween 20 in PBS). After incubation with primary antibodies, the cells were washed with PBS containing 0.1% Tween 20 and then incubated with appropriate secondary antibodies conjugated with fluorescent dyes. The cells were washed and mounted in medium with or without 4′,6′-diamidino-2-phenylindole (DAPI). The processed cells were examined with a Zeiss Axiophot microscope.
Luciferase reporter gene assays.
Ad5-transformed human embryonic kidney cell line 293, osteosarcoma cell line Saos2, and p53-deficient HCT116 cells (2) were cultured in Dulbecco modified Eagle medium with 10% fetal bovine serum. The luciferase reporter construct PG13 contains multiple copies of the p53 DNA-binding site upstream of a TATA box cloned into pGL3-Basic (Promega), and pWAF1-Luc contains the promoter fragment of the p21 gene as previously described (5). Bax-Luc contains a part of the bax promoter that encompasses the p53-binding sites in pGL3-Basic (Science Reagents). Daxx-Luc contains a 1.7-kb DNA fragment of the Daxx gene promoter spanning the CpG island, exon 1 (noncoding), intron 1, and the 5′ portion of exon 2 (coding) in pGL3-Basic. The reporter plasmids were transiently transfected into cells alone or with other plasmids with SuperFect transfection reagent (Qiagen). The transfected cells were harvested 24 to 48 h posttransfection and processed for dual-luciferase assays (Promega). Firefly luciferase activity was normalized against sea pansy luciferase activity.
RESULTS
Identification of Daxx as an E1B-binding protein.
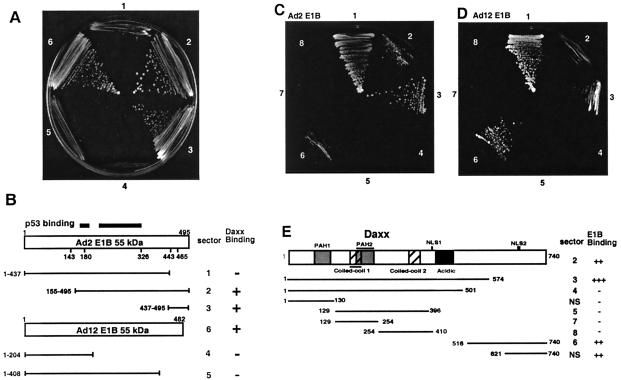
With Ad2 E1B aa 155 to 495 as bait, we have screened approximately 106 yeast transformants. Among 11 positive clones, two identical clones encoding Daxx C-terminal aa 621 to 740 were isolated. This Daxx fragment did not bind to the Gal4 BD (data not shown) or its fusion constructs with various Ad2 and Ad12 E1B fragments lacking the C-terminal domain (Fig. 1A, sectors 1, 4, and 5). By contrast, this Daxx fragment bound to Ad2 E1B aa 155 to 495 (sector 2) and the full-length Ad12 E1B protein (sector 6), as well as the Ad2 C-terminal aa 437 to 495 fragment (sector 3). These results indicate that the Daxx C terminus interacts specifically with the C-terminal domain of the E1B 55-kDa oncoprotein (Fig. 1B).
FIG. 1.
The E1B 55-kDa protein interacts with Daxx in yeast two-hybrid assays. (A) Domain of the E1B 55-kDa protein required for binding to Daxx. The C-terminal fragment (aa 621 to 740) of Daxx fused to the Gal4 activation domain was assayed for interaction with various E1B constructs fused with the Gal4 BD in two-hybrid assays. Sectors 1 to 6: Ad2 E1B 55-kDa protein aa 1 to 437, 155 to 495, and 437 to 495 and Ad12 E1B 55-kDa protein aa 1 to 204 and 1 to 408 and the full-length protein. Yeast cells were plated on medium lacking histidine but containing 5 mM 3-aminotriazole. (B) Summary of E1B-Daxx interaction based on panel A. (C to E) Mapping of the E1B-binding domains of Daxx. Various Daxx fragments fused with the Gal4 BD as depicted in panel E were cotransformed into yeast with the Ad2 E1B 55-kDa protein (aa 155 to 495, panel C) or the Ad12 E1B 55-kDa protein (full-length protein, panel D), and yeast growth was scored in selective medium as in panel A. Sector 1 in panels C and D represents a positive control with cotransformation of SV40 T antigen fused with the Gal4 activation domain and mouse p53 aa 70 to the C terminus fused with the Gal4 BD. NS, not shown.
We then tested various Daxx fragments for interaction with the E1B 55-kDa protein. Full-length Daxx (Fig. 1C and D, sector 2), aa 1 to 574 (sector 3), and aa 516 to 574 (sector 6), as well as aa 621 to 740 (see Fig. 1A) bind to both the Ad2 and Ad12 E1B 55-kDa proteins. These results suggest that two segments (aa 500 to 574 and 621 to 740) within Daxx can bind to the E1B 55-kDa protein.
The E1B 55-kDa protein binds to Daxx in vivo and in vitro.
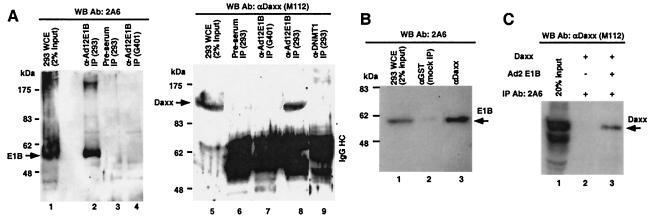
To confirm the Daxx-E1B interaction observed in yeast two-hybrid assays, we performed immunoprecipitation (IP) experiments. The human embryonic kidney cell line 293 expresses Ad5 proteins encoded in the early 1A and 1B regions (9). The whole-cell extract (WCE) of 293 cells was subjected to IP with rabbit polyclonal antibodies raised against the Ad12 E1B 55-kDa protein N-terminal region (aa 1 to 158) (16), and the immunoprecipitates were probed with mouse anti-Ad2/5 E1B 55-kDa protein monoclonal antibody 2A6 (36). The Ad5 E1B 55-kDa protein was effectively precipitated (Fig. 2A, lane 2). Rabbit preimmunization serum did not recognize Ad5 E1B (lane 3). In a different control IP, the anti-Ad12 E1B 55-kDa protein antibody was used for immunoprecipitation from G401 WCE that does not contain the E1B 55-kDa protein, and as expected, this antibody did not precipitate cellular proteins that might cross-react with 2A6 (lane 4). Thus, the rabbit anti-Ad12 E1B polyclonal antibodies specifically recognized the Ad5 E1B 55-kDa protein in IP experiments. The immunoprecipitates were then analyzed in WB with anti-Daxx antibody M112. As shown in Fig. 2A, Daxx was present in the anti-E1B immunocomplexes (lane 8) but not in preimmune serum (lane 6). Daxx was not precipitated by either anti-E1B antibody in cells without E1B 55-kDa protein expression (lane 7) or in the precipitates of anti-DNMT1 rabbit polyclonal antibodies (lane 9), indicating a specific Daxx-E1B 55-kDa protein interaction. In a reciprocal IP, the E1B 55-kDa protein was precipitated by anti-Daxx M112 antibody (Fig. 2B, lane 3) but not by mock antibody (lane 2). Thus, the E1B 55-kDa protein binds to Daxx in vivo.
