Abstract
The product of the UL11 gene of herpes simplex virus type 1 (HSV-1) is a 96-amino-acid tegument protein that accumulates on the cytoplasmic face of internal membranes. Although it is thought to be important for nucleocapsid envelopment and egress, the actual function of this protein is unknown. Previous studies focused on the characterization of sequence elements within the UL11 protein that function in membrane binding and trafficking to the Golgi apparatus. Binding was found to be mediated by two fatty acyl groups (myristate and palmitate), while an acidic cluster and a dileucine motif were identified as being important for the recycling of UL11 from the plasma membrane to the Golgi apparatus. The goal of the experiments described here was to identify and characterize binding partners (viral or cellular) of UL11. Using both immunoprecipitation and glutathione S-transferase (GST) pull-down assays, we identified a 40-kDa protein that specifically associates with UL11 from infected Vero cells. Mutational analyses revealed that the acidic cluster and the dileucine motif are required for this association, whereas the entire second half of UL11 is not. In addition, UL11 homologs from pseudorabies and Marek's disease herpesviruses were also found to be capable of binding to the 40-kDa protein from HSV-1-infected cells, suggesting that the interaction is conserved among alphaherpesviruses. Purification and analysis of the 40-kDa protein by mass spectrometry revealed that it is the product of the UL16 gene, a virion protein reported to be involved in nucleocapsid assembly. Cells transfected with a UL16-green fluorescent protein expression vector produced a protein that was of the expected size, could be pulled down with GST-UL11, and accumulated in a Golgi-like compartment only when coexpressed with UL11, indicating that the interaction does not require any other viral products. These data represent the first steps toward elucidating the network of tegument proteins that UL11 links to membranes.
During herpes simplex virus type 1 (HSV-1) assembly, over 30 different proteins come together to form three major structures: the nucleocapsid, the glycoprotein-containing envelope, and the collection of proteins located between the capsid and the envelope, known as the tegument (33). As with other herpesviruses, the most recent model for HSV-1 envelopment suggests that assembled nucleocapsids are shuttled out of the nucleus by budding and fusion events on the inner and outer nuclear membranes, respectively, and then travel through the cytoplasm until reaching trans-Golgi network (TGN)-derived vesicles (reviewed in reference 27). At this site, nucleocapsids are thought to acquire their final lipid bilayer in a process that also results in the acquisition of the tegument and glycoproteins. The mature virions subsequently follow the secretory pathway out to the cell surface, where they are released into the extracellular medium. Although several lines of evidence support this model, the specific molecular mechanisms by which the different components arrive and interact at the TGN have yet to be defined. As a start to understanding these processes, we have been studying the small tegument protein encoded by the UL11 gene of HSV-1.
The UL11 protein contains 96 amino acids and accumulates on the cytoplasmic faces of both nuclear and Golgi apparatus-derived membranes within infected cells (1, 24). Like other alphaherpesvirus homologs, HSV-1 UL11 is thought to play a role in nucleocapsid envelopment and egress; however, the specific functions of this protein are unknown (3, 21, 25, 35). Consistent with a role in budding at cytoplasmic locations, UL11 localizes primarily to the Golgi apparatus when expressed in the absence of other viral proteins (5). Mutational analyses have revealed that N-terminal myristylation and palmitylation provide both membrane-binding strength and Golgi-targeting specificity (5, 22), while a conserved acidic cluster motif is required for UL11 to recycle from the plasma membrane to the Golgi apparatus (22).
A major hindrance to understanding how UL11 functions during virus assembly and budding is the lack of knowledge concerning its interaction with other proteins. The difference between its localization within infected cells (nucleus and Golgi apparatus) and that within noninfected cells (Golgi apparatus only) suggests that UL11 interacts with either a viral protein or a virus-induced cellular protein; however, as with many other HSV-1 structural proteins, the binding partner(s) of this protein is unknown. As a start to elucidating the network of tegument interactions that emanate from UL11 on membranes, we set out to identify its binding partner(s) (viral or cellular). The experiments described in this report show that UL11 associates with several infected-cell proteins, including a 40-kDa protein identified as the product of the HSV-1 UL16 gene.
MATERIALS AND METHODS
Viruses and cells.
The viruses used in this study were strains KOS (37) and 17(+) (8) of HSV-1, a green fluorescent protein (GFP)-VP22-expressing HSV-1 strain (166v) (12), strain Becker of pseudorabies virus (PRV), and Marek's disease virus (MDV). Vero cells (ATCC CCL-81) and A7 (human melanoma) cells (41) were maintained in Dulbecco's modified Eagle's medium (DMEM; GIBCO) supplemented with 10% fetal bovine serum, penicillin (65 μg/ml), and streptomycin (131 μg/ml). Infected cells were grown in DMEM supplemented with 2% fetal bovine serum, 25 mM HEPES buffer, glutamine (0.3 μg/ml), penicillin (65 μg/ml), and streptomycin (131 μg/ml).
Construction of UL11-GFP and UL16-GFP chimeras.
All UL11 alleles were cloned into the multiple cloning site of pEGFP-N2 (Clontech), thereby producing vectors that encode UL11-GFP fusions. Plasmids encoding UL11 from HSV-1 fused to GFP (here designated UL11H.GFP) and an acidic cluster mutant of this protein [UL11H.AC(−).GFP] were described previously (22). A mutant in which leucine and isoleucine at residues 18 and 19, respectively, are replaced with alanine (UL11H.L18A/I19A.GFP) was constructed by changing the corresponding codons from CTC ATC to GCC GCC. A mutant in which the second half of UL11 is deleted (UL11H.d51-96.GFP) was produced by removing codons 51 to 97, thereby leaving a truncated UL11 gene in frame with the GFP coding sequence.
