Abstract
The VP6 protein of bluetongue virus possesses a number of activities, including nucleoside triphosphatase, RNA binding, and helicase activity (N. Stauber, J. Martinez-Costas, G. Sutton, K. Monastyrskaya, and P. Roy, J. Virol. 71:7220-7226, 1997). Although the enzymatic functions of the protein have been documented, a detailed structure and function study has not been completed and the oligomeric form of the protein in solution has not been described. In this study, we have characterized VP6 activity by creating site-directed mutations in the putative functional helicase domains. Mutant proteins were expressed at high levels in an insect cell by using recombinant baculoviruses purified and analyzed for ATP binding, ATP hydrolysis, and RNA unwinding activities. UV cross-linking experiments indicated that the lysine residue in the conserved motif AXXGXGK110V is directly involved in ATP binding, whereas mutant R205Q in the arginine-rich motif ER205XGRXXR bound ATP at a level comparable to that of the wild-type protein. The RNA binding activity was drastically altered in the R205Q mutant and was also affected in the K110N mutant. Helicase activity was altered in both mutants. The mutation E157N in the DEXX sequence, presumed to act as a Walker B motif, showed an intermediate activity, implying that this motif does not play a crucial role in VP6 function. Purified protein demonstrated stable oligomers with a ring-like morphology in the presence of nucleic acids similar to those shown by other helicases. Gel filtration chromatography, native gel electrophoresis, and glycerol gradient analysis clearly indicated multiple oligomeric forms of VP6.
Helicase proteins catalyze the unwinding of the double-stranded nucleic acids in order to initiate replication and transcription (34). The two processes, translocation and strand separation, occur simultaneously during nucleic acid unwinding and use energy from ATP hydrolysis (3, 11, 35, 37, 58). The exact mechanism of helicase action is still unclear, although a number of models have been proposed. In these models, the helicase protein could be active as a monomer, dimer, or hexamer and the nucleic acid binding site could reside at multiple sites on a monomer or on separate subunits of an oligomeric helicase (1, 4, 33). Experiments that address the active state of the helicase enzyme represent the first step towards a total understanding of the mechanism of helicase unwinding for this virus.
Previously, we identified a helicase activity in the VP6 protein, a nucleocapsid-associated protein of bluetongue virus (BTV). BTV, which is in the Reoviridae family, has a genome of 10 double-stranded RNA (dsRNA) segments, each of which has two cRNA strands with end-to-end complementarities (12). Purified VP6 in isolation has the ability to unwind BTV dsRNA in vitro (52). Like most other nucleic acid-unwinding proteins, VP6 binds a single-stranded region for initial attachment; however, it could also bind to and initiate helicase activity on a perfect duplex RNA (14, 16, 47, 52). Sequence comparisons of a large number of helicases from different organisms have identified various common functional motifs, including putative sequences for RNA binding, ATP binding, and helicase function as well as a Walker A motif commonly present in nucleoside triphosphate (NTP) binding and hydrolyzing enzymes (5, 7, 8, 20, 27, 28, 31, 50, 54). A number of similar, though not identical, functional motifs can be recognized in the BTV VP6 protein when it is compared with other helicases for which crystal structures are available (12, 24, 29, 56, 57). The 35-kDa VP6 protein is one of three minor proteins of BTV located within the core of BTV along with the dsRNA segments and two other proteins (VP1 and VP4) that are involved in viral RNA replication (for a review, see reference 46). The core is mainly composed of two major proteins, VP3 and VP7, apart from the three minor proteins and viral genome. In the mature virions, the core is enclosed by an outer capsid formed by two other major proteins, VP2 and VP5, both of which are lost during the initial stages of virus entry and penetration into the host cells. Within the cytoplasm, the BTV core is transcriptionally active and repeatedly transcribes and caps viral mRNA from each of the 10 dsRNA segments. The newly synthesized RNA transcripts subsequently extrude from the core into the cytoplasm to initiate viral protein translation (46). There is a considerable body of information available regarding the BTV core structure, determined both by cryoelectron microscopy (cryo-EM) and by X-ray crystallography (17, 18, 43). However, although the structures of VP3 and VP7, the two major components of the viral core, have been resolved in detail, the structural organization of the three minor proteins that constitute the transcriptase complex, which is closely associated with the dsRNA genome, have not been defined at atomic resolution. Recent cryo-EM studies of core-like particles have suggested the location and the overall structural organizations of the two minor proteins, VP1 and VP4 (P. Roy and B. V. V. Prasad, unpublished data). The functions of these two proteins, namely the RNA polymerase and the mRNA capping enzyme, have also been established previously (44, 45, 55). It was also possible to assign a role to the smallest minor protein, VP6, in the transcription complex, in that it has RNA helicase activity (52). However, neither the mechanism of helicase activity nor and the oligomeric nature of VP6 has been firmly established. In this study, we map the functional domains of the various individual activities associated with the VP6 helicase. In the absence of structural information, we have identified the putative ATPase, ATP hydrolysis, RNA binding, and RNA unwinding domains through alignment with those of known helicases. Site-specific mutagenesis was then used on each of the putative helicase motifs to confirm a functional role. To produce sufficient protein for analysis, each VP6 mutant was expressed by using the baculovirus expression system and the recombinant mutant protein was purified. The effects of these altered residues on VP6 activities were subsequently determined by in vitro assays. To determine the oligomeric nature of VP6, protein was purified at homogeneity and analyzed for its molecular form in solution both alone and following incubation with single-stranded RNA (ssRNA) or dsRNA. We show that, in common with other known helicases, the protein is hexameric and forms a discrete ring-like structure when complexed with its substrate.
MATERIALS AND METHODS
Virus and cells.
Autographa californica nuclear polyhedrosis virus and the recombinant baculoviruses were propagated in the Spodoptera frugiperda (Sf9) insect cell line at 28°C in serum free SF900 II medium (Gibco) as described elsewhere (25).
Site-directed mutagenesis and isolation of recombinant baculoviruses expressing mutant VP6 proteins.
The plasmid pAcBTV10.9 (47), which contains the DNA sequence encoding the BTV serotype 10 VP6 protein, was employed directly for site-directed mutagenesis with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) according to the vendor's instructions. The oligonucleotides used for mutagenesis and the resulting amino acids changes are as follows: K110N, AGAATGCTAACAGAGGAGATGGAAACGTTGGAGGAGGAGG; D157N, GCATGCTAAAGCCGTTGAGCAAGGAGGACGCAAGCAGAGG; R205Q, GAAGATTGATGTTTACAGGAATGAGGTTCCAGCTCAGATC. Mutated sequences in the recombinant plasmid were confirmed by sequence analysis (48). The Lipofectin technique (10) was used to construct a monolayer of Sf9 cells with recombinant transfer vectors and BSU361 triple-cut A. californica nuclear polyhedrosis virus DNA (26). Recombinant baculoviruses were selected on the basis of their LacZ-negative phenotype and plaque purified as described elsewhere (42).
Expression and purification of BTV-10 VP6 wild-type and mutant proteins.
S. frugiperda (Sf9) insect cells were infected with recombinant baculoviruses at a multiplicity of infection of 5 PFU/cell and incubated in serum-free SF900 (Gibco) medium for 48 h at 28°C. The recombinant wild-type (47) and mutant VP6 proteins were purified as described previously (52).
Gel filtration chromatography.
The molecular weight of each recombinant baculovirus expressing VP6 was determined on a Pharmacia Hi-load 16/60 Superdex-200 column in a buffer containing 20 mM Tris-HCl (pH 7.5) and 20 mM NaCl. The flow rate was 0.3 ml/min, and 1-ml fractions were collected and analyzed by sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE). Molecular mass standards were obtained from Pharmacia.
Protein cross-linking.
