Abstract
Varicella-zoster virus (VZV), a neurotropic alphaherpesvirus, causes childhood chickenpox (varicella), becomes latent in dorsal root and autonomic ganglia, and reactivates decades later to cause shingles (zoster) and other neurologic complications. Although the sequence and configuration of VZV DNA have been determined, relatively little is known about viral gene expression in productively infected cells. This is in part because VZV is highly cell associated, and sufficient titers of cell-free virus for use in synchronizing infection do not develop. PCR-based transcriptional arrays were constructed to simultaneously determine the relative abundance of the ≈70 predicted VZV open reading frames (ORFs). Fragments (250 to 600 bp) from the 5′ and 3′ end of each ORF were PCR amplified and inserted into plasmid vectors. The virus DNA inserts were amplified, quantitated, and spotted onto nylon membranes. Probing the arrays with radiolabeled cDNA synthesized from VZV-infected cells revealed an increase in the magnitude of the expressed VZV genes from days 1 to 3 after low-multiplicity virus infection but little change in their relative abundance. The most abundant VZV transcripts mapped to ORFs 9/9A, 64, 33/33A, and 49, of which only ORF 9 corresponded to a previously identified structural gene. Array analysis also mapped transcripts to three large intergenic regions previously thought to be transcriptionally silent, results subsequently confirmed by Northern blot and reverse transcription-PCR analysis. Array analysis provides a formidable tool to analyze transcription of an important ubiquitous human pathogen.
Varicella-zoster virus (VZV) is propagated by cocultivating infected cells with uninfected cells. Because VZV is cell associated and high titers of cell-free virus that can be used to synchronize infection do not develop, analysis of VZV gene transcription in infected cells in tissue culture has been limited. Three reports described global analysis of VZV transcription in cultures harvested at advanced stages of infection; Northern blot analysis of RNA extracted from late-stage VZV-infected cultures identified 58 to 67 discrete transcripts mapping throughout the virus genome (26, 32), while single-stranded DNA probes used in Northern blots revealed the direction of transcription of 57 of the 58 previously identified VZV transcripts and also identified 20 novel virus transcripts (35). Because the RNA in those studies was sampled at the height of cytopathic effect, the transcripts were presumed to reflect VZV structural genes.
To better understand the pattern of virus gene transcription during lytic infection, we constructed PCR-based VZV transcriptional arrays to analyze the magnitude of expression and relative abundance of each predicted VZV gene as well as detect transcription for three large intergenic regions of the virus genome.
MATERIALS AND METHODS
Virus and cells.
VZV (strain Ellen) was propagated as described previously (13) by cocultivation of infected and uninfected BSC-1 cells, a continuous cell line derived from African green monkey kidney cells. Since VZV is highly cell associated, and infection in vitro is achieved by cocultivation of infected cells with uninfected cells (unlike herpes simplex virus, where high titers of cell-free virus can be used to infect cells), accurate assessment of multiplicity of infection is not feasible. Instead, virus-infected cells were mixed with uninfected cells, resulting in a typical multiplicity of 8.6 × 103 infected cells and 1.2 × 106 uninfected cells (multiplicity of infection = 0.007).
VZV DNA was extracted from isolated nucleocapsids (44) by digestion with 50 μg of pronase K (Roche, Indianapolis, Ind.) per ml in 2% sodium dodecyl sulfate (SDS) and spooling on glass rods upon the addition of ethanol. The integrity of virus DNA was determined by restriction endonuclease digestion and agarose gel electrophoresis.
Array construction.
VZV transcription arrays were modeled after similar arrays developed to analyze human cytomegalovirus (2) and herpes simplex virus type 1 (39, 36) transcription. PCR primers (Integrated DNA Technologies, Coralville, Iowa) were selected to amplify 250- to 600-bp DNA segments from both the 5′ start and 3′ terminus of each predicted unique VZV open reading frame (ORF) (GenBank accession number NC 001348). For ORFs < 350 bp, one set of primers was selected to amplify the entire predicted ORF. Table 1 lists the oligonucleotide primers, the ORF to which the primers map, the 5′ location of each 20- to 25-bp primer, and the size and molar G+C content of each amplified fragment.
TABLE 1.
VZV oligonucleotide primers
| Clone | ORF | Forward | 5′ location | Reverse | 5′ location | Size (bp) | G + C (%) |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 1FA | 915 | 1RA | 588 | 327 | 50.0 |
| 2 | 2 | 2FA | 1134 | 2RA | 1503 | 369 | 48.9 |
| 3 | 2 | 2FB | 1483 | 2RB | 1850 | 367 | 47.1 |
| 4 | 3 | 3FA | 2452 | 3RA | 2146 | 306 | 50.4 |
| 5 | 3 | 3FB | 2198 | 3RB | 1907 | 291 | 53.0 |
| 6 | 4 | 4FA | 4142 | 4RA | 3836 | 306 | 47.0 |
| 7 | 4 | 4FE | 3088 | 4RE | 2781 | 307 | 43.3 |
| 8 | 5 | 5FA | 5275 | 5RA | 4972 | 303 | 39.8 |
| 9 | 5 | 5FD | 4553 | 5RD | 4252 | 301 | 39.6 |
| 10 | 6 | 6FA | 8577 | 6RA | 8271 | 306 | 41.2 |
| 11 | 6 | 6FL | 5640 | 6RL | 5325 | 315 | 43.9 |
| 12 | 7 | 7FA | 8605 | 7RA | 8940 | 335 | 50.7 |
| 13 | 7 | 7FB | 9078 | 7RB | 9387 | 309 | 40.3 |
| 14 | 8 | 8FA | 10667 | 8RA | 10324 | 343 | 41.9 |
| 15 | 8 | 8FE | 9783 | 8RE | 9476 | 307 | 39.2 |
| 16 | 9 | 9FA | 11009 | 9RA | 11484 | 475 | 49.5 |
| 17 | 9 | 9FB | 11464 | 9RB | 11917 | 453 | 48.5 |
| 18 | 10 | 10FA | 12160 | 10RA | 12600 | 440 | 41.4 |
| 19 | 10 | 10FC | 12995 | 10RC | 13392 | 397 | 48.2 |
| 20 | 11 | 11FA | 13590 | 11RA | 13919 | 329 | 45.4 |
| 21 | 11 | 11FH | 15752 | 11RH | 16049 | 297 | 43.4 |
| 22 | 12 | 12FA | 16214 | 12RA | 16551 | 337 | 47.9 |
| 23 | 12 | 12FF | 17854 | 12RF | 18199 | 345 | 42.7 |
| 24 | 13 | 13FA | 18440 | 13RA | 18743 | 303 | 42.3 |
| 25 | 13 | 13FC | 19043 | 13RC | 19346 | 303 | 40.1 |
| 26 | 14 | 14FA | 21113 | 14RA | 20668 | 445 | 65.0 |
| 27 | 14 | 14FF | 19731 | 14RF | 19431 | 300 | 42.8 |
| 28 | 15 | 15FA | 22479 | 15RA | 22177 | 302 | 45.8 |
| 29 | 15 | 15FE | 21554 | 15RE | 21258 | 296 | 35.9 |
| 30 | 16 | 16FA | 23794 | 16RA | 23467 | 327 | 43.4 |
| 31 | 16 | 16FD | 22885 | 16RD | 22568 | 317 | 39.7 |
| 32 | 17 | 17FA | 24149 | 17RA | 24443 | 294 | 45.5 |
| 33 | 17 | 17FE | 25233 | 17RE | 25518 | 285 | 39.0 |