Plasmids encoding PRV UL11-GFP (UL11P.GFP) and MDV UL11-GFP (UL11M.GFP) fusions were made by PCR. For the PRV construct, the UL11 gene was amplified from the PRV Becker genome with a forward primer (5′-GCCTGCGAGATCTCCGTGCTGCTGATCGTCACG-3′) complementary to the sequence 100 bp upstream of the start codon and a reverse primer (5′-CAAACGGGTTTATTGGAATTCGTACGCCCGCGAG-3′) complementary to the 3′ end of the UL11 gene. The product was cloned into pEGFP-N2 with BglII-EcoRI. The MDV construct was made by PCR amplification of the MDV UL11 gene with a forward primer (5′-ACTCCCGAACAGAGCTCCCTTGTCATTCTAATC-3′) complementary to the sequence 100 bp upstream of the start codon and a reverse primer (5′-ATGATTGTCGAATTCTTTATTAAACATCATAAC-3′) complementary to the 3′ end of the UL11 gene. The product was cloned into pEGFP-N2 with SstI-EcoRI.
The construct encoding an HSV-1 UL16-GFP fusion was made by PCR amplification of the UL16 gene from the HSV-1 KOS genome with a forward primer (5′-CCTTAGGAATTCTCGCAAGGTGTCGTCCGGGA-3′) complementary to the sequence 120 bp upstream of the start codon and a reverse primer (5′-GGGCACTGGGATCCATTCGGGATCGCTTGAGGA-3′) complementary to the 3′ end of the UL16 gene. The product was cloned into the EcoRI-BamHI sites of pEGFP-N2.
Construction of GST-UL11 chimeras.
UL11 alleles were amplified by PCR and cloned into pGEX-4T-3 (Amersham Biosciences), thereby producing vectors that encode glutathione S-transferase (GST)-UL11 fusions. For all constructs except GST.UL11H.d51-96, a BglII site was inserted immediately upstream of the UL11 start codon, while a NotI site was inserted immediately downstream of the native stop codon. To make the plasmid that encodes GST.UL11H.d51-96, the NotI site was inserted along with a stop codon which was placed after codon 50. PCR products were cut with BglII-NotI and cloned into the BamHI-NotI sites of pGEX-4T-3.
Construction of untagged UL11 expression vectors.
To produce wild-type UL11 and UL11.AC(−) constructs that have no GFP tag, UL11 alleles which contain stop codons at their 3′ ends were inserted in place of the GFP gene cassette (with SstI and NotI) within vector pEGFP-N2.
Antibodies.
UL11- and VP22-specific antisera, which were produced by Cocalico Biologicals, Inc., were obtained from rabbits that had been injected with purified GST-UL11 and GST-VP22 fusions, respectively. The specificity of the antibodies for their respective antigens was shown by their ability to detect UL11-GFP and VP22-GFP fusions in immunoblot and immunoprecipitation experiments (data not shown). The GFP-specific antibody was obtained from Clontech (product number 8367-1).
Coimmunoprecipitation assay.
Confluent monolayers of Vero cells (60-mm plates) were infected with HSV-1 KOS at a multiplicity of infection of 30. At 5 h postinfection, the cells were starved for 15 min in methionine-free DMEM (GIBCO) and metabolically labeled with EXPRE35S35S protein labeling mixture (Perkin-Elmer NEG-072) (85 μCi/ml; >1,000 Ci/mmol) for 3 h (42). The cells were mixed with NP-40 lysis buffer (0.5% NP-40, 150 mM NaCl, 50 mM Tris-HCl [pH 8.0]) containing protease inhibitors (Sigma product number P8340), and the infected-cell lysates were precleared overnight with protein A-agarose beads (Roche). UL11 and its associated proteins were immunoprecipitated with polyclonal anti-UL11 serum and separated by electrophoresis in sodium dodecyl sulfate (SDS)-10% polyacrylamide gels. The gels were subsequently dried and subjected to autoradiography with Kodak X-Omat AR5 film for 1 to 2 days to detect the radiolabeled proteins.
GST pull-down assay.
GST fusion proteins were purified from 100-ml cultures by using glutathione-Sepharose beads (Amersham Biosciences product number 27-4574-01) according to the manufacturer’s instructions. Vero cells were infected with herpesviruses and then labeled and lysed as described above for the coimmunoprecipitation assay. The lysates were precleared with glutathione-Sepharose 4B beads overnight and then incubated with purified GST proteins on glutathione-Sepharose beads for 2 h at room temperature. The beads were washed three times with NP-40 lysis buffer and one time with 10 mM Tris-HCl (pH 7.6). Proteins bound to the GST constructs were separated by SDS-10% polyacrylamide gel electrophoresis (PAGE), and radiolabeled proteins were detected by autoradiography as described above.
To analyze the UL11-UL16 interaction within transfected cells, a similar GST pull-down assay was used. The calcium phosphate method was used to transfect A7 melanoma cells (10) with a plasmid that encodes a UL16-GFP fusion. A7 cells were used instead of Vero cells because they are more easily transfected and have been used elsewhere to express HSV-1 tegument protein-GFP chimeras (7, 22). At 24 h posttransfection, the cells were harvested in NP-40 lysis buffer. After the lysates were precleared for 2 h with glutathione-Sepharose beads, purified GST-UL11 fusions were added and the beads were washed as described above. Proteins bound to the GST constructs were separated by SDS-9% PAGE and electrotransferred to nitrocellulose membranes. To detect UL16-GFP, standard enhanced chemiluminescence-based Western blot assays were performed with anti-GFP serum.
Confocal microscopy.
A7 cells were transfected as described above. At 18 h posttransfection, cells were washed once with Tris-buffered saline and immediately viewed by using a Zeiss laser scanning microscope with a helium-argon laser (488-nm peak excitation).
In-gel trypsin digestion and MS analysis of the 40-kDa protein.