Purified VP6 protein was cross-linked with 0.1% glutaraldehyde as follows. Cross-linking reactions were performed at room temperature at a final concentration of 2 μM VP6 in cross-linking reaction buffer (20 μl) containing 20 mM Tricine (pH 8.3), 50 mM NaCl, 20% glycerol, 5 mM MgCl2, and 3 mM dithiothreitol (DTT). Samples were preincubated on ice for 15 min prior to the addition of glutaraldehyde (EM grade; Electron Microscopy Science) to a final concentration of 0.1%. Reactions were quenched after 30 min by adding 2 M Tris-HCl (pH 6.8). Samples were resolved on SDS-9% PAGE and stained with Coomassie brilliant blue.
Preparation of in vitro transcripts for RNA binding.
RNA transcripts representing full-length plus-strand RNA of BTV segment 7 were prepared by in vitro transcription of plasmid pEC-10B7-H(δ) DNA which contains a full-length cDNA copy of BTV segment 7 in pUC-119 flanked by an upstream T7 promoter and downstream hepatitis δ ribozyme and T7 polymerase terminator (41). The T7 transcription reaction mixture was formed by using the Ribomax large-scale transcription kit (Promega, Madison, Wis.) under the conditions indicated by the vendor with the following modifications. Each transcription reaction mixture contains the supplied buffer, 4 mM concentrations of each NTP, 1 to 2 μg of DNA, and 2 μl of the supplied enzyme mix in a 20-μl final volume. The reaction mixtures were incubated at 37°C for 3 h. Two units of RNase-free DNase were added, and incubation continued for a further 15 min at 37°C. Following extraction with phenol-chloroform, the RNA was passed through a G-25 spin column (Pharmacia) to remove unincorporated NTPs and quantified by a spectrophotometer (Beckman DU530). The 5′ end of the RNA was radiolabeled with 32P by using [γ-32P]ATP (50 μCi) and T4 polynucleotide kinase. Excess [γ-32P]ATP was removed by passing through a G-25 column (Pharmacia).
Velocity sedimentation analysis.
Purified VP6-bound RNA was applied onto a 15 to 35% (vol/vol) glycerol gradient made in 20 mM Tris-HCl (pH 8), 100 mM NaCl, 3 mM DTT, and 3 mM MgCl2 and sedimented at 60,000 rpm for 3 h in a Beckman VTI 65 rotor at 4°C. Molecular mass markers in this gradient were from Pharmacia high-molecular-mass calibration kits. The standard consists of the following proteins: catalase (4 × 58 = 232 kDa), lactate dehydrogenate (4 × 36 = 140 kDa), and ovalbumin (43 kDa). Fractions were collected from the bottom of the tube and analyzed by SDS-PAGE and by Western blotting.
ATPase activity assays.
ATPase activity was assayed by measuring the extent of [γ-32P]ATP hydrolysis by using polyethyleneimine-cellulose chromatography followed by autoradiogram essentially as described previously (52). The efficiency of the activity of each mutant was calculated from the intensity of the spot as measured by phosphorimager compared with that of wild-type VP6, which was set at 100%.
Cross-linking of the oxidized ATP with VP6.
A cross-linking experiment was performed according to the previously established protocol (52). Briefly, [α-32P]ATP (400 Ci/mmole) was oxidized with 0.5 mM sodium periodate and 0.5 mM HCl, excess periodate was removed, and 0.5 μg of protein was incubated overnight with 0.5 μM oxidized ATP in 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, and 1 mM DTT as described previously (52). Protein complexes were analyzed by SDS-10% PAGE followed by autoradiography. The efficiency of each mutant was calculated by phosphorimager scanning and compared to that of the wild type, which was arbitrarily set at 100%.
Labeling of RNA oligonucleotide.
RNA oligonucleotides were purchased from Dharmacon Research (Lafayette, Colo.). The oligos were constructed by using standard methods (13) and are the following: 5′-AGAGAGAGAGGUUGAGAGAGAGAGAGUUUGAGAGAGAGAG-3′ (40-mer, template strand) and 5′-CAAACUCUCUCUCUCUCAAAAAAAAA-3′ (26-mer, release strand). The RNA oligos were subsequently 5′-end labeled with [γ-32P]ATP by using T4 polynucleotide kinase (New England Biolabs) according to the manufacturer's protocol. Unlabeled nucleotides were removed by using Sephadex G-50 spin columns (Pharmacia) and stored at −20°C in sterile H20 containing 0.25 U of RNasin (Promega) per μl.
RNA binding assays.
A UV cross-linking (61) method was employed to measure the RNA binding ability of the protein. Binding of small, labeled RNA oligonucleotides to the BTV VP6 and the mutant proteins was compared by gel mobility shift assay. One-half microgram of the wild-type or mutant helicase protein was incubated with 1.3 pmol of radiolabeled ssRNA (26-mer) oligonucleotides in 20 μl of helicase reaction buffer (30 mM Tris-HCl [pH 7.5], 3 mM MgCl2, 10 mM DTT, and 1 μl of RNasin [Promega]). The binding reaction mixture was incubated at 37°C for 15 min, and then the reaction was terminated by adding 5× RNA loading dye containing 0.5% Nonidet P-40 and subsequently analyzed on a 4% polyacrylamide-0.5× Tris-borate-EDTA gel. The bound complex was visualized by autoradiography.
Preparation of partial dsRNA substrate.
Two RNA oligos were combined at a molar ratio of release strand (labeled) to template strand (unlabeled) of approximately 1:10 in a solution containing 50 mM Tris-HCl (pH 7.5), 0.5 mM NaCl, and 1 mM EDTA. The mixture was boiled for 10 min, transferred to 65°C for 30 min, and incubation continued overnight at 25°C. The hybridized product was further purified through a Sephadex G-75 spin column (Pharmacia). Eluted duplex RNA was stored at −20°C for further use.
Helicase assays.
The helicase assay was done as described previously (52). Briefly, a duplex unwinding reaction was performed in a 16-μl volume with 30 mM Tris-HCl (pH 7.5), 3 mM MgCl2, 10 mM DTT, 5 mM ATP, 1 μl of RNasin (Promega), 1 nM VP6 protein, and 1 nM 32P-labeled RNA duplex. The reaction mixture was incubated at 37°C for 30 min, and the reaction was terminated by the addition of 4 μl of 5× SDS-PAGE sample buffer and analyzed by 12% (wt/vol) nondenaturing PAGE in Tris-glycine buffer followed by autoradiography as described previously (52).
Electron microscopy analysis.
VP6 was incubated with RNA and diluted to 0.1 mg/ml, adsorbed to a carbon-coated copper grid, and stained with a 1% solution of uranyl acetate. Images were visualized by an electron microscope (Hitachi Instruments, Pleasanton, Calif.).
RESULTS
Rationale for selection of residues within VP6 to generate VP6 mutants: generation and purification of recombinant VP6 proteins.
Purified VP6 possesses a number of properties associated with its helicase function (52). Our laboratory has previously shown that it binds ATP, exhibits an RNA-dependent ATPase activity in the presence of Mg2+, which involves removal of the terminal phosphate, and in vitro unwinding of duplex RNA in the presence of ATP. For helicase activity, an essential prerequisite is the hydrolysis of NTP to generate the required energy for nucleic acid unwinding. In addition, helicases must possess an ATP binding domain and at least two nucleic acid binding sites in order to maintain contact with the substrate before and after unwinding (33). These features have not yet been confirmed on VP6 by identification followed by mutagenesis. We used the published crystal structure data of hepatitis C virus (HCV) NS3 helicase as the predominant guide for the mutational analysis of VP6 (6, 22, 24, 54, 59).