| 34 | 18 | 18FA | 26493 | 18RA | 26153 | 340 | 38.1 |
| 35 | 18 | 18FD | 25876 | 18RD | 25573 | 303 | 39.6 |
| 36 | 19 | 19FA | 28845 | 19RA | 28547 | 298 | 46.0 |
| 37 | 19 | 19FH | 26826 | 19RH | 26517 | 309 | 40.6 |
| 38 | 20 | 20FA | 30475 | 20RA | 30160 | 315 | 40.4 |
| 39 | 20 | 20FE | 29371 | 20RE | 29023 | 348 | 44.4 |
| 40 | 21 | 21FA | 30755 | 21RA | 31083 | 328 | 38.6 |
| 41 | 21 | 21FJ | 33567 | 21RJ | 33875 | 308 | 43.0 |
| 42 | 22 | 22FA | 34082 | 22RA | 34402 | 320 | 45.7 |
| 43 | 22 | 22FZ | 42050 | 22RZ | 42374 | 324 | 44.6 |
| 44 | 23 | 23FA | 43119 | 23RA | 42763 | 356 | 52.1 |
| 45 | 23 | 23FB | 42774 | 23RB | 42430 | 344 | 56.5 |
| 46 | 24 | 24FA | 44021 | 24RA | 43678 | 343 | 45.2 |
| 47 | 24 | 24FB | 43560 | 24RB | 43211 | 349 | 44.8 |
| 48 | 25 | 25FA | 44618 | 25RA | 44303 | 315 | 41.0 |
| 49 | 25 | 25FB | 44448 | 25RB | 44148 | 300 | 39.4 |
| 50 | 26 | 26FA | 44506 | 26RA | 44816 | 310 | 41.4 |
| 51 | 26 | 26FF | 45966 | 26RF | 46263 | 297 | 46.6 |
| 52 | 27 | 27FA | 46127 | 27RA | 46483 | 356 | 49.0 |
| 53 | 27 | 27FC | 46792 | 27RC | 47128 | 336 | 42.8 |
| 54 | 28 | 28FA | 50636 | 28RA | 50318 | 318 | 44.6 |
| 55 | 28 | 28FL | 47356 | 28RL | 47051 | 305 | 42.2 |
| 56 | 29 | 29FA | 50857 | 29RA | 51165 | 308 | 47.7 |
| 57 | 29 | 29FM | 54168 | 29RM | 54471 | 303 | 42.9 |
| 58 | 30 | 30FA | 54650 | 30RA | 54973 | 323 | 39.9 |
| 59 | 30 | 30FG | 56631 | 30RG | 56963 | 332 | 47.9 |
| 60 | 31 | 31FA | 57008 | 31RA | 57278 | 270 | 47.6 |
| 61 | 31 | 31FH | 59315 | 31RH | 59615 | 300 | 46.8 |
| 62 | 32 | 32FA | 59766 | 32RA | 59993 | 227 | 48.9 |
| 63 | 32 | 32FB | 59974 | 32RB | 60198 | 224 | 48.4 |
| 64 | 33 | 33FA | 62138 | 33RA | 61799 | 339 | 42.4 |
| 65 | 33 | 33FG | 60674 | 33RG | 60321 | 353 | 45.6 |
| 66 | 34 | 34FA | 63911 | 34RA | 63597 | 314 | 47.2 |
| 67 | 34 | 34FF | 62496 | 34RF | 62170 | 326 | 45.3 |
| 68 | 35 | 35FA | 64753 | 35RA | 64440 | 313 | 46.9 |
| 69 | 35 | 35FB | 64283 | 35RB | 63977 | 306 | 44.4 |
| 70 | 36 | 36FA | 64806 | 36RA | 65082 | 276 | 48.5 |
| 71 | 36 | 36FD | 65566 | 36RD | 65833 | 267 | 47.7 |
| 72 | 37 | 37FA | 66074 | 37RA | 66382 | 308 | 41.1 |
| 73 | 37 | 37FH | 68317 | 37RH | 68601 | 284 | 47.1 |
| 74 | 38 | 38FA | 70293 | 38RA | 69951 | 342 | 50.8 |
| 75 | 38 | 38FF | 68979 | 38RF | 68668 | 311 | 44.0 |
| 76 | 39 | 39FA | 70633 | 39RA | 70894 | 261 | 46.1 |
| 77 | 39 | 39FC | 71055 | 39RC | 71355 | 300 | 42.5 |
| 78 | 40 | 40FA | 71540 | 40RA | 71835 | 295 | 40.1 |
| 79 | 40 | 40FO | 75421 | 40RO | 75730 | 309 | 50.4 |
| 80 | 41 | 41FA | 75846 | 41RA | 76145 | 299 | 48.6 |
| 81 | 41 | 41FC | 76478 | 41RC | 76798 | 320 | 43.6 |
| 82 | 42 | 42FA | 78038 | 42RA | 77729 | 309 | 35.5 |
| 83 | 42 | 42FD | 77182 | 42RD | 76849 | 333 | 39.3 |
| 84 | 43 | 43FA | 78170 | 43RA | 78468 | 298 | 43.0 |
| 85 | 43 | 43FF | 79883 | 43RF | 80178 | 295 | 41.8 |
| 86 | 44 | 44FA | 80337 | 44RA | 80612 | 275 | 47.4 |
| 87 | 44 | 44FD | 81129 | 44RD | 81429 | 300 | 47.5 |
| 88 | 45 | 45FA | 82574 | 45RA | 82271 | 303 | 49.6 |
| 89 | 45 | 45FC | 81854 | 45RC | 81522 | 332 | 39.6 |
| 90 | 46 | 46FA | 82717 | 46RA | 83028 | 311 | 47.2 |
| 91 | 46 | 46FB | 83008 | 46RB | 83319 | 311 | 47.4 |
| 92 | 47 | 47FA | 83168 | 47RA | 83456 | 288 | 48.6 |
| 93 | 47 | 47FG | 84405 | 47RG | 84702 | 297 | 47.8 |
| 94 | 48 | 48FA | 84667 | 48RA | 84967 | 300 | 44.7 |
| 95 | 48 | 48FG | 86029 | 48RG | 86322 | 293 | 49.8 |
| 96 | 49 | 49FA | 86226 | 49RA | 86472 | 246 | 46.5 |
| 97 | 50 | 50FA | 87883 | 50RA | 87556 | 327 | 48.0 |
| 98 | 50 | 50FE | 86886 | 50RE | 86575 | 311 | 49.2 |
| 99 | 51 | 51FA | 87880 | 51RA | 88182 | 302 | 52.1 |
| 100 | 51 | 51FI | 90090 | 51RI | 90388 | 298 | 46.6 |
| 101 | 52 | 52FA | 90493 | 52RA | 90854 | 361 | 51.5 |
| 102 | 52 | 52FI | 92512 | 52RI | 92808 | 296 | 40.2 |
| 103 | 53 | 53FA | 93850 | 53RA | 93526 | 324 | 48.4 |
| 104 | 53 | 53FC | 93153 | 53RC | 92854 | 299 | 42.8 |
| 105 | 54 | 54FA | 95984 | 54RA | 95662 | 322 | 42.2 |
| 106 | 54 | 54FI | 93983 | 54RI | 93675 | 308 | 46.1 |
| 107 | 55 | 55FA | 95996 | 55RA | 96295 | 299 | 43.1 |
| 108 | 55 | 55FI | 98343 | 55RI | 98642 | 299 | 40.2 |
| 109 | 56 | 56FA | 98568 | 56RA | 98871 | 303 | 45.2 |
| 110 | 56 | 56FC | 99006 | 56RC | 99303 | 297 | 49.3 |
| 111 | 57 | 57FA | 99628 | 57RA | 99411 | 217 | 46.5 |
| 112 | 58 | 58FA | 100272 | 58RA | 99953 | 319 | 46.6 |
| 113 | 58 | 58FB | 99925 | 58RB | 99607 | 318 | 43.7 |
| 114 | 59 | 59FA | 101219 | 59RA | 100909 | 310 | 48.8 |
| 115 | 59 | 59FC | 100605 | 59RC | 100301 | 304 | 51.8 |
| 116 | 60 | 60FA | 101651 | 60RA | 101324 | 327 | 44.9 |
| 117 | 60 | 60FB | 101466 | 60RB | 101168 | 298 | 47.4 |
| 118 | Int | 6F | 101650 | 6R | 102019 | 369 | 38.8 |
| 119 | Int | 7F | 102000 | 7R | 102232 | 232 | 45.3 |
| 120 | Int | 8F | 102221 | 8R | 102680 | 459 | 40.3 |
| 121 | Int | 9F | 102661 | 9R | 103082 | 421 | 42.9 |
| 122 | 61 | 61FA | 104486 | 61RA | 104156 | 330 | 50.4 |
| 123 | 61 | 61FF | 103381 | 61RF | 103082 | 299 | 48.1 |
| 124 | Int | 10F | 104486 | 10R | 104924 | 438 | 42.6 |
| 125 | Int | 11F | 104905 | 11R | 105013 | 108 | 65.1 |
| 126 | Int | 12F | 104996 | 12R | 105182 | 186 | 64.7 |
| 127 | 62/71 | 71FA | 109133 | 71RA | 108834 | 299 | 58.8 |
| 128 | 62/71 | 71FB | 108834 | 71RB | 108426 | 408 | 60.1 |
| 129 | 62/71 | 71FC | 108409 | 71RC | 108014 | 395 | 58.2 |
| 130 | 62/71 | 71FD | 108015 | 71RD | 107657 | 358 | 59.8 |
| 131 | 62/71 | 71FE | 107656 | 71RE | 107177 | 479 | 64.3 |
| 132 | 62/71 | 71FF | 107178 | 71RF | 106759 | 419 | 65.8 |
| 133 | 62/71 | 71FG | 106762 | 71RG | 106242 | 520 | 67.2 |
| 134 | 62/71 | 71FH | 123654 | 71RH | 124086 | 432 | 66.8 |
| 135 | 62/71 | 71FI | 105815 | 71RI | 105204 | 611 | 70.8 |
| 136 | 62/71 | 71FK | 105476 | 71RK | 105200 | 276 | 69.3 |
| 137 | Int | 1F | 109134 | 1R | 109625 | 491 | 45.0 |
| 138 | Int | 2F | 109626 | 2R | 109895 | 269 | 68.2 |
| 139 | Int | 3F | 109894 | 3R | 110077 | 183 | 46.4 |
| 140 | Int | 4F | 110085 | 4R | 110328 | 243 | 40.8 |
| 141 | Int | 5F | 110347 | 5R | 110580 | 233 | 47.4 |
| 142 | 63/70 | 63FA | 110580 | 63RA | 110880 | 300 | 57.4 |
| 143 | 63/70 | 63FC | 111118 | 63RC | 111417 | 299 | 59.7 |
| 144 | 64/69 | 64FA | 118332 | 64RA | 118034 | 298 | 62.3 |
| 145 | 64/69 | 64FB | 111809 | 64RB | 112107 | 298 | 55.6 |
| 146 | 65 | 65FA | 112640 | 65RA | 112332 | 308 | 41.9 |
| 147 | 66 | 66FA | 113037 | 66RA | 113344 | 307 | 42.3 |
| 148 | 66 | 66FC | 113911 | 66RC | 114220 | 309 | 40.7 |