The 40-kDa species was pulled out of unlabeled, infected-cell lysates (two 60-mm plates) by using GST.UL11H.d51-96 as described above and then separated by SDS-10% PAGE. After visualization of proteins with a negative zinc stain (Bio-Rad), the 40-kDa band was cut from the gel and placed into a siliconized 0.65-ml Eppendorf tube. The gel slice was fixed and destained overnight in a 5% methanol-7% acetic acid solution. The fixative was removed, and the slice was dehydrated twice for 40 min each time in 200 mM NH4HCO3 (pH 8.0)-50% acetonitrile (AcN) (15, 34). After the gel slice was dried in a Speed-vac for 20 min, the 40-kDa protein was reduced for 15 min with 2 mM Tris(2-carboxyethyl)phosphine (TCEP) in 25 mM NH4HCO3 (pH 8.0). TCEP was removed, and the protein was alkylated for 30 min (in the dark) with 20 mM iodoacetamide in 25 mM NH4HCO3 (pH 8.0). The gel slice was washed three times with 25 mM NH4HCO3 (pH 8.0), dried in the Speed-vac for 25 min, and rehydrated in 25 mM NH4HCO3 (pH 8.0) containing 0.02 μg of sequencing-grade modified trypsin (Promega)/μl. After 1 h, unabsorbed trypsin was removed, and the gel slice was covered with 100 μl of 25 mM NH4HCO3 (pH 8.0). To allow for efficient trypsin cleavage, the gel slice was incubated overnight at 37°C with agitation. The solution was saved, and the gel slice was washed with 50 μl of 50% AcN-50% 25 mM NH4HCO3 (pH 8.0) and with 50 μl of 50% AcN- 2% trifluoroacetic acid (two times) in order to fully elute the tryptic peptides from the gel. These last three washes were saved and added to the first solution, and the sample was placed in a Speed-vac until only 35% of the original volume remained. The tryptic peptides were purified and concentrated by using a strong-cation-exchange ZipTip cleanup kit (Millipore) according to the manufacturer's instructions. The purified peptides were spotted onto a stainless steel plate, overlaid with an alpha-cyanohydroxycinnamic acid (Sigma) matrix, and analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) with an Applied Biosystems 4700 proteomics analyzer at the core facility at the College of Medicine, The Pennsylvania State University. The 10 most abundant peptide fragments on the spectra were identified and further analyzed by tandem MS-MS ion fragment analysis. The resulting MS-MS spectra of a 1,646-Da fragment which was present in duplicate trypsin digests were examined for matches to theoretical spectra from all known proteins by using the Applied Biosystems GPS Explorer program. The search was carried out against all SwissProt entries, with no limitations as to species, protein size, or pI.
RESULTS
Identification of UL11-binding partners by a coimmunoprecipitation assay.
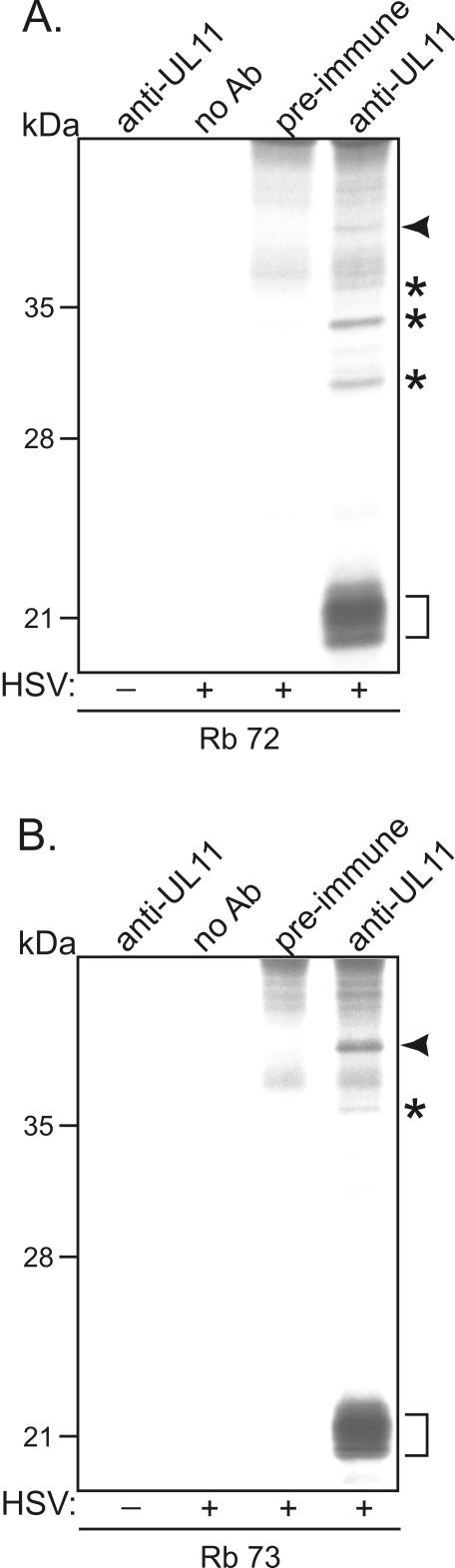
As a start to identifying its binding partners (viral or cellular), UL11 was immunoprecipitated from HSV-1-infected cell lysates (Fig. 1). As in previous reports (1, 24), UL11 was found to migrate as a triplet of bands between 20 and 22 kDa, suggesting that it exists in multiple modified forms (Fig. 1A). At least four additional proteins appeared to coimmunoprecipitate with UL11. These proteins, which have apparent molecular masses of 31, 34, 37, and 40 kDa, were not pulled out of mock-infected cell lysates or HSV-1-infected cell lysates when no antibody or preimmune serum was used. Any larger coimmunoprecipitating proteins that may have been present were obscured by background bands. A second immunoprecipitation assay was carried out with UL11-specific antiserum from a different rabbit (Rb73). While the 37- and 40-kDa proteins were again detected, the 31- and 34-kDa proteins failed to coimmunoprecipitate with UL11 (Fig. 1B), suggesting that the first antiserum (Rb72) may contain antibodies that happen to cross-react with two irrelevant proteins. Alternatively, the second antiserum may contain antibodies that block interactions between UL11 and the 31- and 34-kDa proteins.