Three mutants were constructed by site-directed mutagenesis. One of these was designed to disrupt the ATP binding activity of VP6 by replacing a conserved positively charged lysine residue within the putative ATP binding domain (Fig. 1) with an uncharged residue. The G/AXXGXGKS/T sequence, known as the Walker A motif, is found in nearly all NTP hydrolyzing enzymes and is also responsible for NTP binding activity (32, 54, 57). Previous structural studies demonstrated that the conserved lysine residue in the Walker A motif is involved in binding to the β phosphate of the NTP (53, 56). This domain is represented by ANRGDGKV in BTV VP6 (conserved residues are in boldface). In the NS3 helicase of HCV, it was shown that changing a lysine (K) in the NTP binding domain to E or N changed drastically the ability of NS3 to bind ATP and unwind RNA duplexes (23, 54). Therefore, the lysine at position 110 of the VP6 protein (K110N) was replaced by an asparagine (K110N), creating AXXGXGN110V. In addition to G/AXXGXGKS/T, two other motifs (DEAD or DEXH and TAT or SAT) have also been identified for the ATP hydrolysis activity of helicases (54, 57). The glutamic acid (E) of the Walker B (DEXH) motif may serve as a Lewis base during the hydrolysis of ATP, as previously demonstrated in PcrA DNA helicase (56). In VP6 there are neither obvious DEAD or DEXH domains nor SAT or TAT domains, but a DEVP domain, very similar domain to DEAD, exists at residues of amino acids (aa) 156 to 159 (Fig. 1). To ascertain whether the conserved DE residues within this DEVP domain of VP6 have a role like the Walker B motif and are involved in the ATP hydrolysis, the glutamic acid of this domain was replaced by an asparagine (E157N).
FIG. 1.
Alignment of BTV with known helicases. The top section shows the general structure of typical DEAD box proteins and the three conserved sequence motifs ascribed to the activities shown. The alignment of several known RNA helicases with BTV VP6 are shown in the middle. Sequences were taken from GenBank with the following accession numbers: HCV-1, M62321; human RNA helicase A (HuHelA), L13848; Drosophila melanogaster maleless protein (DroMLE), M74121; plum pox virus CI protein (PpvHel), M92280; yeast PRP2 protein (PRP2), X55936; eIF-4A, X03040; vaccinia virus NPH II protein, M35027; reovirus λ1 protein, NP-694679; BTV-10 VP6, U55801. Amino acid substitutions made in the study to assess the role of BTV motifs in function are shown at the bottom.
The RXGRXXR motif is important for the RNA binding and unwinding and is known as conserved motif VI of helicases (20, 21, 39). This highly positively charged motif (Fig. 1) is easily identifiable in VP6 and is represented by RGGRKQR (aa 205 to 211), consistent with the role of RNA binding (conserved residues are in boldface). This sequence is conserved in the VP6 of other BTV serotypes. To investigate whether this domain constitutes the main RNA binding site, a substitution mutation at amino acid position 205 (R205Q) was introduced.
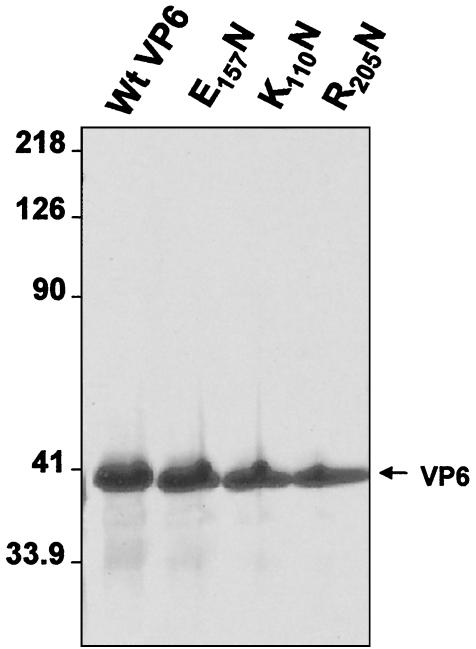
Each mutant construct was used to generate a recombinant baculovirus, and each mutant protein was expressed in an insect cell line. The expression of each recombinant VP6 was determined by SDS-10% PAGE followed by Coomassie blue staining and Western analysis with an anti-BTV10 antiserum (data not shown). All recombinant proteins were subsequently purified by using our established purification methods (see Materials and Methods). SDS-PAGE analysis of the purified proteins showed that the purity of each was ∼90%, which was suitable for direct biochemical analysis (Fig. 2). Moreover, the solubility and level of expression of mutant VP6 proteins were equivalent to that of wild-type VP6, indicating that the mutagenesis did not grossly affect the physical properties of the protein.
FIG. 2.
Purification of wild type (Wt) and recombinant BTV-10 VP6 from the baculovirus-infected Sf9 insect cells. Protein samples were resolved by SDS-10% PAGE and stained with Coomassie brilliant blue dye. Positions of molecular weight markers (left) and purified VP6 for each preparation (right) are indicated. Three recombinant mutant proteins (E157N, K110N, and R205Q) and wild-type VP6 in each lane are indicated.
ATPase activity and ATP binding activity of the VP6 mutants.
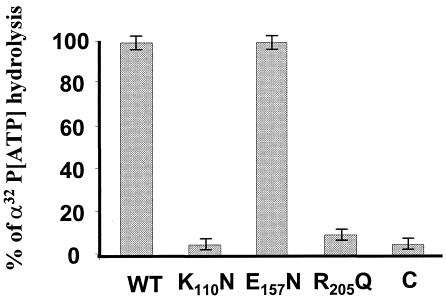
The NTP hydrolysis activity of the helicase enzyme hydrolyzes NTP to nucleoside diphosphate and phosphate, and this step is important for helicase movement and RNA unwinding. Our previous data demonstrated that ATP was the preferred NTP for VP6-catalyzed RNA unwinding. To map the ATPase activity in VP6, we employed the in vitro ATPase assay system established previously (52). All mutant proteins displayed reduced activity when compared to wild-type VP6 protein. Mutation in motif I (AXXGXGK110V) affected ATPase activity drastically and showed only 10% of the wild-type activity (Fig. 3). Mutation at putative motif II (DE157XX), in contrast, had very little effect, and approximately 90% of the activity was retained by the mutant, suggesting that this domain does not play a major role in VP6-catalyzed ATP hydrolysis (Fig. 3). Mutation R205Q at the RNA binding motif (RXGRXXR) also significantly reduced ATPase activity and showed only 10% of that achieved by the wild-type protein (Fig. 3), indicating that RNA binding activity may have a role in ATPase activity, either directly or indirectly. The results clearly demonstrate that the K110 of conserved motif I plays a crucial role in ATP hydrolysis and indicates that the activity is likely to be RNA dependent.
FIG. 3.
ATPase activities of the wild-type (WT) and mutant VP6 proteins. One-half microgram of wild-type VP6 or each VP6 mutant was incubated at 37°C for 15 min in a reaction mixture (20 μl) containing 30 mM Tris-HCl (pH 7.5), 3 mM MgCl2, 1 mM MnCl2, 10 mM DTT, 50 mg of bovine serum albumin per ml, 2 μg of poly(U), and 1 μCi of [γ-32P]ATP. The reaction product was analyzed by spotting 2-μl samples onto polyethyleneimine-cellulose plates, and the chromatograms were developed in 0.75 M potassium phosphate (pH 3.5). The plates were subsequently dried and autoradiographed. The amount of radioactivity at each position was estimated by phosphorimager, and the efficiencies of the ATPase activities of the three mutants were compared, with that of the wild type arbitrarily assigned a value of 100%. The control (C) represents a reaction mixture without VP6. The figure shows the mean values of the results of three different experiments.