| 149 | 67 | 67FA | 114496 | 67RA | 114796 | 300 | 40.2 |
| 150 | 67 | 67FC | 115336 | 67RC | 115562 | 226 | 43.3 |
| 151 | 68 | 68FA | 115808 | 68RA | 116105 | 297 | 44.8 |
| 152 | 69 | 68FG | 117371 | 68RG | 117679 | 308 | 48.3 |
Initially, PCR analysis of VZV-infected and control BSC-1 cell DNA showed that each primer pair was specific for VZV DNA (data not shown). Each primer pair was then used to amplify 106 copies of purified VZV DNA. To facilitate cloning, the primers contained restriction endonuclease sites (EcoRI, PstI, SstII, KpnI, SphI, or HindIII) 5′ proximal to the virus DNA sequence. The amplified product was digested at the engineered restriction endonuclease site, gel purified, and inserted into the multiple cloning site of pGEM3zf− (Promega, Madison, Wis.). Plasmid construction was confirmed by PCR, restriction endonuclease digestion, agarose gel electrophoresis, and, when required, DNA sequencing (Macrogene, Inc., Seoul, Korea). Recombinant clones were propagated, plasmid DNA was extracted, and the VZV DNA insert was PCR amplified with universal plasmid primers. The integrity and quantity of each PCR product were determined by agarose gel electrophoresis, and all DNA targets samples were diluted to 10 ng/μl.
With the aid of multichannel pipettes, each target DNA (4 μl) was spotted onto nylon-based filters (Sigma, St. Louis, Mo.), air dried, denatured in 0.5 M NaOH for 12 min, neutralized in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 4 min, again air dried, and UV cross-linked. Control targets on each array consisted of a 284-bp fragment of human actin cDNA, an 84-bp fragment of human glyceraldehyde phosphate dehydrogenase (GAPdH) cDNA, a 284-bp fragment from the chloramphenicol acetyl transferase (CAT) gene PCR amplified from the pCAT (Gibco-BRL, Carlsbad, Calif.) plasmid (plasmid DNA), and the PCR mixture consisting of vector primers and lacking template plasmid DNA (no DNA). Each set of four control spots was spotted in quadruplet on each array. Each membrane (15.5 by 11 cm) contained one spot each for the VZV targets and four spots for each control target. Each experiment consisted of duplicate arrays, and all experiments were repeated at least thrice with independently obtained probes. Computer analysis of DNA sequences was performed with DNAMax (MiraiBio, Inc., Alameda, Calif.).
RNA extraction and cDNA synthesis.
VZV-infected and control BSC-1 cells were scraped, pelleted, and frozen in liquid nitrogen. Cell pellets were thawed in Tri-Reagent (Molecular Research Center, Cincinnati, Ohio) and briefly sonicated to reduce viscosity, and after addition of 1-bromo-3-chloropropane for phase separation, RNA was precipitated, washed, and dissolved in water. Residual DNA was digested with RNase-free DNase (DNA-free; Ambion, Austin, Tex.) at 37°C for 20 min and poly(A)-containing RNA was extracted by batch-chromatography on oligo(dT)-cellulose (NucleoTrap, Clontech, Palo Alto, Calif.). Poly(A)+ RNA (1 μg) was primed with oligo(dT), and first-strand cDNA was synthesized with modified Moloney murine leukemia virus reverse transcriptase (SuperScript II; Invitrogen, Carlsbad, Calif.). After incubation at 37°C for 90 min, the enzyme was inactivated at 65°C for 5 min and cDNA was cleaned by filtration (Microcon-PCR; Millipore, Bedford, Mass.).
Array probe synthesis.
Array probes were synthesized either by directly incorporating [32P]dCTP during cDNA synthesis or by labeling the cDNA with [32P]dCTP by nick translation. To directly label cDNA, 50 μCi of [32P]dCTP was lyophilized and added to the cDNA reaction modified to contain 500 ng of poly(A)+ RNA and a 10% concentration of unlabeled dCTP. For post-cDNA synthesis, nick translation probe labeling, five reactions (Prediprime II; Amersham Pharmacia Biotech, Piscataway, N.J.), each containing 100 ng of cDNA and 10 μCi of [32P]dCTP, were incubated at 37°C for 60 min. Unincorporated isotope was removed from all labeling reactions by size exclusion gel filtration (Micro Bio-Spin P30; Bio-Rad, Hercules, Calif.). The array probes were heat denatured, quenched on ice, and applied to prehybridized arrays in 20 ml of Perfecthyb (Sigma-Aldrich, St. Louis, Mo.). Hybridization was for 48 h at 65°C, after which the blots were washed three times at 65°C at low stringency (2× SSC-0.1% SDS), twice at high stringency (0.5× SSC-0.1% SDS) and once at ultrahigh stringency (0.1× SSC-0.1% SDS). Arrays were air dried and exposed for uniform times to phosphor-storage screens (Molecular Dynamics, Piscataway, N.J.).
Data analysis.
Arrays were scanned (Storm; Molecular Dynamics) and target spot intensities were determined (Image-Qaunt; Molecular Dynamics). All experiments consisted of replicate arrays and were repeated at least three times. Each experiment also contained arrays hybridized to probes synthesized from uninfected control cells. To enable comparison of results from independent arrays, data were normalized according to the formula:
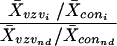 |
where X̄VZVi is average VZV array spot intensity (i), X̄coni is average control array spot intensity, X̄VZVnd is average VZV array no-DNA spot intensity, and X̄connd is average control array no-DNA spot intensity.
Northern blot analysis.
RNA was denatured in formaldehyde at 65°C for 15 min and resolved in 1.8% formamide-denaturing agarose gels, transferred to nylon-based membranes (Zeta Probe; Bio-Rad), and probed with either PCR-generated fragments labeled by nick translation or 32P-end-labeled oligonucleotides (11).
3′-RACE.
First-strand cDNA was synthesized by priming the RNA with a modified oligonucleotide containing a PCR primer pad at the 5′ end of the oligo(dT) (CDS-primer; Clontech, Palo Alto, Calif.). Amplification of the cleaned cDNA with a VZV gene-specific primer located ≈500 nucleotides from the 3′ end of the selected ORF and the 3′ rapid amplification of cDNA ends (RACE) PCR primer resulted in the 3′-terminal sequences of the selected VZV transcript, including the poly(A) tract. Products were cleaned and the DNA sequence wasdetermined.
PCR.
Each 100-μl reaction mixture consisted of 0.3 μg of plasmid DNA or 106 VZV DNA molecules in 1× PCR buffer, 1.2 pmol of each primer, 1.5 μmol of MgCl2 and 0.5 to 1.0 units of Taq (PerkinElmer Life Sciences, Boston, Mass.). Reaction cycles consisted of denaturation for 1 min at 95°C, 40 cycles of amplification at 95°C for 30 s, 55°C for 30 s, and 72°C for 1.5 min, and one extension cycle (94°C for 30 s, 52°C for 30 s, and 72°C for 7 min).