FIG. 1.
Coimmunoprecipitation assay with anti-UL11 serum. Vero cells infected with HSV-1 KOS for 5 h were metabolically labeled with [35S]methionine-cysteine for 3 h and harvested with NP-40 lysis buffer. The UL11 protein (indicated by a bracket) was immunoprecipitated with anti-UL11 serum, separated by SDS-PAGE, and visualized by autoradiography. Specific coimmunoprecipitated proteins are indicated by arrowheads (40 kDa) and asterisks (31, 34, and 37 kDa). Plus and minus signs denote HSV-1-infected cells and noninfected cells, respectively. (A) Rabbit anti-UL11 serum Rb72. (B) Rabbit anti-UL11 serum Rb73.
Identification of UL11-binding partners by a GST pull-down assay.
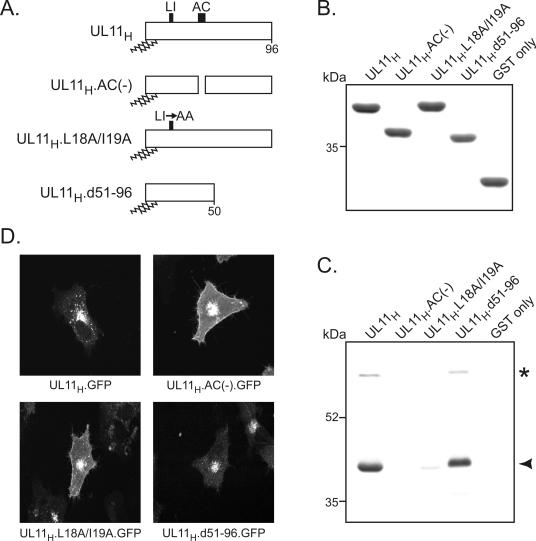
To further identify UL11-binding partners, a pull-down assay was performed with a purified GST-UL11 fusion protein (Fig. 2A and B). Unlike GST by itself, the GST.UL11H construct reproducibly pulled down 40- and 69-kDa proteins from HSV-1-infected cell lysates (Fig. 2C). Treatment with 1 M NaCl and 100 mM EDTA had no effect on either interaction, indicating that both are relatively stable (data not shown). Because a 40-kDa protein was identified by both the coimmunoprecipitation assay and the GST pull-down assay, it was chosen for further characterization.
FIG. 2.
Identification of UL11-binding partners by a GST pull-down assay. (A) Diagrams of UL11 constructs. The 96-amino-acid UL11 sequence from HSV-1 is shown; the positions of the acidic cluster (AC) and the dileucine motif (LI) are indicated. N-terminal myristylation and palmitylation are denoted by wavy lines. (B) Coomassie blue-stained gel of GST fusion proteins. The UL11 constructs shown in panel A were fused to the C terminus of the GST protein and purified from Escherichia coli cells. Approximately equal amounts of proteins were used in all reactions. (C) Proteins pulled down by the GST-UL11 constructs. Purified GST fusion proteins were incubated with radiolabeled, HSV-1-infected cell lysates and washed with NP-40 lysis buffer. Proteins bound to the GST constructs were separated by SDS-10% PAGE, and radiolabeled proteins were detected by autoradiography. The asterisk and the arrowhead denote the 69- and 40-kDa proteins, respectively. (D) Subcellular localization. The UL11 constructs shown in panel A were fused to the N terminus of GFP, expressed in A7 cells, and visualized 18 h later by live-cell confocal microscopy.
To map the region of UL11 that is required for the interaction with the 40-kDa protein, several mutants were examined. The same GST pull-down assay was used to test whether removal of the acidic cluster [GST.UL11H.AC(−)] or replacement of the dileucine motif with alanines (GST.UL11H.L18A/I19A) would have any effect on the interaction with the 40-kDa protein (Fig. 2A). These mutants were chosen because both acidic clusters and dileucine mutants function as binding sites for a variety of proteins, including members of the clathrin adaptor family (14). When equal amounts of mutant GST fusion proteins were used (Fig. 2B), neither the acidic cluster nor the dileucine motif associated with the 40-kDa protein (Fig. 2C). Surprisingly, the acidic clusters of furin and HIV-1 Nef could not replace the native acidic cluster in this assay (data not shown), in spite of their ability to restore Golgi apparatus recycling to an UL11 acidic cluster mutant (22). In contrast, deletion of the entire second half of UL11 (GST.UL11H.d51-96) had no effect on the interaction (Fig. 2C), indicating that the site important for the interaction with the 40-kDa protein is within the first half.
To determine where the different UL11 mutants localize when transiently expressed as GFP fusions, live-cell confocal microscopy was performed. Like the previously characterized UL11H.AC(−).GFP mutant (22), the UL11H.L18A/I19A.GFP mutant localized to the Golgi apparatus but also accumulated abnormally on the plasma membrane (Fig. 2D), suggesting that it fails to be internalized from the cell surface during retrieval to the Golgi apparatus. In contrast, the UL11H.d51- 96.GFP mutant localized to the Golgi apparatus in a manner similar to that of wild-type UL11H.GFP, consistent with a previous study suggesting that all of the trafficking information is contained within the first half of the protein (5).
UL11 homologs from other herpesviruses can associate with the 40-kDa protein.
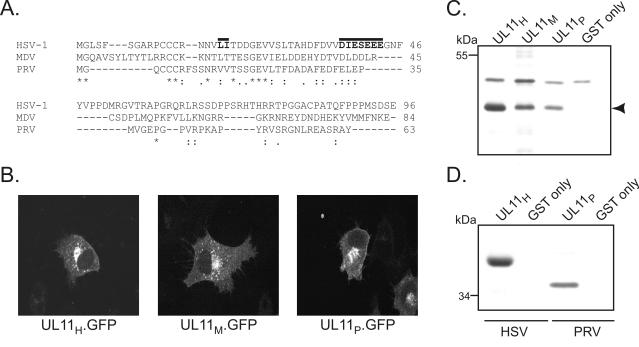
To further address the specificity of the UL11-40-kDa protein interaction, UL11 proteins from other herpesviruses were tested for their ability to pull down the 40-kDa protein from HSV-1-infected cell lysates. Homologs from the alphaherpesviruses PRV and MDV were chosen because they have divergent primary sequences (Fig. 3A) but still traffic to the Golgi apparatus in a manner similar to that of HSV-1 UL11 (Fig. 3B). Despite these sequence differences, GST-UL11 fusion proteins derived from both PRV (GST.UL11P) and MDV (GST.UL11M) bound to the 40-kDa protein (Fig. 3C).