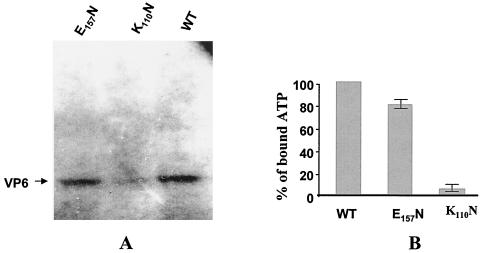
The ATP binding activity of the VP6 protein was examined by cross-linking with [α-32P]ATP as described previously. The labeled ATP-bound protein was resolved by SDS-10% PAGE and autoradiographed (Fig. 4); protein that displayed the ATP binding activity was covalently bound to ATP and radiolabeled. Wild-type VP6 was efficiently labeled by this assay as reported previously (52); however, mutant K110N had completely lost ATP binding activity, as expected (Fig. 4). The VP6 E157N mutant within the DEVP motif, believed to be a DEAD/DEXH Walker B motif, did bind ATP but did so less efficiently (∼75 to 80%) than the wild-type protein (Fig. 4B). The third mutant, R205Q, bound ATP with similar efficiency to that of the wild type (data not shown), indicating that this residue is not involved in the ATP binding activity of the protein.
FIG. 4.
ATP binding activities of the wild-type (WT) and mutant VP6 proteins. One-half microgram of protein was mixed with 0.5 μM oxidized [α-32P]ATP (400 Ci/mmol) in 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, and 1 mM DTT. The reaction was allowed to proceed overnight on ice in the presence of 7.5 mM NaBH3CN. The reaction was stopped by the addition of 5× SDS sample buffer, the mixture was boiled, and protein complexes were loaded onto an SDS-10% PAGE gel. After electrophoresis, the gel was dried and autoradiographed to determine the distribution of labeled proteins. (A) Autoradiogram of the E157N and K110N mutants and wild-type VP6 bound to [α-32P]ATP. (B) The efficiencies of the ATP binding activities of the mutant proteins were compared with that of the wild-type VP6 as described in Materials and Methods. The figure represents the mean values of the results of three different experiments.
RNA binding and helicase activity of the mutant proteins.
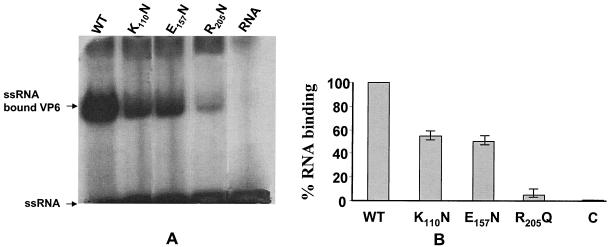
We analyzed the RNA binding activity of VP6 and the mutants by using optimized conditions for helicase enzyme activity (in the presence and absence of ATP) with a 26-mer ssRNA. Previously, in our laboratory, it was shown that VP6 binds nucleic acids in the presence of SDS with no apparent specificity (47). Here, we used non-BTV RNA to study the in vitro binding capacity of the VP6 and the mutant proteins. RNA binding efficiency was measured by gel mobility shift assay (Fig. 5). The data indicate that VP6 is able to form a stable complex with the ssRNA probe in absence of ATP. Among the three mutants tested (K110N, E157N, and R205Q), the mutant R205Q, a mutation at the putative RNA binding site, essentially eliminated binding activity (Fig. 5B), retaining less than 5% of the activity shown by the parental sequence. In contrast, at least 50% of the RNA binding capacity was retained by the other two mutants, K110N and E157N, that are involved in ATP binding and ATP hydrolysis activities, confirming the crucial role played by R205 in RNA binding.
FIG. 5.
RNA binding activities of wild-type (WT) and mutant helicase proteins. The wild-type or mutant protein was incubated with labeled short ssRNA under standard reaction conditions. One-half microgram of the wild-type or mutant helicase protein was incubated with 1.3 pmol of radiolabeled 26-mer ssRNA oligonucleotides in 20 μl of helicase reaction buffer (30 mM Tris-HCl [pH 7.5], 3 mM MgCl2, 10 mM DTT, and 1 μl of RNasin [Promega]). As a control, labeled RNA without VP6 was incubated in one reaction mixture. The binding reaction mixture was incubated at 37°C for 15 min; then the reaction was terminated by the addition of 5× RNA loading dye containing 0.5% Nonidet P-40, and the mixture was subsequently electrophoresed on a 4% polyacrylamide-0.5× Tris-borate-EDTA gel buffer. The bound complex was visualized by autoradiography. (A) Autoradiogram of PAGE showing the positions of bound RNA of the wild-type and three mutant VP6 proteins and free unbound RNA. An RNA probe without VP6 was also resolved as a control (indicated as RNA). (B) The radioactivity of labeled proteins in each reaction mixture was estimated by phosphorimager as described in Materials and Methods, and the efficiencies of the RNA binding activities of the three mutant proteins were compared with that of the wild-type VP6. The RNA probe control is indicated as C. The figure shows the mean values of the results of three different experiments.
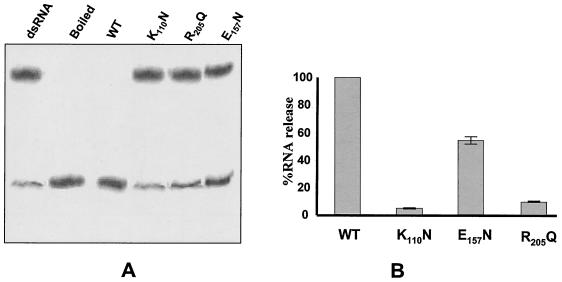
Helicase activity was assessed by the capacity of each purified protein to unwind a labeled partial duplex made from short 26-mer 32P-labeled oligonucleotide annealed to a 40-mer unlabeled template as described in Materials and Methods. The data that we have obtained from these experiments indicate that mutant K110N and R205Q both lost 90% of their ability to unwind the partial duplex RNA, whereas mutant E115N was only partially affected (Fig. 6). These data suggest that E115 is not directly involved in helicase activity.
FIG. 6.
RNA helicase activity associated with wild-type (WT) and mutant helicase protein. Helicase assays were done by using a partial duplex RNA substrate containing a 20-base double-stranded region with 5′ (10 nucleotides) and 3′ (10 nucleotides) single-stranded overhangs as described in Materials and Methods. The reaction was done in a 16-μl volume containing 1 nM VP6 protein and 1 nM 32P-labeled RNA duplex in 30 mM Tris-HCl (pH 7.5), 3 mM MgCl2, 10 mM DTT, 5 mM ATP, and 1 μl of RNasin (Promega). The reaction mixture was incubated at 37°C for 30 min, and the reaction was terminated in 4 μl of 5× SDS-PAGE sample buffer (100 mM Tris-HCl [pH 7.5], 50 mM EDTA, 0.5% SDS, 0.1% Nonidet P-40, 25% [vol/vol] glycerol, 5% [vol/vol] β-mercaptoethanol, 0.01% bromophenol blue). Samples were loaded onto a 12% (wt/vol) nondenaturing PAGE gel in Tris-glycine buffer and electrophoresed at 100 V. Gels were dried and autoradiographed. (A) Autoradiogram of PAGE showing the positions of labeled duplex RNA and ssRNA for each reaction mixture. Two controls, labeled duplex RNA without VP6 (indicated as dsRNA) and boiled labeled duplex RNA (indicated as Boiled), are included in addition to helicase reactions performed with each of the three mutants and wild-type VP6 proteins. (B) The efficiencies of the RNA binding activities of the three mutant proteins were compared with that of the wild-type VP6 as described for Fig. 3 to 5.