RESULTS
Specificity of VZV arrays.
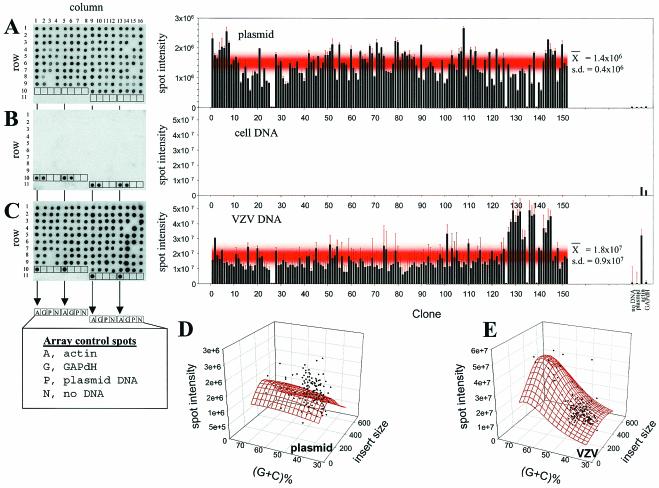
Figure 1 illustrates the array specificity. Each segment of cloned VZV DNA was flanked by ≈50 bp of common plasmid DNA. 32P-end-labeled oligonucleotide probes mapping to this common plasmid sequence detected all amplified targets on the array (panel A), while probes made from cell DNA detected only the actin and GAPdH targets (panel B). Probes made from VZV DNA enriched from virus-infected cells revealed all cloned VZV targets and the cellular actin target (panel C). VZV DNA extracted from enriched nucleocapsids contained small amounts of cellular DNA resulting in detectable levels of cellular actin sequences in the array analysis. The intensity of the actin spots was greater than the intensity of GAPdH spots when the array probe consisted of cellular DNA (panel B), explaining the lack of GAPdH DNA hybridization signal in panel C.
FIG. 1.
Specificity of VZV arrays. VZV arrays were incubated with 32P-end-labeled vector primer (A), 32P-nick-translated control cell DNA (B), or VZV DNA (C) probes. All arrays were configured the same; Table 2 lists the location (column and row) of the 152 VZV DNA ORF targets. Each array also contained a quadruplet set of four control targets. Control targets in each array consisted of actin (row 10, columns 1 and 5; row 11, columns 9 and 13), GAPdH (row 10, columns 2 and 6; row 11, columns 10 and 14), plasmid DNA (row 10, columns 3 and 7; row 11, columns 11 and 15), and no DNA (row 10, columns 4 and 8; row 11, columns 12 and 16). After hybridization, the intensity of each spot was quantitated by phosphoimaging. The graph to the right of each array shows the spot intensity (arbitrary units) associated with each of the 152 VZV target clones, along with the average spot intensity associated with the array controls. Error bars indicate the standard error of the mean for each data point. The red shaded regions in the graphs show the mean ± 1 standard deviation for the VZV target DNAs. The three-dimensional Lorentzian transformation of spot intensity versus insert molar G+C and insert size (in base pairs) revealed no statistical correlation when the array was probed with the plasmid oligonucleotide (D), but a slight correlation between spot intensity and insert G+C content when the array was probed with VZV DNA (E).
Quantitation of spot intensity from the array in panel A revealed no clustering of targets with intensities of >1 standard deviation from the mean (red-shaded region in the plasmid graph), indicating the absence of geometric bias in the array. No statistical correlation between spot intensity and insert G+C molar content (R2 = 0.07) or target size (R2 < 0.01) was detected in arrays probed with the plasmid-specific oligonucleotide (panel D). Similarly, quantitation of spot intensity from the array probed with VZV DNA (panel C) showed that most hybridization signals were within 1 standard deviation from the mean (red shaded region). However, a cluster of targets corresponding to clones 129 to 144 showed ≈2-fold higher than average signal intensity. These targets mapped to VZV ORFs 62, 63, and 64 and the intergenic region separating VZV ORFs 62 and 63, and all of these targets mapped within the inverted repeat of the unique short segment (IRS) of the VZV genome. Thus, a randomly labeled, nick-translated probe synthesized from VZV DNA would be expected to overrepresent (by twofold) the amount of probe accessible to targets within the IRS. Alternatively, the IRS contain areas of high G+C molar content, which could result in a higher hybridization signal. Panel E illustrates this minimal association of spot intensity with (G+C)% (R2 = 0.57), and the absence of any significant association between spot volume and VZV DNA insert size (R2 = 0.08).
Sensitivity of VZV arrays.
The relative abundance of individual transcripts within an RNA preparation is most accurately determined by array analysis when the probe is labeled during first-strand cDNA synthesis (17, 40). Whereas direct cDNA labeling of VZV-infected cell mRNA yielded probe with activity sufficient to detect numerous VZV transcripts, labeling the synthesized cDNA by nick translation yielded probes with higher activity. With the single exception of clone 136, which contains a 276-bp insert mapping to the 3′ terminus of ORF 62, the relative abundance of the virus transcripts was maintained independent of the labeling method (data not shown).
Transcription of VZV genes during lytic infection.
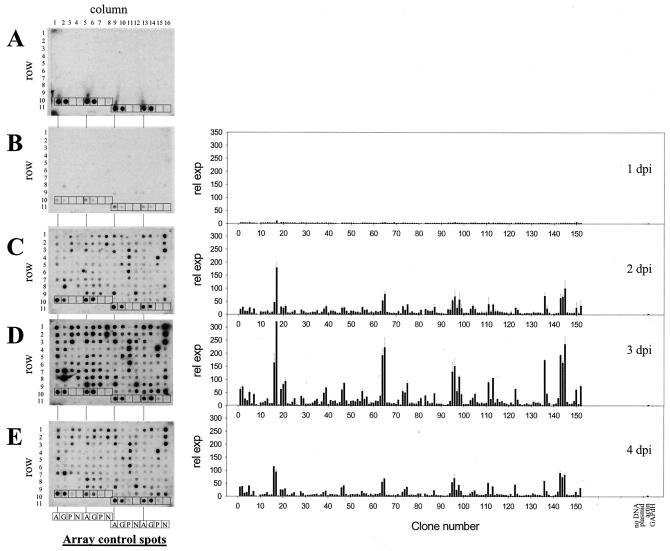
VZV is highly cell associated, which precludes the ability to obtain high titers of cell-free virus, a requirement for synchronized high-multiplicity infections and one-step growth analysis. Instead, virus infection is initiated by cocultivation of infected cells with uninfected cells. Such pseudo-one-step analysis has been used to compare the growth kinetics of wild-type and mutant VZVs containing site-directed mutations (4, 9, 30, 34). Thus, sequential array analysis reveals virus gene transcription as the infection progresses through the culture (Fig. 2). Nick-translated cDNA array probes were synthesized from VZV-infected cell RNA harvested at 1 to 4 days postinfection and compared to probes synthesized from control cells harvested after 2 days in culture (Fig. 2). Probe synthesized from control cells detected only actin and GAPdH transcripts (panel A), whereas probe synthesized from VZV-infected cells harvested at 1 to 4 days postinfection detected, along with actin and GAPdH, numerous VZV targets (panels B to E). Quantitative analysis of individual spot intensities revealed increased expression of VZV genes as a function of time but no significant variation in the pattern of virus gene expression (Fig. 2, graphs).
FIG. 2.
Array analysis of VZV transcripts during lytic infection. Arrays were hybridized to probes synthesized from control cell RNA (A) or RNA extracted from VZV-infected cells harvested 1 day postinfection (dpi) (B), 2 days postinfection (C), 3 days postinfection (D), or 4 days postinfection (E). VZV DNA target and control spots are configured on each array as described in the Fig. 1 legend. After hybridization, spot intensities were determined and normalized to the values obtained from the array hybridized with the control cell probe. Graphs show the relative expression (rel. exp.) associated with each VZV DNA target clone and the control spots in the arrays. The overall abundance of VZV transcription peaked at 3 days postinfection, when targets mapping to all possible virus genes were detected.