FIG. 3.
Analysis of UL11 homologs. (A) Sequence alignment of UL11 homologs from HSV-1, MDV, and PRV. Amino acids that are identical are indicated by asterisks, whereas residues that are conserved are denoted by a period (lower conservation) or a colon (higher conservation). The locations of the dileucine motif and the acidic cluster within HSV-1 UL11 are indicated by solid lines. (B) Subcellular localization. A7 cells expressing UL11-GFP chimeras were visualized 18 h posttransfection by confocal microscopy. (C) GST pull-down assay with HSV-1-infected cell lysates. Purified GST-UL11 fusion proteins were incubated with radiolabeled, HSV-1-infected cell lysates. Bound proteins were separated by SDS-10% PAGE and detected by autoradiography. The arrowhead denotes the position of the 40-kDa protein. (D) GST pull-down assay with PRV-infected cell lysates. The assay was performed as described for panel C, except that GST fusion proteins were incubated with either HSV-1- or PRV-infected cell lysates.
Identification of the 40-kDa protein.
Three observations suggested that the 40-kDa protein is of viral origin. First, it is never observed in mock-infected cell lysates (Fig. 1; GST-UL11 pull-down data, not shown). Second, the radiolabeled 40-kDa protein is readily obtained from infected cells even though most cellular proteins are down-regulated by certain HSV-1 gene products (32). Finally, UL11 from PRV, which binds to the 40-kDa protein from HSV-1-infected Vero cells (Fig. 3C), pulls down a slightly smaller (36-kDa) protein from the same type of cells when they are infected instead with PRV (Fig. 3D).
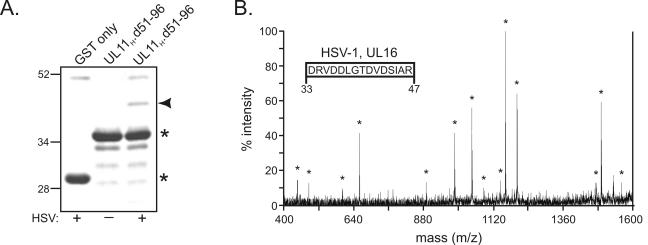
To directly identify the 40-kDa protein, it was first pulled out of HSV-1-infected-cell lysates by using the GST.UL11H.d51- 96 construct and then separated from other proteins by SDS-PAGE. (GST.UL11H.d51-96 was used instead of full-length GST.UL11H because the latter nearly comigrates with the unknown 40-kDa protein [Fig. 2B].) The 40-kDa species (Fig. 4A) was cut from the gel and digested with trypsin, and the tryptic peptides were analyzed by MS. The 10 most abundant peptide fragments on the spectra were identified and further analyzed by tandem MS-MS ion fragment analysis. In particular, a 1,646-Da peptide was chosen for further characterization because it was clearly present in duplicate trypsin digests. The resulting MS-MS spectrum of the 1,646-Da fragment (Fig. 4B) was used to determine its peptide sequence (Fig. 4B, inset), which was then examined for matches with the sequences of all known proteins. With over 99% confidence (data not shown), the 40-kDa protein was identified as the product of the HSV-1 UL16 gene, a virion protein that has been reported to be involved in nucleocapsid assembly and maturation (29, 31). Consistent with these findings, the 36-kDa protein that was pulled out of PRV-infected cell lysates by PRV UL11 (Fig. 3D) was about the size predicted for PRV UL16 (Lynn Enquist, personal communication).
FIG. 4.
Direct analysis of the 40-kDa protein by MS. (A) Visualization of the 40-kDa band by zinc staining. The 40-kDa protein (denoted by an arrowhead) was pulled out of unlabeled, HSV-1-infected cell lysates with GST.UL11H.d51-96 and separated by SDS-PAGE. The gel was negatively stained with a Bio-Rad zinc stain kit; the isolated 40-kDa band was cut from the gel, digested with trypsin, and analyzed by MALDI-TOF MS. Asterisks denote the positions of the GST derivatives on the gel. Plus or minus signs indicate that GST proteins were mixed with HSV-1-infected cell lysates or not, respectively, prior to being loaded on the gel. Numbers at left indicate kilodaltons. (B) MS-MS spectrum of the 1,646-Da tryptic peptide. The 10 most abundant peptide fragments on the MS spectrum were identified and further analyzed by tandem MS-MS ion fragment analysis. The resulting MS-MS spectrum of the 1,646-Da peptide was used to determine its sequence (inset), which was then examined for matches with the sequences of all known proteins. Residue numbers within the HSV-1 UL16 protein are indicated below the inset. Asterisks denote MS-MS peaks that were used in elucidating the 1,646-Da peptide sequence.
Interaction between UL11 and UL16 in the absence of other viral proteins.