The overall results clearly indicate that there is a strong correlation between ATPase activity and RNA helicase activity of VP6. Unwinding of the dsRNA is an ATP-dependent process, and ATP hydrolysis is essential for the helicase activity. Both of these characteristic features are shown by VP6, and the putative domains identified by homology with known helicases appear to be functionally significant domains of VP6.
Purified VP6 forms a multimer in solution.
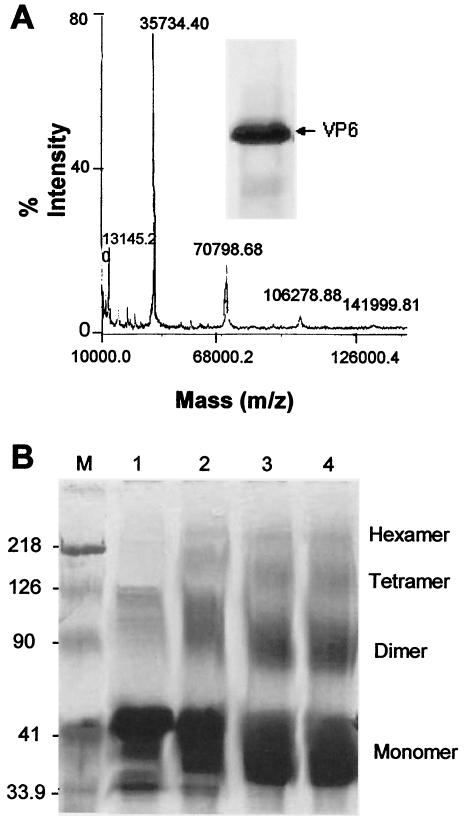
Helicase proteins generally function as hexamers. However it is not clear whether VP6 exists in a multimeric form consistent with the expectations of the confirmed biochemical analysis. We have addressed this issue by investigating the oligomeric nature of VP6 by using recombinant wild-type VP6 expressed by a previously generated recombinant baculovirus (47). Recombinant protein was purified to 90% (Fig. 7A, inset) homogeneity by ammonium sulfate precipitation followed by high-performance liquid chromatography, and the molecular weight in solution was initially investigated by mass spectrophotometer (MS). When approximately 5 μg of protein was subjected to MS analysis, both dimeric and tetrameric forms of VP6 were detectable (Fig. 7A), but the majority of the protein remained as a monomer, suggesting that the multimers formed by VP6 are not stable under these conditions.
FIG. 7.
Analysis of the oligomeric nature of purified VP6 protein. Wild-type BTV-10 VP6 was purified as described in Materials and Methods from the baculovirus-infected Sf9 insect cells. (A) Mass spectrometry of purified VP6 analyzed by matrix-assisted laser desorption ionization-time of flight (mass spectrometry). The ranges of the mass spectra containing the single mass ionization peaks for VP6 are displayed. The inset shows Coomassie blue-stained SDS-10% PAGE of purified VP6. (B) Purified VP6 was chemically cross-linked and analyzed by SDS-PAGE as described in Materials and Methods. Lanes 1 to 4, VP6 (3 μg) incubated with glutaraldehyde (0.1%) for 5, 10, 40, and 60 min, respectively. Molecular mass in kilodaltons is indicated on the side of the marker (M). The VP6 oligomers cross-linked by glutaraldehyde were identified based on their positions with respect to the marker position. Note that VP6 migrates as dimers, trimers, and hexamers in addition to monomers in the PAGE gel.
To stabilize the VP6-VP6 interactions, protein cross-linking was used by incubation of purified VP6 with 0.1% glutaraldehyde. When the products of this reaction were analyzed by SDS-10% PAGE, a number of protein species ranging in size from 35 to 200 kDa were detectable by Coomassie blue staining (Fig. 7B) and the molecular weight increments of the visualized bands would be consistent with a range of VP6 oligomers from monomer up to hexamer, although the majority of the species were dimeric (Fig. 7B). The identities of the oligomeric forms as VP6 were confirmed by Western blotting with anti-BTV antisera (data not shown). It is noteworthy that unmodified VP6 generally migrates slightly slower than its predicted molecular mass in SDS-10% PAGE but that, after glutaraldehyde treatment, the protein migrates near to the 37-kDa position (Fig. 7B).
MS analysis and a protein-protein cross-linking experiment demonstrated that the VP6 protein was capable of forming oligomers in solution, but VP6 dimers predominated and no uniform oligomeric state was defined.
VP6 forms defined hexamers in the presence of BTV transcripts and assembled into distinct virus-like structures.
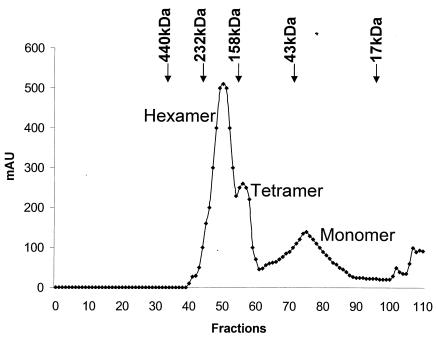
It is likely that multimerization of VP6 could be induced and stabilized by the addition of BTV RNA. Indeed, it has been shown previously that certain helicase proteins form specific oligomeric structures only in the presence of nucleic acid or NTPs (2, 11, 15, 38, 40). Such a possibility was therefore investigated for VP6 by incubating purified protein with in vitro-synthesized exact copy RNA transcripts of BTV segment 7. Electrophoresis mobility shift assays showed that with a saturated amount of VP6 protein, 100% RNA binding could be achieved (data not shown). The formation of a stable RNA-protein complex was further confirmed by gel filtration. RNA-bound VP6 protein was analyzed by a Hi Load Superdex-200 column. The elution profile when compared with the molecular markers clearly indicated the presence of higher oligomers of VP6 bound with RNA (Fig. 8).
FIG. 8.
Analysis of VP6-RNA complexes by gel filtration chromatography. Purified VP6 was incubated with S7 transcript in 30 mM Tris-HCl (pH 7.5), 10 mM DTT, 3 mM MgCl2, and 2.5 U of RNasin. The reaction mixture was incubated at 37°C for 30 min. Products were loaded on to an equilibrated Hi-Load Superdex-200 gel filtration column and eluted with same buffer at a flow rate of 0.3 ml/min as described in Materials and Methods. The column was calibrated with a set of globular protein standards (Amersham Pharmacia Biotech) consisting of ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), albumin (43 kDa), and myoglobulin (17 kDa) prior to the VP6 loading. The arrows point to the positions of their elution. Note that the three peaks of VP6 eluted near the positions of catalase, aldolase, and albumin, which are equivalent to the hexamer, tetramer, and monomer of VP6, respectively.
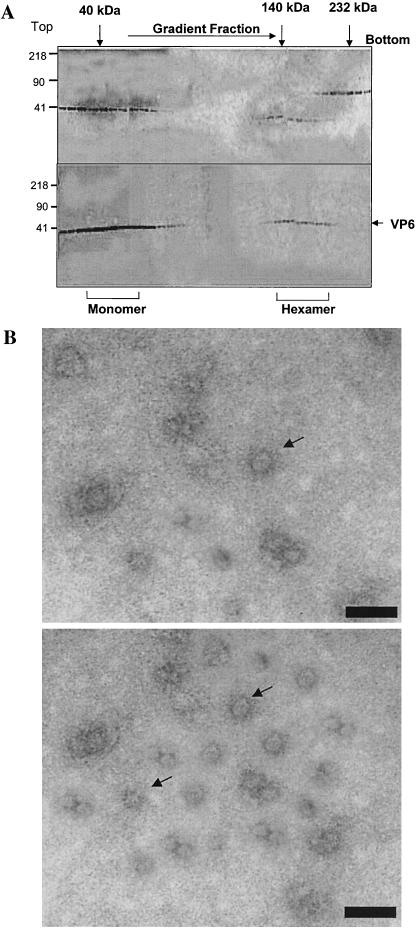
The oligomeric status of VP6 following RNA binding was investigated by glycerol gradient sedimentation as previously described (30). Gradients were fractionated, and the presence of VP6 was assessed by analysis by SDS-10% PAGE (Fig. 9A).