Table 2 presents the quantitative analysis of VZV gene transcription. Columns 1 to 5 correspond to the array images and list the clone number, predicted VZV ORF, and region to which the cloned VZV DNA insert maps and the physical coordinates that locate each clone on the array by column and row. Other columns list the relative expression and standard deviation of each array target generated from probe synthesized from VZV-infected cells harvested at 1 to 4 days postinfection and the order (in descending relative expression) of each VZV target resulting from the probe at 1 to 4 days postinfection. For example, the highest relative expression obtained with the 1-day postinfection probe was associated with clone 17, the VZV insert mapping to the 3′ terminus of ORF 9. Since DNA corresponding to clones 26, 27, 126, 135, and 139 (indicated in italics) was not applied to the arrays, the order number of these spots was used to determine the threshold for detection of VZV gene expression. Thus, transcripts mapping to a particular target VZV clone are listed as present (indicated in bold) if the order number was less than the minimum order number corresponding to clones 26, 27, 126, 135, or 139.
TABLE 2.
Quantitative analysis of VZV gene transcriptiona
| A1 clone | A2 ORF | A3 region | A4 column | A5 row | 1 dpi
|
2 dpi
|
3 dpi
|
4 dpi
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B1 rel exp | B2 std | B3 order | C1 rel exp | C2 std | C3 order | D1 rel exp | D2 std | D3 order | E1 rel exp | E2 std | E3 order | |||||
| 1 | 1 | all | 1 | 1 | 3.6 | 0.7 | 36 | 19.3 | 3.8 | 39 | 62.5 | 8.9 | 24 | 36.0 | 1.5 | 19 |
| 2 | 2 | 5′ | 1 | 2 | 3.7 | 0.7 | 25 | 28.2 | 0.6 | 22 | 71.9 | 6.6 | 19 | 38.0 | 0.7 | 17 |
| 3 | 2 | 3′ | 1 | 3 | 3.4 | 0.5 | 56 | 13.2 | 0.3 | 55 | 26.5 | 1.0 | 47 | 13.1 | 0.4 | 43 |
| 4 | 3 | 5′ | 1 | 4 | 3.5 | 0.2 | 49 | 12.8 | 0.4 | 59 | 19.3 | 0.6 | 59 | 15.0 | 0.6 | 41 |
| 5 | 3 | 3′ | 1 | 5 | 4.1 | 0.7 | 8 | 25.5 | 1.2 | 28 | 51.3 | 4.9 | 29 | 39.8 | 2.4 | 15 |
| 6 | 4 | 5′ | 1 | 6 | 2.7 | 0.1 | 117 | 5.7 | 0.9 | 119 | 7.8 | 0.4 | 118 | 5.3 | 0.1 | 116 |
| 7 | 4 | 3′ | 1 | 7 | 2.7 | 0.1 | 122 | 21.8 | 0.3 | 34 | 42.7 | 3.8 | 36 | 19.2 | 0.8 | 33 |
| 8 | 5 | 5′ | 1 | 8 | 1.5 | 0.7 | 147 | 2.5 | 0.5 | 148 | 4.8 | 0.0 | 141 | 1.9 | 0.1 | 150 |
| 9 | 5 | 3′ | 1 | 9 | 0.3 | 0.1 | 152 | 0.4 | 0.1 | 152 | 0.5 | 0.0 | 152 | 0.5 | 0.0 | 152 |
| 10 | 6 | 5′ | 2 | 1 | 3.3 | 0.2 | 59 | 10.8 | 0.3 | 68 | 11.4 | 0.1 | 91 | 5.5 | 0.1 | 114 |
| 11 | 6 | 3′ | 2 | 2 | 3.6 | 0.3 | 35 | 9.4 | 0.7 | 77 | 12.1 | 0.2 | 85 | 6.8 | 0.3 | 90 |
| 12 | 7 | 5′ | 2 | 3 | 3.5 | 0.3 | 46 | 9.7 | 1.2 | 74 | 17.3 | 0.1 | 69 | 8.8 | 0.1 | 70 |
| 13 | 7 | 3′ | 2 | 4 | 4.5 | 0.3 | 5 | 14.0 | 3.2 | 51 | 26.6 | 1.2 | 46 | 14.8 | 1.7 | 42 |
| 14 | 8 | 5′ | 2 | 5 | 3.2 | 0.6 | 81 | 6.1 | 0.3 | 111 | 8.6 | 0.0 | 106 | 5.8 | 0.0 | 110 |
| 15 | 8 | 3′ | 2 | 6 | 3.1 | 0.2 | 96 | 9.3 | 1.3 | 79 | 8.5 | 0.0 | 107 | 6.7 | 0.1 | 91 |
| 16 | 9 | 5′ | 2 | 7 | 2.9 | 0.7 | 109 | 46.5 | 1.0 | 11 | 164.5 | 35.3 | 8 | 114.3 | 2.3 | 1 |
| 17 | 9 | 3′ | 2 | 8 | 10.8 | 0.7 | 1 | 179.5 | 20.7 | 1 | 321.0 | 2.7 | 1 | 93.9 | 9.0 | 2 |
| 18 | 10 | 5′ | 2 | 9 | 1.7 | 0.3 | 145 | 2.4 | 0.4 | 149 | 3.8 | 0.5 | 145 | 2.5 | 0.2 | 146 |
| 19 | 10 | 3′ | 3 | 1 | 4.0 | 1.5 | 11 | 28.8 | 1.9 | 21 | 63.3 | 0.7 | 22 | 26.1 | 0.3 | 24 |
| 20 | 11 | 5′ | 3 | 2 | 4.4 | 1.0 | 6 | 24.6 | 4.7 | 30 | 82.3 | 2.2 | 17 | 25.3 | 0.7 | 25 |
| 21 | 11 | 3′ | 3 | 3 | 3.7 | 0.5 | 30 | 31.4 | 4.9 | 17 | 93.8 | 13.9 | 13 | 29.8 | 0.2 | 22 |
| 22 | 12 | 5′ | 3 | 4 | 3.3 | 0.2 | 63 | 6.0 | 2.0 | 112 | 8.2 | 0.3 | 113 | 6.0 | 0.2 | 102 |
| 23 | 12 | 3′ | 3 | 5 | 3.2 | 0.1 | 87 | 5.4 | 1.5 | 126 | 5.8 | 0.3 | 137 | 4.8 | 0.0 | 124 |
| 24 | 13 | 5′ | 3 | 6 | 3.2 | 0.9 | 89 | 9.3 | 1.3 | 78 | 14.3 | 0.0 | 78 | 10.0 | 0.3 | 56 |
| 25 | 13 | 3′ | 3 | 7 | 3.2 | 0.6 | 82 | 14.6 | 2.4 | 49 | 27.8 | 0.5 | 44 | 15.3 | 0.1 | 40 |
| 26 | 14 | 5′ | 3 | 8 | 2.4 | 0.3 | 130 | 2.7 | 0.0 | 145 | 3.2 | 0.3 | 148 | 2.5 | 0.2 | 147 |
| 27 | 14 | 3′ | 3 | 9 | 2.4 | 0.2 | 131 | 2.6 | 0.0 | 146 | 2.4 | 0.2 | 149 | 2.4 | 0.2 | 148 |