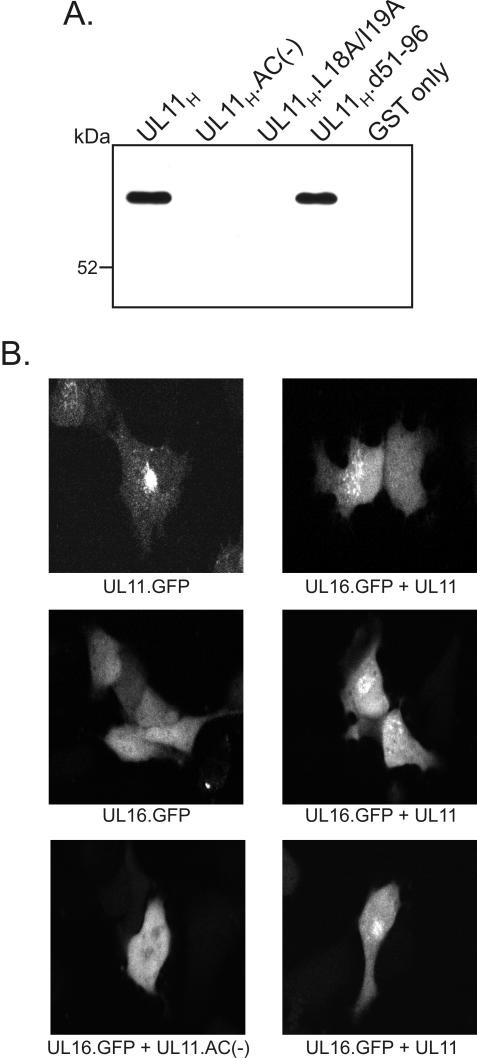
To test whether UL11 and UL16 can associate with one another in the absence of other viral proteins, a GST pull-down assay was performed with lysates derived from cells transfected with a UL16-GFP expression vector. Similar to what was found with infected cells, the UL16-GFP chimera was pulled down by the GST.UL11H and GST.UL11H.d51-96 constructs but not by GST alone or by the acidic cluster and dileucine motif mutants (Fig. 5A). These findings show that the UL11-UL16 interaction does not require other viral proteins; however, it remains possible that cellular proteins play some role in mediating the interaction. Further evidence for the interaction comes from cotransfection experiments in which the UL16-GFP chimera was expressed in the presence or absence of an untagged UL11 protein (Fig. 5B). When UL16-GFP was expressed by itself, it was present throughout cells; however, when it was coexpressed with wild-type UL11, a portion of UL16 was relocalized to a punctate, perinuclear compartment that is reminiscent of the TGN. This relocalization was abolished when the acidic cluster of UL11 was deleted, as predicted by the mapping experiments. Thus, the interaction of UL11 and UL16 seen in vitro also occurs within intact cells.
FIG. 5.
Association of UL11 and UL16 within transfected cells. (A) GST pull-down assay with transfected-cell lysates. A7 cells expressing a UL16-GFP fusion were harvested in NP-40 lysis buffer at 24 h posttransfection. After the lysates were precleared for 2 h with glutathione-Sepharose beads, purified GST-UL11 fusion proteins were added. Proteins bound to the GST constructs were separated by SDS-9% PAGE and electrotransferred to nitrocellulose membranes. Western blot analysis was then performed with a GFP-specific antiserum. (B) Subcellular localization of UL16-GFP in the presence or absence of UL11. A7 cells were transfected with the indicated constructs and visualized 18 h later by live-cell confocal microscopy.
DISCUSSION
Although many tegument proteins have been found to be important for viral replication, very little has been done to elucidate the network of interactions that connect them. Of the ∼20 HSV-1 proteins that constitute the tegument, only three capsid-tegument (26, 31, 40), four tegument-tegument (11, 36, 44), and two tegument-glycoprotein (30, 45) interactions have been identified. Similarly, very few interactions have been identified among the tegument proteins of other alphaherpesviruses, such as PRV (13, 20) and varicella-zoster virus (23, 38, 39). As a result, the structural organization and function of most herpesvirus virion proteins still remain a mystery.
The goal of the present study was to identify and characterize binding partners for membrane-bound tegument protein UL11. In addition to several minor protein species, an abundant 40-kDa protein appeared to strongly associate with UL11 in both immunoprecipitation and GST pull-down assays. Based only on its size and evidence that suggested a viral origin, the 40-kDa protein could have been any of the following eight HSV-1 proteins: VP22, glycoprotein K, thymidine kinase, UL16, glycoprotein L, UL50, VP22a, and UL40 (4, 6, 16, 17, 19, 29). Of these proteins, we initially considered VP22 to be the likeliest candidate because it is a very abundant tegument protein that associates with membranes and localizes to a cellular compartment similar to where UL11 localizes (7). However, when the GST-UL11 pull-down assay was carried out with a mutant HSV-1 strain (166v) that expresses a 65-kDa GFP-VP22 fusion protein instead of native VP22, the 40-kDa protein was still detected, indicating that it is not VP22 (data not shown). Instead of testing each of the other seven candidates individually, the 40-kDa protein was analyzed directly by MS and shown to be the product of the HSV-1 UL16 gene.
The herpesvirus UL16 gene family encodes a group of small (33- to 40-kDa), conserved proteins that are produced late in the viral life cycle and packaged into virions (9, 29, 31, 43). Members of this protein family, which includes HSV-1 UL16, cytomegalovirus UL94, Epstein-Barr virus BGLF2, and varicella-zoster virus ORF44, contain potential Zn finger domains in the conserved C terminus and a potential nuclear localization signal in the N terminus (43). Within infected cells, HSV-1 UL16 localizes to both the cytoplasm (diffuse) and discrete intranuclear inclusions referred to as assemblons (29, 31). Although the actual function of the protein is still unknown, UL16 has been predicted to play a role in genome packaging and nucleocapsid maturation due to its weak association with mature, DNA-containing type C capsids (31), its ability to bind to single-stranded DNA in vitro (31), and its potential Zn finger domains (43). Consistent with a role in viral replication, cells infected with an HSV-1 UL16 deletion mutant produce 10-fold fewer virions than those infected with wild-type virus (2).