FIG. 2.
Daxx binds to the E1B 55-kDa protein in vivo and in vitro. (A) Coimmunoprecipitation of E1B and Daxx with anti-Ad12 E1B 55-kDa protein antibodies (Ab). The WCE of 293 cells was subjected to IP with a rabbit polyclonal antibody raised against the Ad12 E1B 55-kDa protein (lanes 2 and 8), and the precipitates were analyzed with an anti-Ad5 E1B 55-kDa protein monoclonal antibody (2A6; left side) or rabbit polyclonal anti-Daxx antibodies (M112; right side) in an immunoblot analysis. Control IPs included rabbit preimmune serum (lanes 3 and 6), 293 WCE with rabbit polyclonal anti-DNMT1 (lane 9), and G401 WCE that did not contain the E1B 55-kDa protein with anti-Ad12 E1B 55-kDa protein antibodies (lanes 4 and 7). (B) Coimmunoprecipitation of E1B and Daxx with anti-Daxx antibodies. The WCE of 293 cells was subjected to IP with M-112 (lane 3) or mock antibody to GST (lane 2). The precipitates were analyzed in a Western blot assay with 2A6. (C) Direct interaction between the E1B 55-kDa protein and Daxx in vitro. Full-length Daxx was expressed in Sf9 cells with a baculovirus vector, and the purified protein was incubated with either an antibody to E1B alone (2A6; lane 2) or 2A6 plus the purified Ad2 E1B 55-kDa protein from Sf9 cells (lane 3). The mixtures were then subjected to IP, and the precipitates were analyzed in a Western blot assay with an antibody to Daxx M-112.
We then examined the in vitro interaction of purified Daxx and the Ad2 E1B 55-kDa protein. Both proteins were expressed in Sf9 insect cells with a baculovirus expression system and purified with Ni2+ affinity chromatography. The purified proteins were mixed and subjected to IP with anti-E1B 2A6. As shown in Fig. 2C, Daxx was coprecipitated with the E1B 55-kDa protein, indicating a direct interaction in vitro.
Daxx and the E1B 55-kDa protein colocalize in the nucleus.
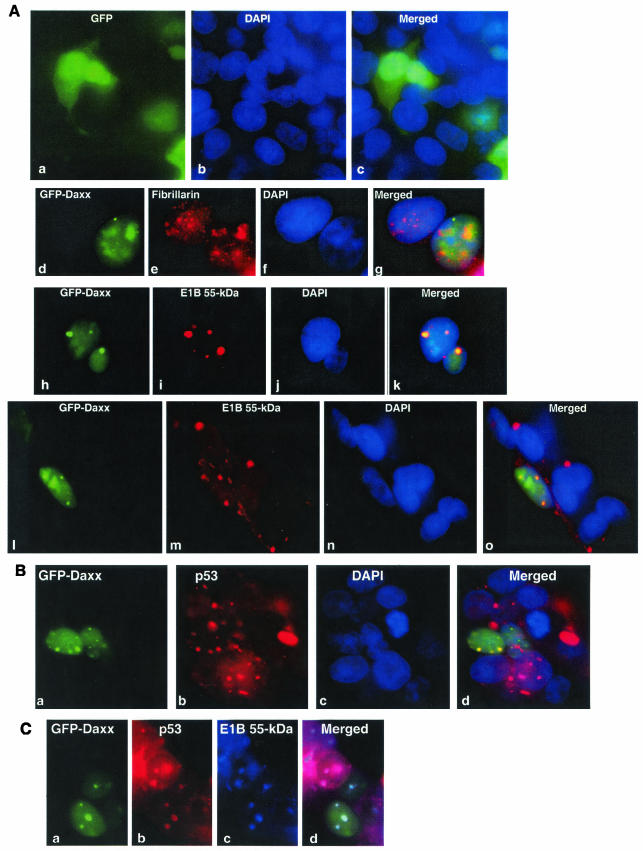
To further substantiate the Daxx-E1B 55-kDa protein interaction, we examined the potential colocalization of Daxx and the E1B 55-kDa oncoprotein. GFP and GFP-Daxx fusion constructs were transfected into 293 cells. GFP was present in the nuclei and cytoplasm of the transfected cells (Fig. 3, a to c). The GFP-Daxx fusion protein localizes the same way as wild-type Daxx, including predominantly nuclear localization and appearance in the nucleolus along with the nucleolar protein fibrillarin (Fig. 3A, d to g). Nucleolar localization of Daxx was also previously reported (18). We thus concluded that GFP-Daxx behaved identically to wild-type Daxx in terms of subcellular distribution.
FIG.3.
Colocalization of the E1B 55-kDa protein, p53, and Daxx in 293 cells. (A) Colocalization of the E1B 55-kDa protein and Daxx. GFP (parts a to c) or GFP-Daxx fusion (parts d to o) constructs were transfected into 293 cells. The cells were stained with monoclonal antifibrillarin (part e) or anti-Ad5 E1B 55-kDa protein antibody 2A6 (parts i and m). Nuclei were stained with DAPI (parts b, f, j, and n). Merged images are also shown. Yellow color indicates colocalization of proteins. (B) Daxx and p53 colocalized in discrete nuclear structures. The GFP-Daxx fusion construct was transfected into 293 cells. The cells were stained with a monoclonal antibody to p53 (DO-1; part b) and counterstained with DAPI (part c). (C) Colocalization of the E1B 55-kDa protein, Daxx, and p53. The GFP-Daxx construct was transfected into 293 cells, which were stained with a goat anti-p53 polyclonal antibody (FL-393; Santa Cruz; part b) or a mouse anti-Ad5 E1B 55-kDa protein monoclonal antibody (2A6; part c). The donkey secondary antibodies were anti-goat immunoglobulin G conjugated with rhodamine (for detection of p53) and anti-mouse immunoglobulin G conjugated with Alexa-Fluor 350 (highly cross-absorbed, for detection of the E1B 55-kDa protein; Molecular Probes).