| 28 | 15 | 5′ | 4 | 1 | 3.8 | 0.8 | 23 | 17.5 | 1.9 | 44 | 36.1 | 3.9 | 39 | 18.7 | 0.5 | 36 |
| 29 | 15 | 3′ | 4 | 2 | 3.3 | 0.8 | 61 | 12.1 | 3.1 | 64 | 27.4 | 3.6 | 45 | 11.5 | 0.2 | 48 |
| 30 | 16 | 5′ | 4 | 3 | 3.3 | 0.3 | 67 | 6.4 | 1.5 | 106 | 6.9 | 0.2 | 126 | 3.9 | 0.0 | 137 |
| 31 | 16 | 3′ | 4 | 4 | 3.3 | 0.1 | 74 | 5.5 | 1.5 | 122 | 6.7 | 0.0 | 132 | 5.1 | 0.0 | 119 |
| 32 | 17 | 5′ | 4 | 5 | 3.3 | 0.0 | 66 | 7.0 | 1.5 | 94 | 7.3 | 0.0 | 124 | 5.3 | 0.0 | 117 |
| 33 | 17 | 3′ | 4 | 6 | 3.3 | 0.2 | 69 | 8.9 | 2.9 | 82 | 11.2 | 0.3 | 94 | 6.9 | 0.1 | 89 |
| 34 | 18 | 5′ | 4 | 7 | 2.8 | 0.1 | 114 | 8.9 | 1.0 | 84 | 18.0 | 2.4 | 64 | 7.9 | 0.2 | 76 |
| 35 | 18 | 3′ | 4 | 8 | 3.7 | 0.3 | 28 | 15.4 | 0.5 | 48 | 24.8 | 2.7 | 52 | 9.4 | 0.4 | 60 |
| 36 | 19 | 5′ | 4 | 9 | 2.4 | 0.3 | 128 | 3.8 | 0.8 | 138 | 5.7 | 0.4 | 138 | 3.9 | 0.4 | 136 |
| 37 | 19 | 3′ | 5 | 1 | 3.7 | 0.0 | 26 | 9.6 | 1.1 | 76 | 15.2 | 0.5 | 76 | 7.5 | 0.1 | 80 |
| 38 | 20 | 5′ | 5 | 2 | 3.4 | 0.1 | 52 | 21.6 | 7.2 | 35 | 48.7 | 2.3 | 31 | 15.9 | 0.1 | 39 |
| 39 | 20 | 3′ | 5 | 3 | 3.7 | 0.4 | 29 | 30.7 | 6.1 | 19 | 69.2 | 8.2 | 20 | 21.5 | 0.6 | 29 |
| 40 | 21 | 5′ | 5 | 4 | 3.2 | 0.1 | 90 | 5.8 | 2.5 | 117 | 6.6 | 0.2 | 133 | 4.3 | 0.1 | 133 |
| 41 | 21 | 3′ | 5 | 5 | 3.6 | 0.2 | 39 | 13.8 | 2.7 | 52 | 21.1 | 0.0 | 56 | 8.7 | 0.0 | 71 |
| 42 | 22 | 5′ | 5 | 6 | 3.0 | 0.2 | 103 | 13.3 | 4.4 | 54 | 16.9 | 0.9 | 72 | 8.8 | 0.1 | 69 |
| 43 | 22 | 3′ | 5 | 7 | 2.5 | 0.0 | 126 | 10.4 | 1.4 | 70 | 18.5 | 1.5 | 61 | 7.1 | 0.0 | 85 |
| 44 | 23 | 5′ | 5 | 8 | 0.9 | 0.2 | 149 | 6.0 | 0.0 | 114 | 8.3 | 0.6 | 111 | 6.6 | 0.3 | 92 |
| 45 | 23 | 3′ | 5 | 9 | 0.4 | 0.1 | 150 | 5.7 | 0.8 | 118 | 12.2 | 1.6 | 83 | 3.3 | 0.2 | 143 |
| 46 | 24 | 5′ | 6 | 1 | 3.6 | 0.2 | 40 | 25.0 | 3.9 | 29 | 61.0 | 5.6 | 25 | 33.4 | 1.4 | 20 |
| 47 | 24 | 3′ | 6 | 2 | 3.8 | 0.1 | 20 | 26.1 | 3.0 | 26 | 87.2 | 2.0 | 15 | 41.2 | 2.4 | 14 |
| 48 | 25 | 5′ | 6 | 3 | 3.7 | 0.3 | 32 | 12.3 | 2.6 | 63 | 19.4 | 0.9 | 58 | 9.2 | 0.1 | 65 |
| 49 | 25 | 3′ | 6 | 4 | 3.0 | 0.4 | 101 | 5.3 | 0.7 | 128 | 6.8 | 0.4 | 129 | 4.3 | 0.1 | 132 |
| 50 | 26 | 5′ | 6 | 5 | 3.4 | 0.7 | 58 | 13.8 | 2.6 | 53 | 17.0 | 0.8 | 71 | 7.8 | 0.1 | 78 |
| 51 | 26 | 3′ | 6 | 6 | 3.1 | 0.2 | 94 | 12.9 | 2.2 | 58 | 19.4 | 0.0 | 57 | 8.9 | 0.1 | 68 |
| 52 | 27 | 5′ | 6 | 7 | 2.3 | 0.1 | 133 | 9.0 | 3.0 | 81 | 16.0 | 0.4 | 74 | 7.4 | 0.4 | 82 |
| 53 | 27 | 3′ | 6 | 8 | 1.9 | 0.1 | 143 | 9.7 | 1.6 | 75 | 25.5 | 0.5 | 49 | 9.4 | 0.4 | 61 |
| 54 | 28 | 5′ | 6 | 9 | 1.8 | 0.2 | 144 | 7.3 | 0.6 | 93 | 17.5 | 0.1 | 68 | 6.0 | 0.7 | 101 |
| 55 | 28 | 3′ | 7 | 1 | 3.8 | 0.3 | 17 | 27.6 | 2.2 | 23 | 56.1 | 7.1 | 27 | 20.8 | 0.8 | 32 |
| 56 | 29 | 5′ | 7 | 2 | 3.5 | 0.7 | 45 | 17.4 | 0.5 | 45 | 25.3 | 0.7 | 51 | 12.7 | 0.0 | 44 |
| 57 | 29 | 3′ | 7 | 3 | 3.4 | 0.4 | 50 | 18.5 | 0.1 | 41 | 34.4 | 2.0 | 40 | 11.2 | 0.1 | 51 |
| 58 | 30 | 5′ | 7 | 4 | 3.3 | 0.1 | 75 | 5.9 | 1.6 | 115 | 7.3 | 0.0 | 123 | 4.7 | 0.0 | 126 |
| 59 | 30 | 3′ | 7 | 5 | 3.2 | 0.0 | 80 | 10.8 | 2.1 | 69 | 12.3 | 0.1 | 82 | 9.9 | 0.2 | 57 |
| 60 | 31 | 5′ | 7 | 6 | 2.8 | 0.4 | 111 | 6.7 | 1.9 | 100 | 11.7 | 0.1 | 88 | 5.9 | 0.2 | 105 |
| 61 | 31 | 3′ | 7 | 7 | 2.6 | 0.2 | 124 | 12.5 | 3.5 | 61 | 18.3 | 1.1 | 62 | 9.4 | 0.5 | 64 |
| 62 | 32 | 5′ | 7 | 8 | 1.9 | 0.1 | 142 | 8.9 | 2.9 | 83 | 17.6 | 0.4 | 66 | 9.4 | 0.4 | 63 |
| 63 | 32 | 3′ | 7 | 9 | 3.1 | 0.3 | 92 | 13.0 | 1.6 | 57 | 38.4 | 0.3 | 38 | 12.3 | 0.2 | 45 |
| 64 | 33 | 5′ | 8 | 1 | 4.0 | 0.8 | 14 | 53.7 | 0.5 | 10 | 192.9 | 22.6 | 5 | 55.6 | 2.2 | 9 |
| 65 | 33 | 3′ | 8 | 2 | 4.7 | 1.5 | 4 | 78.7 | 11.2 | 3 | 222.9 | 39.4 | 3 | 68.3 | 2.9 | 7 |
| 66 | 34 | 5′ | 8 | 3 | 3.4 | 0.2 | 51 | 10.0 | 0.6 | 72 | 11.8 | 0.9 | 87 | 5.8 | 0.0 | 108 |
| 67 | 34 | 3′ | 8 | 4 | 3.3 | 0.1 | 71 | 8.1 | 0.2 | 90 | 11.9 | 0.6 | 86 | 5.8 | 0.3 | 109 |
| 68 | 35 | 5′ | 8 | 5 | 2.8 | 0.2 | 112 | 5.4 | 1.0 | 127 | 8.2 | 0.2 | 112 | 4.5 | 0.2 | 130 |
| 69 | 35 | 3′ | 8 | 6 | 4.0 | 1.8 | 13 | 5.9 | 1.6 | 116 | 9.9 | 0.1 | 100 | 6.0 | 0.2 | 103 |
| 70 | 36 | 5′ | 8 | 7 | 2.7 | 0.7 | 119 | 4.8 | 2.2 | 135 | 7.4 | 0.4 | 122 | 4.6 | 0.2 | 128 |