The finding that UL16 interacts with UL11 may provide clues as to how both proteins function during viral replication. Based on the evidence that UL16 associates with nucleocapsids (31) and UL11 peripherally binds to TGN membranes (5), it is possible that the interaction between these two proteins functions to target nucleocapsids for tegument acquisition and envelopment at the site of assembly. Consistent with this hypothesis, cells infected with a UL11 knockout mutant produce large amounts of nucleocapsids which enter the cytoplasm but fail to become enveloped at the TGN (3). Furthermore, the relocalization of UL16 to a punctate, perinuclear compartment in the presence of UL11 (Fig. 5B) suggests that UL11 has the ability to recruit UL16 to cytoplasmic membranes. Whether or not UL11 can recruit nucleocapsid-associated UL16 to cytoplasmic membranes within infected cells remains to be determined.
A similar type of recruitment mechanism has been described for other HSV-1 tegument proteins. The membrane-associated VP22 protein can interact with and relocalize the normally diffuse VP16 protein to a perinuclear compartment that resembles the TGN (7, 11). In addition, like UL16, a significant fraction of the VP16 protein within HSV-1-infected cells is associated with nonenveloped, cytoplasmic capsids (28). Thus, it is possible that the UL11-UL16 and VP22-VP16 interactions both play a role (redundantly) in recruiting nucleocapsids to the TGN for tegument formation and envelopment. Redundancy of this function would explain why UL11 knockout mutants still retain a high level of infectivity (3, 25).
Another hypothesis for the role of UL16 is that it serves to regulate the trafficking and localization of UL11. The acidic cluster and the dileucine motif within UL11 are required for its trafficking from the plasma membrane to the Golgi apparatus (22). These types of sequence motifs are known to serve as binding sites for the PACS-1 protein (41) as well as members of the clathrin adaptor family (e.g., AP-1 and AP-2) (18), which function to link cargo membrane proteins to clathrin coats at specific compartments. Since these trafficking signals within UL11 also appear to be required for its interaction with UL16 (Fig. 2), it is possible that UL16 acts as an adaptor-like protein and competes with cellular proteins for binding to UL11 at these sites. In doing so, UL16 may alter the subcellular localization of UL11. If UL16 is an adaptor-like protein, one may expect it to have sequence similarity to one or more of the adaptor proteins. Primary sequence alignments of UL16 with the PACS-1 protein and the cargo interaction subunits of the AP-1, AP-2, and AP-3 proteins have revealed no obvious homology (data not shown); however, functional homology may exist.
Equally important to understanding why the UL11 and UL16 proteins interact is elucidating how they interact. As a start to this goal, it will be essential to determine whether UL11 and UL16 interact directly or are bridged by other proteins. Because the interaction can occur within transfected cells (Fig. 5), bridging proteins, if there are any, cannot be virus encoded. Experiments are in progress to ascertain whether a direct interaction is involved. Another question which remains to be addressed is what regions within the two proteins are responsible for the association. Data presented in this report demonstrate that the binding site for UL16 is within the first 50 residues of UL11 and that the acidic cluster and the dileucine motif within this region are required for the interaction (Fig. 2). However, it is not known whether these motifs serve as the actual binding site for UL16 or whether our mutations somehow altered the structure of UL11, rendering the true binding site nonfunctional. In addition to mapping of the binding site within UL11, experiments are under way to identify the sequences within UL16 that mediate its association with UL11.
Acknowledgments
Special thanks are extended to Michael Brignati for discussions and preparation of the UL11- and VP22-specific antisera. We also thank Lynn Enquist for providing PRV reagents, Bob Silva for providing MDV reagents, and Patric Lundberg for providing the VP22-GFP-expressing virus.
This work was supported by National Institutes of Health (NIH) grant CA42460 to R.J.C. J.S.L. was partially supported by NIH training grant CA60395.
REFERENCES
- 1.Baines, J. D., R. J. Jacob, L. Simmerman, and B. Roizman. 1995. The herpes simplex virus type 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 69:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines, J. D., and B. Roizman. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus type 1 in cell culture. J. Virol. 65:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines, J. D., and B. Roizman. 1992. The UL11 gene of herpes simplex virus type 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66:5168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjornberg, O., A. C. Bergman, A. M. Rosengren, R. Persson, I. R. Lehman, and P. O. Nyman. 1993. dUTPase from herpes simplex virus type 1; purification from infected green monkey kidney (Vero) cells and from an overproducing Escherichia coli strain. Protein Expr. Purif. 4:149-159. [DOI] [PubMed] [Google Scholar]
- 5.Bowzard, J. B., R. J. Visalli, C. B. Wilson, J. S. Loomis, E. M. Callahan, R. J. Courtney, and J. W. Wills. 2000. Membrane targeting properties of a herpesvirus tegument protein-retrovirus Gag chimera. J. Virol. 74:8692-8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, D. K., B. Roizman, and L. Pereira. 1984. Characterization of posttranslational products of herpes simplex virus gene 35 proteins binding to the surfaces of full capsids but not empty capsids. J. Virol. 49:142-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brignati, M. J., J. S. Loomis, J. W. Wills, and R. J. Courtney. 2003. Membrane association of VP22, a herpes simplex virus type 1 tegument protein. J. Virol. 77:4888-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, S. M., D. A. Ritchie, and J. H. Subak-Sharpe. 1973. Genetic studies with herpes simplex virus type 1. The isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map. J. Gen. Virol. 18:329-346. [DOI] [PubMed] [Google Scholar]