In 293 cells, the E1B 55-kDa protein is localized mainly in the cytoplasm, especially in the dense perinuclear aggregates; diffused nuclear staining of E1B was also evident (Fig. 3A, m), consistent with previous observations (40, 45). Strikingly, discrete nuclear dots containing the E1B 55-kDa protein were found and Daxx was colocalized with E1B within such nuclear structures (Fig. 3A, h to o; see also Fig. 5A, g to i). Interestingly, GFP-Daxx was never detectable in the cytoplasm. Conversely, although Daxx was seen in the nucleolus in 293 cells (Fig. 3A, d to g), the E1B 55-kDa protein was not found in the nucleolus. Thus, the Daxx-E1B 55-kDa protein interaction may occur only in the nucleoplasm, mostly concentrated in dot-like structures (Fig. 3A, h to o; see also Fig. 5A, g to i).
FIG. 5.
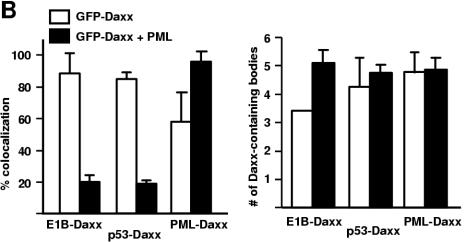
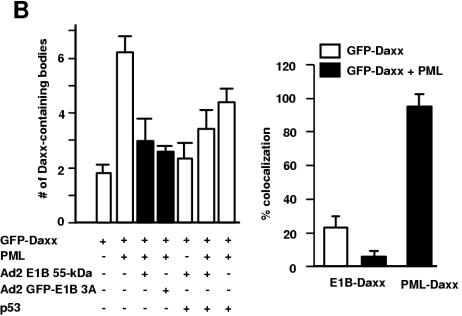
Dynamic redistribution of Daxx in PML bodies and E1B 55-kDa protein-p53 complexes. (A) Effect of PML overexpression on Daxx-E1B 55-kDa protein colocalization. GFP-Daxx alone or together with PML constructs was transfected into 293 cells, as indicated. The transfected cells were stained with appropriate antibodies (anti-PML [part b], anti-Flag [part e], 2A6 [parts h and k], and anti-p53 DO1 [parts n and q]). (B) Quantification of Daxx-E1B 55-kDa protein and Daxx-PML colocalization. The discrete Daxx-containing nuclear bodies were counted, and the bodies that exhibited colocalization with the E1B 55-kDa protein, p53, or PML were separately tallied. The ratio of the number of bodies with colocalization of two proteins as indicated to the total number of Daxx-containing bodies is expressed as a percentage (left). The average number of Daxx-containing bodies per transfected cell is also shown (right). White bar, with GFP-Daxx transfection; solid bar, with GFP-Daxx and PML cotransfection. Error bars represent 1 standard deviation.
p53 is present in nuclear structures containing Daxx and the E1B 55-kDa protein.
The tumor suppressor p53 is found in dense cytoplasmic bodies containing the E1B 55-kDa protein in adenovirus-transformed cells (40, 45). In agreement with this, a high concentration of p53 was found in the cytoplasmic bodies in 293 cells, although a substantial nuclear presence of p53 was also evident (Fig. 3B). To examine whether p53 also colocalizes with Daxx, GFP-Daxx was transfected into 293 cells, which were stained with anti-p53 antibody DO-1. Colocalization of p53 with Daxx in nuclear dots was clearly demonstrated (Fig. 3B, a to d). In addition, simultaneous staining of the transfected 293 cells with goat anti-p53 antibody and 2A6 indicated that Daxx, p53, and the E1B 55-kDa protein were present in the same complex in the nuclear dots (Fig. 3C).
Dynamic redistribution of Daxx between PML bodies and E1B 55-kDa protein-p53 structures.
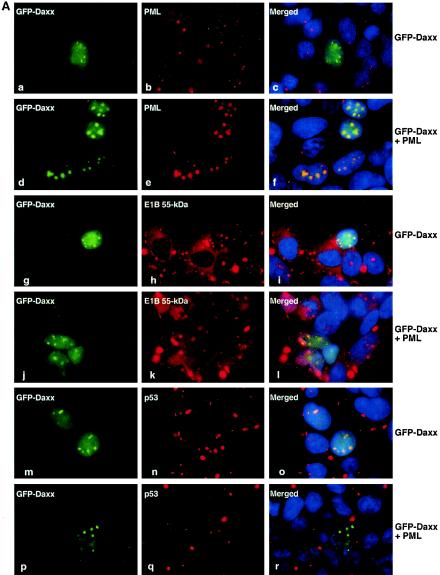
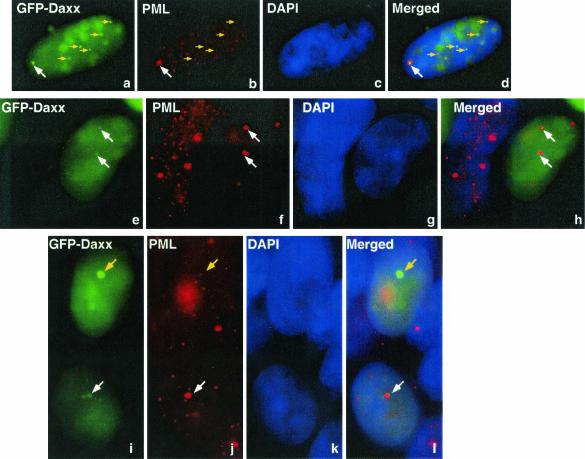
The nuclear dots containing Daxx, the E1B 55-kDa protein, and p53 resemble PML bodies (also called nuclear domain 10 [ND10], the PML oncogenic domain, or POD), and Daxx has been seen frequently in PML bodies (13). To examine whether these structures were indeed PML bodies, a GFP-Daxx construct was transfected into 293 cells, which were stained with an anti-PML antibody (PG-M3). Surprisingly, PML and Daxx only partially colocalized in 293 cells (Fig. 4; see also Fig. 5). In some instances, Daxx and PML appeared to be concentrated in neighboring structures, and colocalization of these two proteins was only evident in the overlapping region (Fig. 4a to d, white arrows; see also Fig. 7B). In other cases, dots with intense PML signals contained diminished amount of Daxx (panels e to l, white arrows). Conversely, structures with a high concentration of Daxx had a reduced presence of PML (panels a to d and i to l, yellow arrows). These results suggest that Daxx may be recruited away from PML bodies in the presence of the E1B 55-kDa oncoprotein.