FIG. 9.
VP6-RNA complexes by glycerol gradient sedimentation and by electron microscope. (A) RNA-bound VP6 was subjected to velocity sedimentation analysis on a 15 to 35% glycerol gradient to separate the oligomeric forms of the protein. Fractions (0.4 ml) collected from bottom to top (right to left) were subjected to SDS-PAGE analysis and stained with Coomassie blue. Upper panel, the high-molecular-mass calibration standard (in kilodaltons) consists of catalase (4 × 58 = 232 kDa), lactate dehydrogenase (4 × 36 = 140 kDa), and ovalbumin (42 kDa). Lower panel, RNA-bound VP6, showing the position of VP6 in two regions, one in the top fractions and one nearthe bottom fractions. (B) Electron microscopy analysis of VP6-RNA complex. A glycerol gradient fraction containing a high-molecular-mass species of VP6 was applied to the glow-discharged carbon grids and stained with 1% uranyl acetate. Grids were examined with a Hitachi 7000 electron microscope at 75 kV. Upper panel, VP6-ssRNA complex; lower panel, VP6-dsRNA complex. Bars, 100 Å.
The two predominate peaks were identified, which, when compared to molecular mass markers analyzed in parallel, suggested that the slowest migrating peak corresponded to VP6 monomers (∼35 kDa) while the second peak represented a hexameric form (>210 kDa).
Glycerol gradient fractions containing the putative hexameric and monomeric forms of VP6 were analyzed by electron microscopy. The images of the putative hexameric fraction showed a ring-like structure (Fig. 9B, upper panel) very similar to the structures reported for other viral helicases (2, 11, 15, 38, 40). Similar apparent ring-like structures of hexamers were also formed when dsRNA was used (Fig. 9B, lower panel).
DISCUSSION
The data presented in this report confirm that VP6 is a typical RNA helicase protein with all the functional criteria for its identification mapped to specific sequences. Helicases identified from different organisms have been grouped into three general super families (SF), SFI, SFII, and SFIII, on the basis of conserved motifs (20, 21, 28, 50). At first glance, VP6 does not appear to belong to any of these families, as it does not possess the conserved sequences typical of the known helicases although partially matching motifs are present. The helicase λ1 protein of reovirus (Bisaillon M1997), the other member of the Reoviridae family, also, like BTV VP6, does not show any sequence similarities with other helicase proteins but possesses the characteristic motifs found in the DEAD subfamily (see sequences of K110N, D157N, and R205Q in Materials and Methods).
Helicase proteins are responsible for unwinding duplex nucleic acids, DNA, RNA, and DNA-RNA hybrids. Two essential features are shared by all helicases: ATP hydrolysis, which releases the energy required for helix destabilization, and nucleic acid binding site(s), which maintain contact with the nucleic acid that is to be unwound (33). RNA helicases also have a particular signature sequence, the DEAD box, or DECH box, which is responsible for forming the protein-ATP complex in the presence of Mg2+ (7, 8, 28). BTV VP6, with 328 aa, has a high proportion of hydrophilic and positively charged residues and inherently binds both ssRNA and dsRNA (47). It was also shown that VP6 possesses dsRNA unwinding activity in the presence of ATP and Mg2+ (52) and is capable of unwinding perfect RNA duplexes in vitro, unlike most helicases, which require a single-stranded region for initial attachment (47, 52). Surprisingly, however, it does not have the canonical DEAD box or DEXH signature motif, although other helicase motifs, such as those for RNA binding, ATP binding, and ATP hydrolysis, are present. Our studies used site-specific mutagenesis together with expression and purification and various biochemical assays to obtain direct evidence of the importance of these putative motifs in VP6 function.
The most N-terminal motif, ANRGDGKV (aa 104 to 111), of VP6 is comparable with the N-terminal conserved motif G/AXXGXGKS/T of other helicases. This motif forms a phosphate binding loop and is required for ATP binding and hydrolysis activities (56). The lysine of the GKS motif is directly involved in binding the β and γ phosphates of the NTP. Our findings that this sequence is also required for VP6 ATP hydrolysis and helicase activity suggests that the VP6 sequence may form similar loops. At least three RNA helicases have been well characterized: the eukaryotic translation initiation factor 4A (eIF-4A) mouse helicase protein, which belongs to the DEAD box family of proteins (39), and two members of the DEXH family, vaccinia virus NPH II protein (19) and HCV NS3 helicase protein (23, 54). For eIF-4A, replacement of the lysine residue of the GKT motif with glutamine severely reduced ATP binding and resulted in the complete loss of ATP hydrolysis and RNA helicase activity (39). Similarly, replacement of the lysine in the GKS motif of HCV NS3 helicase with asparagine (N) completely abolished ATPase activity, ATP binding activity, and RNA unwinding activity (54). In VP6, replacement of lysine (K110) in this motif with asparagine also affected ATP binding, ATP hydrolysis, and RNA unwinding. These data suggest that VP6 helicase activity, like that of all other helicases, is dependent on ATP hydrolysis.
The common conserved helicase RNA binding motif, RXGRXXR, is shared by VP6 protein and is represented by RRGRTGR (aa 205 to 211). RNA binding activity was completely abolished in eIF-4A variants with mutations within this motif (39). Similarly, changes in the first arginine in the RKGRVGR of the vaccinia virus NPH II protein or HCV NS3 helicase NS3 protein severely reduced RNA binding activity (19, 54). Mutational analysis in HCV NS3 strongly suggests that the first arginine residue forms hydrogen bonds with the key residues involved in RNA binding (31, 54). As expected, substitution at a single residue in this motif of BTV VP6 severely affected RNA binding and inhibited RNA unwinding activity. In addition, ATP hydrolysis was impaired, indicating that all three functions are interlinked and depended on the same specific sequences and structure of the protein. Our data indicate that VP6 is able to form a stable complex with ssRNA in the absence of ATP. A similar effect was observed for the NS3 helicase of HCV (13) and also for the helicase domain of the 126-kDa replicase protein of Tobacco mosaic virus (16).
Studies with other helicases have reported a key role for the conserved residues in the DEXD/H motif. Aspartic acid (D) can bind Mg2+ and assist in orienting the Mg2+-ATP complex for ATP hydrolysis (36). Glutamic acid (E) can serve as a Lewis base, initiating the hydrolysis of ATP as previously demonstrated in PerA DNA helicase (56). Changing glutamic acid (E) to glutamine (Q) in the HCV helicase did not alter ATP binding but abolished ATPase activity (54). However, as stated above, VP6 does not seem to have a DEAD-like motif, although there is a DE sequence in the context of DEVP (aa 156 to 159) in which valine may represent the alanine residue of the DEAD motif. Some helicases also have either isoleucine or cysteine residues in this position (Fig. 1). The fourth position of this motif is often replaced by histidine in some helicases, whereas VP6 has a proline residue in this position. It is possible that only the first two residues, DE, are important for maintenance of helicase function. When we mutated E157 to asparagine (DN157), ATP hydrolysis, RNA binding, and RNA unwinding were marginally affected (90% of that shown by the wild-type protein), not at the level reported for the HCV NS3 mutant protein (54), so it is possible that this motif may not be essential for VP6 enzyme activity.