| 71 | 36 | 3′ | 8 | 8 | 2.6 | 0.3 | 123 | 12.1 | 2.4 | 65 | 22.6 | 1.5 | 54 | 8.1 | 0.8 | 74 |
| 72 | 37 | 5′ | 8 | 9 | 2.4 | 0.3 | 129 | 3.8 | 0.9 | 140 | 5.0 | 0.0 | 140 | 3.9 | 0.3 | 135 |
| 73 | 37 | 3′ | 9 | 1 | 4.1 | 0.3 | 10 | 31.0 | 5.0 | 18 | 55.4 | 8.9 | 28 | 16.9 | 0.9 | 38 |
| 74 | 38 | 5′ | 9 | 2 | 3.2 | 0.5 | 76 | 17.9 | 3.6 | 42 | 50.3 | 2.7 | 30 | 27.1 | 1.7 | 23 |
| 75 | 38 | 3′ | 9 | 3 | 3.2 | 0.2 | 85 | 38.8 | 0.8 | 13 | 87.0 | 10.0 | 16 | 36.5 | 1.7 | 18 |
| 76 | 39 | 5′ | 9 | 4 | 3.0 | 0.1 | 102 | 6.2 | 1.6 | 110 | 9.2 | 0.4 | 102 | 5.9 | 0.2 | 106 |
| 77 | 39 | 3′ | 9 | 5 | 3.2 | 0.5 | 78 | 11.3 | 2.8 | 66 | 11.6 | 1.2 | 90 | 7.4 | 0.3 | 81 |
| 78 | 40 | 5′ | 9 | 6 | 3.1 | 0.4 | 97 | 6.2 | 1.5 | 109 | 8.3 | 0.2 | 110 | 5.7 | 0.3 | 111 |
| 79 | 40 | 3′ | 9 | 7 | 2.9 | 0.4 | 108 | 5.0 | 1.3 | 134 | 8.5 | 0.1 | 108 | 5.6 | 0.5 | 113 |
| 80 | 41 | 5′ | 9 | 8 | 2.1 | 0.3 | 138 | 6.7 | 1.4 | 102 | 17.8 | 0.1 | 65 | 11.3 | 0.7 | 50 |
| 81 | 41 | 3′ | 9 | 9 | 1.6 | 0.3 | 146 | 17.3 | 4.2 | 46 | 28.6 | 5.9 | 43 | 10.2 | 0.9 | 55 |
| 82 | 42 | 5′ | 9 | 10 | 0.3 | 0.0 | 151 | 0.5 | 0.1 | 151 | 1.0 | 0.0 | 151 | 0.5 | 0.0 | 151 |
| 83 | 42 | 3′ | 10 | 1 | 3.2 | 0.6 | 84 | 20.8 | 1.2 | 37 | 39.1 | 3.8 | 37 | 10.8 | 0.1 | 54 |
| 84 | 43 | 5′ | 10 | 2 | 3.3 | 0.2 | 62 | 15.5 | 2.2 | 47 | 17.3 | 1.6 | 70 | 7.0 | 0.1 | 88 |
| 85 | 43 | 3′ | 10 | 3 | 2.7 | 0.2 | 120 | 12.4 | 2.2 | 62 | 8.0 | 0.8 | 115 | 4.2 | 0.2 | 134 |
| 86 | 44 | 5′ | 10 | 4 | 3.0 | 0.2 | 104 | 14.1 | 2.6 | 50 | 11.2 | 1.0 | 95 | 5.9 | 0.3 | 107 |
| 87 | 44 | 3′ | 10 | 5 | 3.5 | 0.3 | 47 | 28.8 | 11.1 | 20 | 47.9 | 4.8 | 32 | 23.5 | 1.8 | 27 |
| 88 | 45 | 5′ | 10 | 6 | 3.1 | 0.6 | 93 | 6.4 | 3.0 | 104 | 8.4 | 0.6 | 109 | 5.6 | 0.3 | 112 |
| 89 | 45 | 3′ | 10 | 7 | 2.8 | 0.4 | 113 | 5.0 | 2.3 | 131 | 7.6 | 0.6 | 120 | 4.8 | 0.3 | 125 |
| 90 | 46 | 5′ | 10 | 8 | 2.9 | 0.7 | 105 | 5.5 | 2.9 | 124 | 7.9 | 0.6 | 116 | 5.1 | 0.2 | 122 |
| 91 | 46 | 3′ | 10 | 9 | 2.6 | 0.5 | 125 | 3.8 | 1.6 | 139 | 4.5 | 0.2 | 143 | 3.8 | 0.3 | 141 |
| 92 | 47 | 5′ | 10 | 10 | 2.1 | 0.6 | 136 | 2.6 | 1.1 | 147 | 3.7 | 0.1 | 146 | 3.0 | 0.2 | 144 |
| 93 | 47 | 3′ | 11 | 1 | 3.2 | 1.1 | 83 | 19.8 | 3.2 | 38 | 6.7 | 0.4 | 131 | 5.0 | 0.1 | 123 |
| 94 | 48 | 5′ | 11 | 2 | 3.4 | 1.3 | 57 | 18.7 | 7.0 | 40 | 18.2 | 1.5 | 63 | 12.2 | 0.4 | 46 |
| 95 | 48 | 3′ | 11 | 3 | 3.9 | 0.6 | 15 | 54.4 | 7.0 | 9 | 130.8 | 36.3 | 10 | 54.3 | 2.6 | 10 |
| 96 | 49 | all | 11 | 4 | 5.9 | 0.7 | 3 | 69.5 | 25.1 | 5 | 152.1 | 31.9 | 9 | 70.5 | 13.8 | 6 |
| 97 | 50 | 5′ | 11 | 5 | 3.6 | 0.9 | 37 | 25.9 | 11.8 | 27 | 56.2 | 10.4 | 26 | 25.0 | 1.2 | 26 |
| 98 | 50 | 3′ | 11 | 6 | 3.3 | 1.1 | 60 | 57.4 | 26.6 | 8 | 112.0 | 6.7 | 11 | 43.2 | 1.9 | 13 |
| 99 | 51 | 5′ | 11 | 7 | 3.7 | 0.5 | 31 | 24.1 | 14.6 | 31 | 44.1 | 4.5 | 35 | 21.5 | 0.9 | 30 |
| 100 | 51 | 3′ | 11 | 8 | 2.9 | 0.6 | 107 | 5.6 | 3.4 | 121 | 6.7 | 0.6 | 130 | 5.1 | 0.2 | 121 |
| 101 | 52 | 5′ | 11 | 9 | 2.7 | 0.9 | 116 | 6.3 | 3.4 | 107 | 7.0 | 0.8 | 125 | 5.9 | 0.0 | 104 |
| 102 | 52 | 3′ | 11 | 10 | 2.3 | 0.6 | 132 | 5.2 | 2.6 | 129 | 6.8 | 0.4 | 127 | 4.4 | 0.4 | 131 |
| 103 | 53 | 5′ | 12 | 1 | 3.4 | 1.2 | 54 | 21.9 | 3.1 | 33 | 21.3 | 0.6 | 55 | 11.4 | 0.2 | 49 |
| 104 | 53 | 3′ | 12 | 2 | 3.4 | 1.2 | 55 | 33.5 | 4.7 | 16 | 30.1 | 2.9 | 42 | 17.8 | 0.5 | 37 |
| 105 | 54 | 5′ | 12 | 3 | 3.8 | 1.3 | 19 | 20.9 | 5.5 | 36 | 45.6 | 4.2 | 34 | 23.4 | 0.1 | 28 |
| 106 | 54 | 3′ | 12 | 4 | 3.1 | 0.8 | 91 | 6.9 | 2.4 | 96 | 14.1 | 0.4 | 79 | 9.1 | 0.1 | 67 |
| 107 | 55 | 5′ | 12 | 5 | 2.9 | 0.4 | 106 | 6.7 | 3.3 | 98 | 8.9 | 0.6 | 105 | 5.1 | 0.3 | 120 |
| 108 | 55 | 3′ | 12 | 6 | 3.6 | 0.8 | 41 | 8.5 | 4.4 | 85 | 11.3 | 1.2 | 93 | 6.5 | 0.2 | 93 |
| 109 | 56 | 5′ | 12 | 7 | 3.3 | 0.4 | 68 | 6.7 | 3.7 | 99 | 8.9 | 0.4 | 104 | 6.2 | 0.3 | 97 |
| 110 | 56 | 3′ | 12 | 8 | 3.1 | 0.2 | 100 | 8.4 | 5.8 | 87 | 11.0 | 0.7 | 96 | 9.9 | 0.0 | 58 |
| 111 | 57 | all | 12 | 9 | 4.0 | 1.5 | 12 | 40.0 | 25.4 | 12 | 90.4 | 8.2 | 14 | 52.0 | 2.6 | 11 |
| 112 | 58 | 5′ | 12 | 10 | 2.1 | 0.6 | 139 | 11.2 | 5.5 | 67 | 25.6 | 1.4 | 48 | 9.4 | 0.3 | 62 |
| 113 | 58 | 3′ | 13 | 1 | 3.8 | 1.5 | 18 | 38.5 | 2.8 | 14 | 107.0 | 0.6 | 12 | 39.0 | 0.2 | 16 |