FIG. 4.
Displacement of Daxx from PML bodies by the E1B 55-kDa protein. The GFP-Daxx fusion construct was transfected into 293 cells. The cells were stained with a monoclonal antibody to PML (PG-M3; panels b, f, and j) and counterstained with DAPI (panels c, g, and k). Arrows (white and yellow) pinpoint the same positions in each panel to facilitate visualization of the locations of Daxx and PML.
FIG. 7.
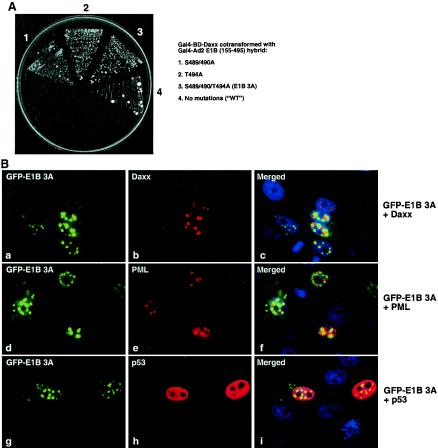
The C-terminal phosphorylation sites of the E1B 55-kDa protein are not required for its binding to Daxx. (A) Yeast two-hybrid assay of interaction between Daxx and mutant E1B proteins. Three potential phosphorylation sites (S489/490 and T494) near the C terminus of the Ad2 E1B 55-kDa protein (aa 155 to 495) were mutated to alanine, and the mutant constructs were fused with the Gal4 activation domain and introduced along with the Gal4 BD-Daxx construct into yeast. The two-hybrid assays were as described in the legend to Fig. 1. WT, wild type. (B) Localization of GFP-mutant E1B 3A (S489/490/T494A) in transfected cells. The GFP-E1B 3A fusion was cotransfected with Flag-Daxx (parts a to c), Flag-PML (parts d to f), and p53 (parts g to i), and the transfected Saos2 cells were stained with rabbit polyclonal anti-Flag (parts b and e) and anti-p53 (part h) antibodies and a rhodamine-conjugated antibody to rabbit immunoglobulin G.
If Daxx is distributed dynamically between the E1B-p53 structure and PML bodies, increased expression of PML might reduce the colocalization of Daxx with E1B-p53 structures. To test this, we transfected 293 cells with a PML expression plasmid and then examined the colocalization of Daxx with E1B, p53, and PML, respectively. Indeed, in cells without exogenous PML expression, GFP-Daxx colocalized with E1B in ∼90% of the Daxx-containing speckles (Fig. 5A and B, g to i). Daxx-E1B 55-kDa protein colocalization was dramatically reduced when PML construct was transfected (Fig. 5A, j to l, and B); only ∼20% of the Daxx-containing bodies had the E1B 55-kDa protein. The patterns of Daxx-p53 colocalization were nearly identical to that of Daxx-E1B 55-kDa protein colocalization (Fig. 5A, m to r, and B), suggesting that p53 and the E1B 55-kDa protein reside in the same structures, consistent with the observations described above (Fig. 3C). The opposite was observed for PML-Daxx colocalization. While about 50% of the Daxx-containing bodies have PML in 293 cells expressing GFP-Daxx, PML was present in nearly all of the Daxx-containing bodies when GFP-Daxx and a PML-expressing plasmid were cotransfected (Fig. 5A, a to f, and B).
We also examined whether PML overexpression could increase the number of Daxx-containing bodies. As shown in Fig. 5B, overexpression of PML did not significantly affect the number of Daxx-containing bodies. On average, the number of Daxx-containing bodies was about five per cell in the >1,000 GFP-Daxx-transfected cells we examined. Collectively, these results suggest that PML and the E1B 55-kDa protein compete in recruiting cellular Daxx to different nuclear structures.
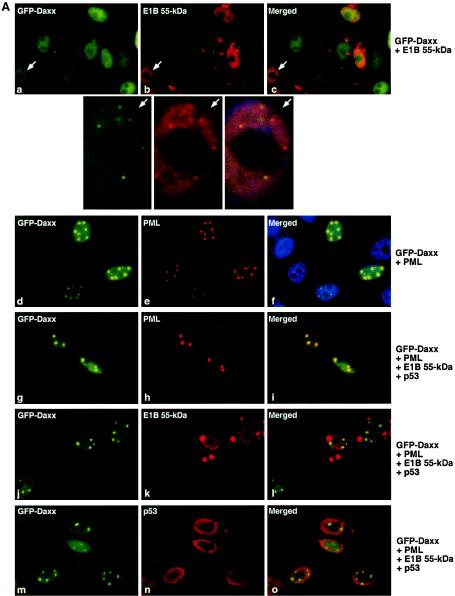
To further examine how the E1B 55-kDa protein influences Daxx distribution in the cell, GFP-Daxx was transfected into p53-null Saos2 osteosarcoma cells. Daxx was diffusely distributed in the nucleus with or without E1B 55-kDa construct cotransfection (Fig. 6A, a to c, and data not shown), although discrete speckles, albeit very small, were also observed (Fig. 6A, a to c, white arrows). Thus, the intracellular distribution of Daxx in Saos2 cells is different from that in 293 cells. This may be due to low expression of PML or lack of p53 in Saos2 cells. Indeed, the endogenous PML was largely undetectable (data not shown). Transfection of PML into Saos2 cells dramatically increased the size and number of PML bodies, and Daxx was found in nearly all PML bodies (Fig. 6A d to f, and B,). Interestingly, expression of the E1B 55-kDa protein significantly reduced the number of PML bodies from about six per cell to about three per cell (Fig. 6A, g to o, and B). Although the Daxx-containing speckles are small in the absence of PML transfection, colocalization of the E1B 55-kDa protein with Daxx in these speckles was observed (Fig. 6A, a to c, white arrows; the cell with Daxx-E1B 55-kDa protein colocalization in parts a to c is enlarged for better visualization). We found that the E1B 55-kDa protein localized in ∼20% of these Daxx-containing bodies, and exogenous expression of PML reduced the colocalization of E1B and Daxx (Fig. 6B). Transfection of p53 into Saos2 cells was not able to increase the size or number of speckles with Daxx and the E1B 55-kDa protein, and p53 was not concentrated in the speckles (Fig. 6A, g to o, and B). Thus, lack of p53 was not the cause of the decreased colocalization of Daxx and the E1B 55-kDa protein observed in Saos2 cells. Since 293 cells express both the Ad E1A and E1B proteins, it is possible that E1A proteins and the E1B 19-kDa protein are involved in the formation of the conspicuous Daxx-p53-E1B 55-kDa protein structures observed in 293 cells (Fig. 3 and 5). However, we failed to reconstitute the frequency and size of Daxx-p53-E1B 55-kDa protein structures in Saos2 cells when an E1A-expressing plasmid or the entire E1 region was transfected in Saos2 cells (data not shown). Therefore, unknown cellular proteins that are present in 293, but not in Saos2, cells may be involved in the formation of Daxx-p53-E1B structures. Nonetheless, the data obtained with Saos2 cells are fully consistent with the finding obtained with 293 cells that Daxx is actively recruited by the E1B 55-kDa protein and PML to distinct nuclear complexes.