Overall, the results obtained from this study clearly demonstrate that unwinding of RNA is an ATP-dependent process. It was instructive to compare the mutagenesis results with other well-characterized helicases like eIF-4A, HCV NS3, and vaccinia virus NPH II helicases, since all of these enzymes share common motifs and more or less similar activities. Although our results show evidence of the roles of individual amino acids in the overall function of VP6, these data do not exclude the possibility that other residues may play direct or indirect roles in the unwinding process and remain to be investigated.
Our studies also indicate that physically VP6 is similar to the other helicases. Generally, helicases are hexamers or at least exist as multimers. Our results demonstrate that the purified protein can form multimers, which, in the presence of RNA, could be stabilized as distinct ring-like particulate structures, a characteristic feature that is shared by other helicases (11, 15, 51). A range of biochemical methods, including MS, gel filtration, protein-protein cross-linking, and density ultracentrifugation, were used to identify the oligomeric nature of VP6. Clearly detectable VP6 oligomers ranging in size from monomer to hexamer could be easily detected, but it was not possible to isolate only the higher multimers of VP6, even in the presence of a cross-linking agent.
Hexameric helicases usually assemble into a ring-shaped structure. Escherichia coli DNAB and rho proteins were the first helicase proteins reported to be hexamers, but many helicases from different organisms are now known to be active as hexamers. Moreover, electron microscopy studies have shown that they share a common ring-like structure (2, 9, 11, 15, 21, 49, 60), a structure shared by the hexameric structures formed by the VP6-RNA complex. It is likely that the active form of VP6 has a similar structural organization, but only high-resolution cryo-EM studies or X-ray crystallography can confirm this view. It is noteworthy that other BTV RNA binding proteins, VP1 (polymerase), VP4 (capping enzyme), and NS2 (role in packaging genome), do not form a similar structure (data not shown). The accumulated data on function and structure of VP6 confirm that it is a helicase with key sequence features shared with other viral and cellular helicase proteins.
Acknowledgments
We thank Ian Jones (Reading University) for constructive comments on the manuscript.
This work was supported by grants from the NIH (United States) and Wellcome Trust fund (United Kingdom).
REFERENCES
- 1.Ahnert, P., and S. S. Patel. 1997. Asymmetric interactions of hexameric bacteriophage T7 DNA helicase with the 5′- and 3′-tails of the forked DNA substrate. J. Biol. Chem. 272:32267-32273. [DOI] [PubMed] [Google Scholar]
- 2.Barcena, M., C. S. Martin, F. Weise, S. Ayora, J. C. Alonso, and J. M. Carazo. 1998. Polymorphic quaternary organization of the Bacillus subtilis bacteriophage SPP1 replicative helicase (G40 P). J. Mol. Biol. 283:809-819. [DOI] [PubMed] [Google Scholar]
- 3.Bayliss, C. D., and G. L. Smith. 1996. Vaccinia virion protein I8R has both DNA and RNA helicase activities: implications for vaccinia virus transcription. J. Virol. 70:794-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird, L. E., J. A. Brannigan, H. S. Subramanya, and D. B. Wigley. 1998. Characterisation of Bacillus stearothermophilus PcrA helicase: evidence against an active rolling mechanism. Nucleic Acids Res. 26:2686-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bisaillon, M., J. Bergeron, and G. Lemay. 1997. Characterization of the nucleoside triphosphate phosphohydrolase and helicase activities of the reovirus lambda1 protein. J. Biol. Chem. 272:18298-18303. [DOI] [PubMed] [Google Scholar]
- 6.Cho, H. S., N. C. Ha, L. W. Kang, K. M. Chung, S. H. Back, S. K. Jang, and B. H. Oh. 1998. Crystal structure of RNA helicase from genotype 1b hepatitis C virus. A feasible mechanism of unwinding duplex RNA. J. Biol. Chem. 273:15045-15052. [DOI] [PubMed] [Google Scholar]
- 7.de la Cruz, J., D. Kressler, and P. Linder. 1999. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 24:192-198. [DOI] [PubMed] [Google Scholar]
- 8.Dillingham, M. S., P. Soultanas, P. Wiley, M. R. Webb, and D. B. Wigley. 2001. Defining the roles of individual residues in the single-stranded DNA binding site of PcrA helicase. Proc. Natl. Acad. Sci. USA 98:8381-8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egelman, E. H., X. Yu, R. Wild, M. M. Hingorani, and S. S. Patel. 1995. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc. Natl. Acad. Sci. USA 92:3869-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felgner, P. L., T. R. Gadek, M. Holm, R. Roman, H. W. Chan, H. Wenz, J. P. Northrop, G. M. Ringold, and M. Danielson. 1987. Lipofectin: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA 84:7413-7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fouts, E. T., X. Yu, E. H. Egelman, and M. R. Botchan. 1999. Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J. Biol. Chem. 274:4447-4458. [DOI] [PubMed] [Google Scholar]
- 12.Fukusho, A., Y. Yu, S. Yamaguchi, and P. Roy. 1989. Completion of the sequence of bluetongue virus serotype 10 by the characterization of a structural protein, VP6, and a non-structural protein, NS2. J. Gen. Virol. 70:1677-1689. [DOI] [PubMed] [Google Scholar]
- 13.Gallinari, P., D. Brennan, C. Nardi, M. Brunetti, L. Tomei, C. Steinkuhler, and R. De Francesco. 1998. Multiple enzymatic activities associated with recombinant NS3 protein of hepatitis C virus. J. Virol. 72:6758-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geider, K., and H. Hoffmann-Berling. 1981. Proteins controlling the helical structure of DNA. Annu. Rev. Biochem. 50:233-260. [DOI] [PubMed] [Google Scholar]
- 15.Gogol, E. P., S. E. Seifried, and P. H. von Hippel. 1991. Structure and assembly of the Escherichia coli transcription termination factor rho and its interaction with RNA. I. Cryoelectron microscopic studies. J. Mol. Biol. 221:1127-1138. [DOI] [PubMed] [Google Scholar]
- 16.Goregaoker, S. P., and J. N. Culver. 2003. Oligomerization and activity of the helicase domain of the tobacco mosaic virus 126- and 183-kilodalton replicase proteins. J. Virol. 77:3549-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimes, J. M., J. N. Burroughs, P. Gouet, J. M. Diprose, R. Malby, S. Zientara, P. P. C. Mertens, and D. I. Stuart. 1998. The atomic structure of the bluetongue virus core. Nature 395:470-478. [DOI] [PubMed] [Google Scholar]
- 18.Grimes, J. M., J. Jakana, M. Ghosh, A. K. Basak, P. Roy, W. Chiu, D. I. Stuart, and B. V. Prasad. 1997. An atomic model of the outer layer of the bluetongue virus core derived from X-ray crystallography and electron cryomicroscopy. Structure 5:885-893. [DOI] [PubMed] [Google Scholar]
- 19.Gross, C. H., and S. Shuman. 1995. Mutational analysis of vaccinia virus nucleoside triphosphate phosphohydrolase II, a DExH box RNA helicase. J. Virol. 69:4727-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadare, G., and A. L. Haenni. 1997. Virus-encoded RNA helicases. J. Virol. 71:2583-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karow, J. K., R. H. Newman, P. S. Freemont, and I. D. Hickson. 1999. Oligomeric ring structure of the Bloom's syndrome helicase. Curr. Biol. 9:597-600. [DOI] [PubMed] [Google Scholar]
- 22.Kim, D. W., J. Kim, Y. Gwack, J. H. Han, and J. Choe. 1997. Mutational analysis of the hepatitis C virus RNA helicase. J. Virol. 71:9400-9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, H., E. Fodor, G. Brownlee, and B. Seong. 1997. Mutational analysis of the RNA-fork model of the influenza A virus vRNA promoter in vivo. J. Gen. Virol. 78:353-357. [DOI] [PubMed] [Google Scholar]
- 24.Kim, J. L., K. A. Morgenstern, J. P. Griffith, M. D. Dwyer, J. A. Thomson, M. A. Murcko, C. Lin, and P. R. Caron. 1998. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6:89-100. [DOI] [PubMed] [Google Scholar]
- 25.King, L. A., and R. D. Possee (ed.). 1992. The baculovirus expression system: a laboratory guide. Chapman and Hall, London, United Kingdom.