- 9.Chen, M. R., T. Y. Hsu, S. W. Lin, J. Y. Chen, and C. S. Yang. 1991. Cloning and characterization of cDNA clones corresponding to transcripts from the BamHI G region of the Epstein-Barr virus genome and expression of BGLF2. J. Gen. Virol. 72:3047-3055. [DOI] [PubMed] [Google Scholar]
- 10.Craven, R. C., A. E. Leure-duPree, R. A. Weldon, Jr., and J. W. Wills. 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 69:4213-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heilker, R., M. Spiess, and P. Crottet. 1999. Recognition of sorting signals by clathrin adaptors. Bioessays 21:558-567. [DOI] [PubMed] [Google Scholar]
- 15.Hellman, U., C. Wernstedt, J. Gonez, and C. H. Heldin. 1995. Improvement of an “in-gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem. 224:451-455. [DOI] [PubMed] [Google Scholar]
- 16.Hutchinson, L., H. Browne, V. Wargent, N. Davis-Poynter, S. Primorac, K. Goldsmith, A. C. Minson, and D. C. Johnson. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchinson, L., K. Goldsmith, D. Snoddy, H. Ghosh, F. L. Graham, and D. C. Johnson. 1992. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J. Virol. 66:5603-5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirchhausen, T., J. S. Bonifacino, and H. Riezman. 1997. Linking cargo to vesicle formation: receptor tail interactions with coat proteins. Curr. Opin. Cell Biol. 9:488-495. [DOI] [PubMed] [Google Scholar]
- 19.Kit, S., G. N. Jorgensen, D. R. Dubbs, D. Trkula, and V. Zaslavsky. 1978. Detection of herpes simplex virus thymidine kinase polypeptides in cells labeled with 35S-methionine. Intervirology 9:162-172. [DOI] [PubMed] [Google Scholar]
- 20.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loomis, J. S., J. B. Bowzard, R. J. Courtney, and J. W. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lynch, J. M., T. K. Kenyon, C. Grose, J. Hay, and W. T. Ruyechan. 2002. Physical and functional interaction between the varicella zoster virus IE63 and IE62 proteins. Virology 302:71-82. [DOI] [PubMed] [Google Scholar]
- 24.MacLean, C. A., B. Clark, and D. J. McGeoch. 1989. Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J. Gen. Virol. 70:3147-3157. [DOI] [PubMed] [Google Scholar]
- 25.MacLean, C. A., A. Dolan, F. E. Jamieson, and D. J. McGeoch. 1992. The myristylated virion proteins of herpes simplex virus type 1: investigation of their role in the virus life cycle. J. Gen. Virol. 73:539-547. [DOI] [PubMed] [Google Scholar]
- 26.McNabb, D. S., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221-232. [DOI] [PubMed] [Google Scholar]
- 27.Mettenleiter, T. C. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed]
- 28.Miranda-Saksena, M., R. A. Boadle, P. Armati, and A. L. Cunningham. 2002. In rat dorsal root ganglion neurons, herpes simplex virus type 1 tegument forms in the cytoplasm of the cell body. J. Virol. 76:9934-9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nalwanga, D., S. Rempel, B. Roizman, and J. D. Baines. 1996. The UL 16 gene product of herpes simplex virus 1 is a virion protein that colocalizes with intranuclear capsid proteins. Virology 226:236-242. [DOI] [PubMed] [Google Scholar]
- 30.Ng, T. I., W. O. Ogle, and B. Roizman. 1998. UL13 protein kinase of herpes simplex virus 1 complexes with glycoprotein E and mediates the phosphorylation of the viral Fc receptor: glycoproteins E and I. Virology 241:37-48. [DOI] [PubMed] [Google Scholar]
- 31.Oshima, S., T. Daikoku, S. Shibata, H. Yamada, F. Goshima, and Y. Nishiyama. 1998. Characterization of the UL16 gene product of herpes simplex virus type 2. Arch. Virol. 143:863-880. [DOI] [PubMed] [Google Scholar]
- 32.Read, G. S., and N. Frenkel. 1983. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of α (immediate-early) viral polypeptides. J. Virol. 46:498-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roizman, B., and A. E. Sears. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 34.Rosenfeld, J., J. Capdevielle, J. C. Guillemot, and P. Ferrara. 1992. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203:173-179. [DOI] [PubMed] [Google Scholar]
- 35.Schimmer, C., and A. Neubauer. 2003. The equine herpesvirus 1 UL11 gene product localizes to the trans-Golgi network and is involved in cell-to-cell spread. Virology 308:23-36. [DOI] [PubMed] [Google Scholar]
- 36.Smibert, C. A., B. Popova, P. Xiao, J. P. Capone, and J. R. Smiley. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, K. O. 1964. Relationship between the envelope and the infectivity of herpes simplex virus. Proc. Soc. Exp. Biol. Med. 115:814-816. [DOI] [PubMed] [Google Scholar]
- 38.Spengler, M., N. Niesen, C. Grose, W. T. Ruyechan, and J. Hay. 2001. Interactions among structural proteins of varicella zoster virus. Arch. Virol. Suppl. 17:71-79. [DOI] [PubMed] [Google Scholar]
- 39.Spengler, M. L., W. T. Ruyechan, and J. Hay. 2000. Physical interaction between two varicella zoster virus gene regulatory proteins, IE4 and IE62. Virology 272:375-381. [DOI] [PubMed] [Google Scholar]
- 40.Takakuwa, H., F. Goshima, T. Koshizuka, T. Murata, T. Daikoku, and Y. Nishiyama. 2001. Herpes simplex virus encodes a virion-associated protein which promotes long cellular processes in over-expressing cells. Genes Cells 6:955-966. [DOI] [PubMed] [Google Scholar]
- 41.Wan, L., S. S. Molloy, L. Thomas, G. Liu, Y. Xiang, S. L. Rybak, and G. Thomas. 1998. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell 94:205-216. [DOI] [PubMed] [Google Scholar]
- 42.Wills, J. W., R. C. Craven, and J. A. Achacoso. 1989. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J. Virol. 63:4331-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wing, B. A., G. C. Lee, and E. S. Huang. 1996. The human cytomegalovirus UL94 open reading frame encodes a conserved herpesvirus capsid/tegument-associated virion protein that is expressed with true late kinetics. J. Virol. 70:3339-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao, F., and P. A. Schaffer. 1994. Physical interaction between the herpes simplex virus type 1 immediate-early regulatory proteins ICP0 and ICP4. J. Virol. 68:8158-8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu, Q., and R. J. Courtney. 1994. Chemical cross-linking of virion envelope and tegument proteins of herpes simplex virus type 1. Virology 204:590-599. [DOI] [PubMed] [Google Scholar]