FIG. 6.
Influence of Daxx distribution by PML and the E1B 55-kDa protein in Saos2 cells. (A) Colocalization of Daxx with the E1B 55-kDa protein, p53, or PML in Saos2 cells. The cells were transfected with different combinations of plasmids as indicated, and the cells were stained with the indicated antibodies as in Fig. 3. Representative micrographs are shown. Each arrow points to a cell where Daxx and the E1B 55-kDa protein colocalized in discrete nuclear bodies, and the enlarged images of this cell are shown below. (B) Quantification of Daxx-E1B 55-kDa protein and Daxx-PML colocalization in Saos2 cells. The average number of Daxx-containing bodies per transfected cell (left) and percent colocalization are shown as in Fig. 5B. Note that because of extremely low levels of endogenous PML, an examination of the colocalization of Daxx and PML without PML transfection was not possible. GFP-E1B 3A, Ad2 aa 155 to 495 with alanine substitutions at S489/490 and T494 with GFP fusion at the N terminus (see Fig. 7).
Mutations of C-terminal phosphorylation sites of the E1B 55-kDa protein do not affect the Daxx-E1B interaction.
Three potential phosphorylation sites in the Ad5 E1B 55-kDa protein (S490/491 and T495) are critical for its repression of transcription and cooperation with the E1A and E1B 19-kDa proteins to transform cells, as mutation of these sites to alanine essentially abolished its repression and transformation activities (39). To test if these mutations would also affect the Daxx-E1B 55-kDa protein interaction, we mutated the equivalent sites in the Ad2 E1B 55-kDa protein (S489/490 and T494) to alanine and assayed the interaction of these mutant forms with Daxx in a yeast two-hybrid system. As shown in Fig. 7A, mutation of S489/490, T494, or all three residues to alanine did not affect the Daxx-E1B interaction. We then assayed if these mutations would affect the distribution of Daxx in the cell. Like the wild-type Ad2 E1B 55-kDa protein, the GFP-E1B 3A mutant protein (S489/490/T494A) reduced the number of Daxx-containing PML bodies in transfected Saos2 cells (Fig. 6B), indicating that the Daxx-E1B interaction might be sufficient for recruitment of Daxx from PML bodies. Interestingly, GFP-E1B 3A was concentrated predominantly in discrete nuclear dots, although this mutant protein was also present in the cytoplasm (Fig. 7B). By contrast, while such nuclear dots were clearly observed for the wild-type E1B 55-kDa protein in 293 cells (Fig. 5A, h and i), they were not present in all of cells in the population. In transfected Saos2 cells, the wild-type Ad2 E1B 55-kDa protein was distributed evenly within the nucleus, with occasional small dots in some cells, and as in 293 cells, it was localized to large cytoplasmic bodies (Fig. 6A; also Fig. 5A). It is worth noting that the Ad2 E1B S489/490A or T494A mutant proteins or their fusion constructs made with GFP were all expressed at a low level in transfected cells, but E1B 3A was highly expressed whether or not it was fused with GFP (data not shown), suggesting that GFP tagging at the N terminus of the 55-kDa protein may not have an obvious effect on the structure and function of this viral protein.
We then examined the colocalization of Daxx and the GFP-E1B 3A mutant protein. As shown in Fig. 7B, Daxx speckles did not completely overlap with the E1B dots, and they were concentrated on closely spaced neighbor structures with a partial overlap (Fig. 7B, a to c). PML exhibited a similar pattern of localization in relation to GFP-E1B 3A (Fig. 7B, d to f). p53 was uniformly distributed in the nuclei of transfected Saos2 cells and was not concentrated within the dots of the GFP-E1B 3A mutant protein (Fig. 7B, g to i). Collectively, these results indicate that interaction between Daxx and E1B is sufficient to reduce the number of PML bodies, consistent with our model in which the E1B 55-kDa protein recruits Daxx from PML bodies.
Daxx enhances p53-mediated transactivation, and the E1B 55-kDa protein inhibits such enhancement.