- 26.Kitts, P. A., and R. D. Possee. 1993. A method for producing recombinant baculovirus expression vectors at high frequency. BioTechniques 14:810-817. [PubMed] [Google Scholar]
- 27.Koonin, E. V. 1993. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 21:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 29.Korolev, S., J. Hsieh, G. H. Gauss, T. M. Lohman, and G. Waksman. 1997. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell 90:635-647. [DOI] [PubMed] [Google Scholar]
- 30.Limn, C.-H., N. Staeuber, K. Monastyrskaya, P. Gouet, and P. Roy. 2000. Functional dissection of the major structural protein of bluetongue virus: identification of key residues within VP7 essential for capsid assembly. J. Virol. 74:8658-8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, C., and J. L. Kim. 1999. Structure-based mutagenesis study of hepatitis C virus NS3 helicase. J. Virol. 73:8798-8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin-Chao, S., C. L. Wei, and Y. T. Lin. 1999. RNase E is required for the maturation of ssrA RNA and normal ssrA RNA peptide-tagging activity. Proc. Natl. Acad. Sci. USA 96:12406-12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lohman, T. M., K. Thom, and R. D. Vale. 1998. Staying on track: common features of DNA helicases and microtubule motors. Cell 93:9-12. [DOI] [PubMed] [Google Scholar]
- 34.Luking, A., U. Stahl, and U. Schmidt. 1998. The protein family of RNA helicases. Crit. Rev. Biochem. Mol. Biol. 33:259-296. [DOI] [PubMed] [Google Scholar]
- 35.McDougal, V. V., and L. A. Guarino. 2001. DNA and ATP binding activities of the baculovirus DNA helicase P143. J. Virol. 75:7206-7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pai, E. F., U. Krengel, G. A. Petsko, R. S. Goody, W. Kabsch, and A. Wittinghofer. 1990. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 9:2351-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paolini, C., R. De Francesco, and P. Gallinari. 2000. Enzymatic properties of hepatitis C virus NS3-associated helicase. J. Gen. Virol. 81:1335-1345. [DOI] [PubMed] [Google Scholar]
- 38.Patel, S. S., and M. M. Hingorani. 1993. Oligomeric structure of bacteriophage T7 DNA primase/helicase proteins. J. Biol. Chem. 268:10668-10675. [PubMed] [Google Scholar]
- 39.Pause, A., N. Methot, Y. Svitkin, W. C. Merrick, and N. Sonenberg. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13:1205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Picha, K. M., and S. S. Patel. 1998. Bacteriophage T7 DNA helicase binds dTTP, forms hexamers, and binds DNA in the absence of Mg2+. The presence of dTTP is sufficient for hexamer formation and DNA binding. J. Biol. Chem. 273:27315-27319. [DOI] [PubMed] [Google Scholar]
- 41.Polkinhorne, I. G. 1994. Towards development of a system for reverse genetic studies on bluetongue virus. Ph.D. dissertation. University of Oxford, Oxford, United Kingdom.
- 42.Possee, R. D., C. J. Thomas, and L. A. King. 1999. The use of baculovirus vectors for the production of membrane proteins in insect cells. Biochem. Soc. Trans. 27:928-932. [DOI] [PubMed] [Google Scholar]
- 43.Prasad, B. V., S. Yamaguchi, and P. Roy. 1992. Three-dimensional structure of single-shelled bluetongue virus. J. Virol. 66:2135-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramadevi, N., J. N. Burroughs, P. P. C. Mertens, I. M. Jones, and P. Roy. 1998. Capping and methylation of mRNA by purified recombinant VP4 protein of bluetongue virus. Proc. Natl. Acad. Sci. USA 95:13537-13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramadevi, N., G. Kochan, and P. Roy. 1999. Polymerase complex of Bluetongue virus, p. 35. In Proceedings of the International Congress of Virology, Sydney, Australia.
- 46.Roy, P. 2001. Orbiviruses and their replication, p.1835-1869. In B. N. Fields (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 47.Roy, P., A. Adachi, T. Urakawa, T. F. Booth, and C. P. Thomas. 1990. Identification of bluetongue virus VP6 protein as a nucleic acid-binding protein and the localization of VP6 in virus-infected vertebrate cells. J. Virol. 64:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.San Martin, M. C., C. Gruss, and J. M. Carazo. 1997. Six molecules of SV40 large T antigen assemble in a propeller-shaped particle around a channel. J. Mol. Biol. 268:15-20. [DOI] [PubMed] [Google Scholar]
- 50.Schmid, S. R., and P. Linder. 1992. D-E-A-D protein family of putative RNA helicases. Mol. Microbiol. 6:283-291. [DOI] [PubMed] [Google Scholar]
- 51.Sedman, J., and A. Stenlund. 1998. The papillomavirus E1 protein forms a DNA-dependent hexameric complex with ATPase and DNA helicase activities. J. Virol. 72:6893-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stauber, N., J. Martinez Costas, G. Sutton, K. Monastyrskaya, and P. Roy. 1997. Bluetongue virus VP6 protein binds ATP and exhibits an RNA-dependent ATPase function and a helicase activity that catalyze the unwinding of double-stranded RNA substrates. J. Virol. 71:7220-7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanya, H. S., L. E. Bird, J. A. Brannigan, and D. B. Wigley. 1996. Crystal structure of a DExx box DNA helicase. Nature 384:379-383. [DOI] [PubMed] [Google Scholar]
- 54.Tai, C. L., W. C. Pan, S. H. Liaw, U. C. Yang, L. H. Hwang, and D. S. Chen. 2001. Structure-based mutational analysis of the hepatitis C virus NS3 helicase. J. Virol. 75:8289-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urakawa, T., D. G. Ritter, and P. Roy. 1989. Expression of largest RNA segment and synthesis of VP1 protein of bluetongue virus in insect cells by recombinant baculovirus: association of VP1 protein with RNA polymerase activity. Nucleic Acids Res. 17:7395-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Velankar, S. S., P. Soultanas, M. S. Dillingham, H. S. Subramanya, and D. B. Wigley. 1999. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97:75-84. [DOI] [PubMed] [Google Scholar]
- 57.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.White, P. W., A. Pelletier, K. Brault, S. Titolo, E. Welchner, L. Thauvette, M. Fazekas, M. G. Cordingley, and J. Archambault. 2001. Characterization of recombinant HPV6 and 11 E1 helicases: effect of ATP on the interaction of E1 with E2 and mapping of a minimal helicase domain. J. Biol. Chem. 276:22426-22438. [DOI] [PubMed] [Google Scholar]
- 59.Yao, N., T. Hesson, M. Cable, Z. Hong, A. D. Kwong, H. V. Le, and P. C. Weber. 1997. Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol. 4:463-467. [DOI] [PubMed] [Google Scholar]
- 60.Yu, X., M. M. Hingorani, S. S. Patel, and E. H. Egelman. 1996. DNA is bound within the central hole to one or two of the six subunits of the T7 DNA helicase. Nat. Struct. Biol. 3:740-743. [DOI] [PubMed] [Google Scholar]
- 61.Yue, V. T., and P. R. Schimmel. 1977. Direct and specific photochemical cross-linking of adenosine 5′-triphosphate to an aminoacyl-tRNA synthetase. Biochemistry 16:4678-4684. [DOI] [PubMed] [Google Scholar]