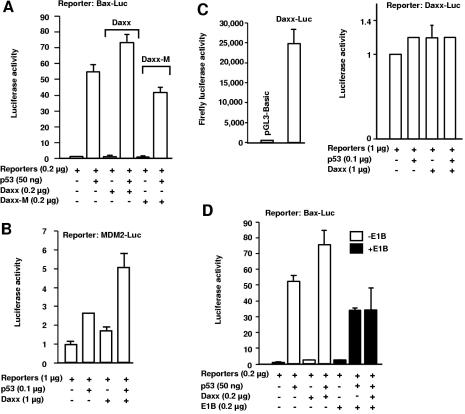
Daxx colocalizes with p53 in 293 cells (Fig. 3 and 5), and a recent yeast two-hybrid screen identified a Daxx-p53 interaction (26). Thus, Daxx could modulate p53 function. We assessed whether Daxx could influence p53-mediated transactivation with luciferase reporter assays. As shown in Fig. 8A, wild-type Daxx, but not a mutant Daxx protein (Daxx-M) that cannot bind to p53, significantly stimulated p53-mediated transcription from the Bax promoter, which contains p53-binding sites, when p53- and Daxx-expressing plasmids were cotransfected into p53-deficient HCT116 cells (2). Specifically, the fold induction was markedly different in the presence of wild-type versus mutant Daxx. In cells cotransfected with p53 and wild-type Daxx, p53 led to a 74-fold increase in reporter activity (in comparison to that in cells transfected with reporters only) or a 45-fold increase relative to that of Daxx-transfected cells. By contrast, in cells transfected with p53 and Daxx-M, the increase in reporter activity was 42-fold greater than that in cells transfected with the reporters only or 34-fold greater than that of cells transfected with Daxx-M. Transfection of a Daxx or Daxx-M construct alone had virtually no effect on Bax promoter-mediated reporter activity (a 1.6-fold increase for wild-type Daxx versus a 1.3-fold increase for Daxx-M transfection above that from cells transfected with reporters only; Fig. 8A).
FIG. 8.
The E1B 55-kDa protein inhibits enhancement of p53's transactivation by Daxx. The indicated p53-responsive luciferase reporter or a control luciferase reporter along with pRL-SV40 (internal control) was transfected with various combinations of expression vectors as indicated. Dual-luciferase assays were performed 24 to 48 h after transfection. The values shown are means of two independent experiments, and the error bars represent 1 standard deviation. (A) Daxx enhances p53-dependent transcription from the Bax promoter in p53−/− HCT116 cells. The Bax promoter (between positions 286 and 656 of GenBank entry U17193) cloned into pGL3-Basic and pRL-SV40 (0.2 μg of each) along with other indicated plasmids was transfected into p53−/− HCT116 cells grown on 24-well plates. Dual-luciferase assays were performed 48 h after transfection. Daxx-M is an internal fragment of Daxx cDNA that encodes aa 129 to 396 (Fig. 1E). This fragment did not bind to p53 (data not shown), consistent with the finding that the acidic domain of Daxx (aa 400 to 500) interacts with p53 (26). Both wild-type Daxx and Daxx-M were tagged with Flag at the N terminus and cloned into pcDNA3.1(−). The vector used for expression of wild-type p53 was pCMV-Neo-Bam. (B) Daxx enhances p53-dependent transcription from the MDM2 promoter in Saos2 cells. The reporter (pGL2-Basic) carries a DNA fragment encompassing the p53-binding sites from the human MDM2 gene. The reporter assays were performed 24 h after transfection. (C) p53 does not activate the Daxx promoter. The promoter of the human Daxx gene was cloned into pGL3-Basic and used for the reporter assay in Saos2 cells as described for panel B. (D) The E1B 55-kDa protein eliminates Daxx's enhancement of p53-mediated transactivation. p53−/− HCT116 cells were transfected with Bax-Luc plus pRL-SV40 (0.2 μg of each) and with the specified amounts of the other plasmids indicated. Dual-luciferase assays were performed as described for panel A. The Ad2 E1B 55-kDa protein open reading frame cloned in vector pcDL-SRα-296 was used for E1B expression.
With a different reporter that contains the p53-binding sequence derived from the MDM2 gene promoter, Daxx similarly enhanced p53-mediated transactivation when p53- and Daxx-expressing vectors were transfected into Saos2 cells (Fig. 8B). In control reporter assays, we examined whether p53 affects the promoter activity of the human Daxx gene. This promoter is highly active in driving the expression of the firefly luciferase reporter in transfected Saos2 cells (Fig. 8C). This promoter does not contain any recognizable p53-binding elements, and as expected, p53 did not affect the reporter activity driven by the Daxx promoter, and cotransfection of expression vectors for Daxx and p53 had no effect (Fig. 8C). Therefore, we concluded that Daxx specifically enhances p53-mediated transcription.
We then assessed how the E1B 55-kDa protein might affect Daxx-mediated enhancement of p53 transactivation. p53-deficient HCT116 cells were transfected with the luciferase reporter driven by the Bax promoter and various combinations of expression vectors for p53, Daxx, and the E1B 55-kDa protein. As shown in Fig. 8D, p53 expression led to a 53-fold increase in reporter activity (compared to that of cells transfected with only reporters), whereas in the presence of the Ad2 E1B 55-kDa protein, the fold induction was reduced to 34 (compared to that of reporter-transfected cells) or only 16-fold relative to that in cells transfected with E1B and a reporter. In the presence of E1B, Daxx expression was no longer able to augment p53-dependent transcription; the fold induction was essentially unchanged whether or not a Daxx expression vector was cotransfected (35-fold when Daxx was transfected above that of reporter-transfected cells or 16-fold versus that of cells transfected with either Daxx plus reporters or E1B plus reporters). Taken together, these data suggest that sequestration of Daxx and p53 in dot-like nuclear structures by the E1B 55-kDa oncoprotein impairs p53-dependent transcription and entirely blocks the enhancement of such activities by Daxx.
DISCUSSION
In this study, we have identified Daxx as a binding protein of the E1B 55-kDa protein through yeast two-hybrid screens. The interaction between these two proteins was confirmed in vivo and in vitro by biochemical and immunological methods. The in vitro binding result indicates that the two proteins directly bind to each other (Fig. 2). Strikingly, Daxx, p53, and the E1B 55-kDa protein colocalize in a few punctate nuclear dots in 293 cells (Fig. 3 and 5). Overexpression of PML drastically reduced the colocalization of Daxx with p53 and E1B, and at the same time, Daxx-PML colocalization markedly increased (Fig. 5). Furthermore, expression of the E1B 55-kDa protein in Saos2 cells significantly reduced the number of PML bodies (Fig. 6 and 7). Thus, there appears to be an active competition between PML and the E1B 55-kDa protein in the recruitment of Daxx between PML bodies and p53-E1B 55-kDa protein complexes.
What could be the biological consequence of dynamic redistribution of Daxx? It is possible that Daxx is required for PML-mediated, as well as E1B 55-kDa protein-mediated, processes. Daxx could tether enzymatic activities to the E1B 55-kDa protein and PML bodies. This possibility is consistent with the fact that Daxx interacts with diverse cellular, as well as viral, proteins that have very different functions and subcellular distributions. What kind of enzymatic activities that Daxx can deliver remains to be determined. One obvious possibility is that Daxx can bring various modifying enzymes. For example, Daxx is found to associate with histone deacetylases (15). Our unpublished data indicated that Daxx associates with several kinases, including casein kinase II (CK2; data not shown). This finding may be significant with respect to the function of the E1B 55-kDa protein. Branton and colleagues found that CK2 can phosphorylate several serine residues in the C-terminal domain of the 55-kDa protein and phosphorylation of these sites is critical for its repression of p53-dependent transcription, inhibition of apoptosis, and cooperation with E1As and the E1B 19-kDa protein in cell transformation (39). With our findings regarding Daxx-E1B interaction, we propose that Daxx may facilitate phosphorylation of the E1B 55-kDa protein by CK2 or other kinases. This facilitation does not necessarily require the phosphorylation sites in the 55-kDa protein for Daxx-E1B interaction. Indeed, mutation of the C-terminal phosphorylation sites of the Ad2 E1B 55-kDa protein (S489/490 and T494) to alanine did not affect the Daxx-E1B interaction (Fig. 7).
On the other hand, control of access to Daxx might have important regulatory roles in the cell. Thus, sequestration of Daxx by PML and the E1B 55-kDa oncoprotein may regulate various cellular processes. We report here that Daxx can enhance p53-mediated transcription (Fig. 8). This enhancement might be explained in the light of our proposal. Daxx may tether kinases to p53 and facilitate its phosphorylation. This might lead to the observed enhancement of p53's transactivation function by Daxx. Importantly, expression of the E1B 55-kDa protein eliminated Daxx's effect on p53 (Fig. 8). As both the p53 and E1B 55-kDa proteins colocalize with Daxx in large nuclear bodies (Fig. 3 to 5), it is plausible that Daxx is removed from sites of p53 action (e.g., p53-responsive promoters) and sequestered in E1B 55-kDa protein complexes.
Recruitment of Daxx from PML bodies and reduction of their number by the E1B 55-kDa protein could compromise PML's role in the regulation of cell growth. It was shown previously that oncogenic Ras upregulates both p53 and PML and induces premature senescence (6, 28). In the same time, Ras stimulates the localization of CBP, a transcriptional coactivator and acetylase, to PML bodies and stabilizes a trimeric complex containing p53, PML, and CBP. Significantly, p53 acetylation appears to be increased in response to Ras expression, but this enhancement of p53 acetylation is lost in PML-null cells (28). Thus, acetylation of p53 by CBP may be facilitated in PML bodies in the context of cell nuclei. We note that Daxx may be recruited away from PML bodies (Fig. 5 and 6) to a structure containing E1B, p53, and Daxx (Fig. 4 to 6). Therefore, one can envision that expression of the E1B 55-kDa protein alters the intracellular interactions of PML, p53, Daxx, and CBP, among others, which could affect p53 acetylation and its transactivation activity. Consistently, our previous results showed that the E1B 55-kDa protein specifically inhibits p53 acetylation (20). Precisely how the E1B 55-kDa protein modulates PML and p53 function through competitive recruitment of Daxx and its ensuing implication in Ad-induced cell transformation remain to be investigated.
Acknowledgments
We thank John Aris, Arnold Berk, Kun-Sang Chang, Ronald Evans, Arnold Levine, Hua Lu, Thomas Shenk, and Bert Vogelstein for reagents and Bill Dunn and Steve Sugrue for reading the manuscript.
This work was supported by National Institutes of Health grant R01 CA92236 (D.L.) and in part by Canadian Institutes of Health Research grants MOP-14109 and MOP-42429 (D.L.). In addition, D. Liao received pilot project grants from the University of Florida Shands Cancer Center and Biomedical Research Support Program for Medical Schools awarded to the University of Florida College of Medicine by the Howard Hughes Medical Institute.
REFERENCES
- 1.Beck, G. R., Jr., B. R. Zerler, and E. Moran. 1998. Introduction to DNA tumor viruses: adenovirus, simian virus 40, and polyomavirus, p. 51-86. In D. J. McCance (ed.), Human tumor viruses. ASM Press, Washington, D.C.
- 2.Bunz, F., A. Dutriaux, C. Lengauer, T. Waldman, S. Zhou, J. P. Brown, J. M. Sedivy, K. W. Kinzler, and B. Vogelstein. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497-1501. [DOI] [PubMed] [Google Scholar]
- 3.Debbas, M., and E. White. 1993. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 7:546-554. [DOI] [PubMed] [Google Scholar]
- 4.Dobbelstein, M., J. Roth, W. T. Kimberly, A. J. Levine, and T. Shenk. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 16:4276-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 6.Ferbeyre, G., E. de Stanchina, E. Querido, N. Baptiste, C. Prives, and S. W. Lowe. 2000. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 14:2015-2027. [PMC free article] [PubMed] [Google Scholar]
- 7.Flint, J., and T. Shenk. 1997. Viral transactivating proteins. Annu. Rev. Genet. 31:177-212. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez, R. A., and S. J. Flint. 2002. Effects of mutations in the adenoviral E1B 55-kilodalton protein coding sequence on viral late mRNA metabolism. J. Virol. 76:4507-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 10.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 11.Hahn, W. C., C. M. Counter, A. S. Lundberg, R. L. Beijersbergen, M. W. Brooks, and R. A. Weinberg. 1999. Creation of human tumour cells with defined genetic elements. Nature 400:464-468. [DOI] [PubMed] [Google Scholar]
- 12.Harada, J. N., A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, H., C. Leo, J. Zhu, X. Wu, J. O'Neil, E. J. Park, and J. D. Chen. 2000. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol. Cell. Biol. 20:1784-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao, D., A. Yu, and A. M. Weiner. 1999. Coexpression of the adenovirus 12 E1B 55 kDa oncoprotein and cellular tumor suppressor p53 is sufficient to induce metaphase fragility of the human RNU2 locus. Virology 254:11-23. [DOI] [PubMed] [Google Scholar]
- 17.Lill, N. L., S. R. Grossman, D. Ginsberg, J. DeCaprio, and D. M. Livingston. 1997. Binding and modulation of p53 by p300/CBP coactivators. Nature 387:823-827. [DOI] [PubMed] [Google Scholar]
- 18.Lin, D. Y., and H. M. Shih. 2002. Essential role of the 58-kDa microspherule protein in the modulation of Daxx-dependent transcriptional repression as revealed by nucleolar sequestration. J. Biol. Chem. 277:25446-25456. [DOI] [PubMed] [Google Scholar]
- 19.Lin, J., J. Chen, B. Elenbaas, and A. J. Levine. 1994. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 8:1235-1246. [DOI] [PubMed] [Google Scholar]
- 20.Liu, Y., A. L. Colosimo, X. J. Yang, and D. Liao. 2000. Adenovirus E1B 55-kilodalton oncoprotein inhibits p53 acetylation by PCAF. Mol. Cell. Biol. 20:5540-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe, S. W., and H. E. Ruley. 1993. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 7:535-545. [DOI] [PubMed] [Google Scholar]
- 22.Marcellus, R. C., J. G. Teodoro, R. Charbonneau, G. C. Shore, and P. E. Branton. 1996. Expression of p53 in Saos-2 osteosarcoma cells induces apoptosis which can be inhibited by Bcl-2 or the adenovirus E1B-55 kDa protein. Cell Growth Differ. 7:1643-1650. [PubMed] [Google Scholar]
- 23.Martin, M. E., and A. J. Berk. 1998. Adenovirus E1B 55K represses p53 activation in vitro. J. Virol. 72:3146-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin, M. E., and A. J. Berk. 1999. Corepressor required for adenovirus E1B 55,000-molecular-weight protein repression of basal transcription. Mol. Cell. Biol. 19:3403-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morales, C. P., S. E. Holt, M. Ouellette, K. J. Kaur, Y. Yan, K. S. Wilson, M. A. White, W. E. Wright, and J. W. Shay. 1999. Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nat. Genet. 21:115-118. [DOI] [PubMed] [Google Scholar]
- 26.Ohiro, Y., A. Usheva, S. Kobayashi, S. L. Duffy, R. Nantz, D. Gius, and N. Horikoshi. 2003. Inhibition of stress-inducible kinase pathways by tumorigenic mutant p53. Mol. Cell. Biol. 23:322-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orlando, J. S., and D. A. Ornelles. 2002. E4orf6 variants with separate abilities to augment adenovirus replication and direct nuclear localization of the E1B 55-kilodalton protein. J. Virol. 76:1475-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson, M., R. Carbone, C. Sebastiani, M. Cioce, M. Fagioli, S. Saito, Y. Higashimoto, E. Appella, S. Minucci, P. P. Pandolfi, and P. G. Pelicci. 2000. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406:207-210. [DOI] [PubMed] [Google Scholar]
- 29.Prives, C., and J. L. Manley. 2001. Why is p53 acetylated? Cell 107:815-818. [DOI] [PubMed] [Google Scholar]
- 30.Punga, T., and G. Akusjarvi. 2000. The adenovirus-2 E1B-55K protein interacts with a mSin3A/histone deacetylase 1 complex. FEBS Lett. 476:248-252. [DOI] [PubMed] [Google Scholar]
- 31.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reid, J. L., A. J. Bannister, P. Zegerman, M. A. Martinez-Balbas, and T. Kouzarides. 1998. E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J. 17:4469-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ricciardi, R. P. 1995. Transformation and tumorigenesis mediated by the adenovirus E1A and E1B oncogenes, p. 195-225. In G. Barbanti-Brodano et al. (ed.), DNA tumor viruses: oncogenic mechanisms. Plenum Press, New York, N.Y.
- 34.Roulston, A., R. C. Marcellus, and P. E. Branton. 1999. Viruses and apoptosis. Annu. Rev. Microbiol. 53:577-628. [DOI] [PubMed] [Google Scholar]
- 35.Ruley, H. E. 1983. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature 304:602-606. [DOI] [PubMed] [Google Scholar]
- 36.Sarnow, P., C. A. Sullivan, and A. J. Levine. 1982. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology 120:510-517. [DOI] [PubMed] [Google Scholar]
- 37.Steegenga, W. T., T. van Laar, N. Riteco, A. Mandarino, A. Shvarts, A. J. van der Eb, and A. G. Jochemsen. 1996. Adenovirus E1A proteins inhibit activation of transcription by p53. Mol. Cell. Biol. 16:2101-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steegenga, W. T., T. Van Laar, A. Shvarts, C. Terleth, A. J. Van der Eb, and A. G. Jochemsen. 1995. Distinct modulation of p53 activity in transcription and cell-cycle regulation by the large (54 kDa) and small (21 kDa) adenovirus E1B proteins. Virology 212:543-554. [DOI] [PubMed] [Google Scholar]
- 39.Teodoro, J. G., and P. E. Branton. 1997. Regulation of p53-dependent apoptosis, transcriptional repression, and cell transformation by phosphorylation of the 55-kilodalton E1B protein of human adenovirus type 5. J. Virol. 71:3620-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van den Heuvel, S. J., T. van Laar, I. The, and A. J. van der Eb. 1993. Large E1B proteins of adenovirus types 5 and 12 have different effects on p53 and distinct roles in cell transformation. J. Virol. 67:5226-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weigel, S., and M. Dobbelstein. 2000. The nuclear export signal within the E4orf6 protein of adenovirus type 5 supports virus replication and cytoplasmic accumulation of viral mRNA. J. Virol. 74:764-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yew, P. R., and A. J. Berk. 1992. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature 357:82-85. [DOI] [PubMed] [Google Scholar]
- 43.Yew, P. R., X. Liu, and A. J. Berk. 1994. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 8:190-202. [DOI] [PubMed] [Google Scholar]
- 44.Yu, A., H. Y. Fan, D. Liao, A. D. Bailey, and A. M. Weiner. 2000. Activation of p53 or loss of the Cockayne syndrome group B repair protein causes metaphase fragility of human U1, U2, and 5S genes. Mol. Cell 5:801-810. [DOI] [PubMed] [Google Scholar]
- 45.Zantema, A., J. A. Fransen, A. Davis-Olivier, F. C. Ramaekers, G. P. Vooijs, B. DeLeys, and A. J. Van der Eb. 1985. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology 142:44-58. [DOI] [PubMed] [Google Scholar]