| 114 | 59 | 5′ | 13 | 2 | 2.8 | 0.9 | 115 | 12.7 | 6.7 | 60 | 11.7 | 1.1 | 89 | 6.1 | 0.1 | 99 |
| 115 | 59 | 3′ | 13 | 3 | 3.6 | 1.4 | 34 | 13.1 | 3.2 | 56 | 25.5 | 2.3 | 50 | 11.0 | 0.1 | 53 |
| 116 | 60 | 5′ | 13 | 4 | 3.3 | 1.4 | 65 | 9.9 | 3.4 | 73 | 14.7 | 0.5 | 77 | 8.3 | 0.2 | 72 |
| 117 | 60 | 3′ | 13 | 5 | 3.3 | 1.4 | 72 | 10.1 | 3.6 | 71 | 17.6 | 0.8 | 67 | 7.9 | 0.5 | 75 |
| 118 | Int 6 | na | 13 | 6 | 3.2 | 0.0 | 86 | 7.3 | 2.9 | 92 | 10.8 | 0.8 | 97 | 7.1 | 0.3 | 84 |
| 119 | Int 7 | na | 13 | 7 | 3.5 | 0.1 | 48 | 6.9 | 3.4 | 97 | 8.1 | 1.2 | 114 | 6.3 | 0.3 | 96 |
| 120 | Int 8 | na | 13 | 8 | 2.5 | 0.2 | 127 | 3.5 | 1.9 | 142 | 4.6 | 0.3 | 142 | 3.8 | 0.2 | 139 |
| 121 | Int 9 | na | 13 | 9 | 3.2 | 0.8 | 79 | 8.4 | 5.2 | 86 | 12.6 | 0.7 | 81 | 9.8 | 0.6 | 59 |
| 122 | 61 | 5′ | 13 | 10 | 1.0 | 0.5 | 148 | 2.9 | 1.5 | 144 | 6.8 | 0.0 | 128 | 2.8 | 0.0 | 145 |
| 123 | 61 | 3′ | 14 | 1 | 3.3 | 1.4 | 73 | 26.5 | 1.5 | 25 | 64.4 | 4.4 | 21 | 45.1 | 0.7 | 12 |
| 124 | Int 10 | na | 14 | 2 | 3.5 | 1.0 | 42 | 17.7 | 6.5 | 43 | 23.4 | 1.1 | 53 | 11.1 | 0.3 | 52 |
| 125 | Int 11 | na | 14 | 3 | 3.9 | 0.9 | 16 | 5.5 | 2.5 | 123 | 7.8 | 0.1 | 117 | 4.6 | 0.0 | 127 |
| 126 | Int 12 | na | 14 | 4 | 3.5 | 0.5 | 44 | 3.7 | 0.7 | 141 | 3.5 | 0.1 | 147 | 3.5 | 0.1 | 142 |
| 127 | 62 | 5′-a | 14 | 5 | 2.7 | 0.5 | 121 | 4.6 | 1.9 | 136 | 6.4 | 0.3 | 134 | 4.5 | 0.0 | 129 |
| 128 | 62 | 5′-b | 14 | 6 | 3.2 | 0.7 | 77 | 6.0 | 2.1 | 113 | 10.4 | 0.0 | 98 | 7.1 | 0.2 | 86 |
| 129 | 62 | 5′-c | 14 | 7 | 3.1 | 0.1 | 95 | 5.0 | 1.7 | 132 | 5.9 | 0.2 | 136 | 6.3 | 0.1 | 95 |
| 130 | 62 | 5′-d | 14 | 8 | 2.7 | 0.0 | 118 | 5.0 | 1.5 | 133 | 5.6 | 0.1 | 139 | 5.2 | 0.2 | 118 |
| 131 | 62 | 5′-e | 14 | 9 | 2.8 | 0.5 | 110 | 9.0 | 3.6 | 80 | 16.4 | 0.2 | 73 | 12.1 | 0.4 | 47 |
| 132 | 62 | 5′-f | 14 | 10 | 2.0 | 0.8 | 141 | 6.4 | 1.6 | 105 | 12.1 | 0.8 | 84 | 8.2 | 0.5 | 73 |
| 133 | 62 | 5′-g | 15 | 1 | 2.3 | 0.9 | 135 | 8.3 | 0.7 | 88 | 12.7 | 0.6 | 80 | 6.1 | 0.3 | 100 |
| 134 | 62 | 5′-h | 15 | 2 | 3.6 | 1.1 | 38 | 8.0 | 0.0 | 91 | 10.3 | 0.2 | 99 | 6.2 | 1.0 | 98 |
| 135 | 62 | 5′-i | 15 | 3 | 3.8 | 0.2 | 22 | 4.0 | 0.8 | 137 | 4.0 | 0.2 | 144 | 3.9 | 0.1 | 138 |
| 136 | 62 | 3′ | 15 | 4 | 3.1 | 0.7 | 99 | 71.6 | 14.4 | 4 | 174.9 | 5.7 | 6 | 60.4 | 1.0 | 8 |
| 137 | Int 1 | na | 15 | 5 | 4.1 | 0.3 | 9 | 23.6 | 12.9 | 32 | 47.0 | 1.0 | 33 | 21.4 | 0.7 | 31 |
| 138 | Int 2 | na | 15 | 6 | 3.3 | 0.3 | 70 | 6.7 | 3.3 | 101 | 11.4 | 0.1 | 92 | 7.8 | 0.0 | 79 |
| 139 | Int 3 | na | 15 | 7 | 2.3 | 0.2 | 134 | 2.0 | 0.2 | 150 | 1.9 | 0.0 | 150 | 2.4 | 0.0 | 149 |
| 140 | Int 4 | na | 15 | 8 | 2.1 | 0.4 | 137 | 3.4 | 1.2 | 143 | 6.3 | 0.9 | 135 | 3.8 | 0.3 | 140 |
| 141 | Int 5 | na | 15 | 9 | 3.1 | 0.5 | 98 | 5.2 | 1.6 | 130 | 9.3 | 0.4 | 101 | 7.9 | 0.0 | 77 |
| 142 | 63 | 5′ | 15 | 10 | 2.0 | 0.4 | 140 | 6.6 | 4.2 | 103 | 30.4 | 0.1 | 41 | 19.0 | 0.4 | 34 |
| 143 | 63 | 3′ | 16 | 1 | 3.7 | 1.7 | 33 | 64.4 | 15.7 | 7 | 194.3 | 3.7 | 4 | 91.2 | 6.4 | 3 |
| 144 | 64 | 5′ | 16 | 2 | 3.8 | 1.2 | 24 | 69.0 | 11.2 | 6 | 165.0 | 8.2 | 7 | 74.3 | 4.0 | 5 |
| 145 | 64 | 3′ | 16 | 3 | 4.3 | 1.4 | 7 | 100.4 | 31.8 | 2 | 236.0 | 31.8 | 2 | 83.7 | 2.3 | 4 |
| 146 | 65 | all | 16 | 4 | 3.8 | 0.5 | 21 | 8.1 | 3.0 | 89 | 15.2 | 1.9 | 75 | 9.2 | 0.0 | 66 |
| 147 | 66 | 5′ | 16 | 5 | 3.7 | 0.2 | 27 | 6.3 | 2.8 | 108 | 7.7 | 0.7 | 119 | 5.5 | 0.2 | 115 |
| 148 | 66 | 3′ | 16 | 6 | 3.5 | 0.1 | 43 | 5.6 | 3.0 | 120 | 8.9 | 0.3 | 103 | 6.4 | 0.1 | 94 |
| 149 | 67 | 5′ | 16 | 7 | 3.4 | 0.4 | 53 | 7.0 | 4.7 | 95 | 19.0 | 0.2 | 60 | 7.3 | 0.2 | 83 |
| 150 | 67 | 3′ | 16 | 8 | 6.4 | 2.4 | 2 | 26.6 | 19.7 | 24 | 62.5 | 8.4 | 23 | 18.8 | 0.2 | 35 |
| 151 | 68 | 5′ | 16 | 9 | 3.2 | 0.4 | 88 | 5.5 | 3.3 | 125 | 7.5 | 0.4 | 121 | 7.0 | 0.3 | 87 |
| 152 | 68 | 3′ | 16 | 10 | 3.3 | 0.3 | 64 | 34.4 | 23.7 | 15 | 76.8 | 6.4 | 18 | 33.0 | 0.8 | 21 |
| Array controls | ||||||||||||||||
| Avg plasmid DNA | 2.3 | 0.2 | 132 | 3.3 | 0.3 | 144 | 4.1 | 0.2 | 144 | 4.7 | 0.1 | 126 | ||||
| Avg no DNA | 1.0 | 0.1 | 149 | 1.0 | 0.0 | 152 | 1.0 | 0.1 | 152 | 1.0 | 0.1 | 152 | ||||
| Avg actin | 0.1 | 0.0 | 155 | 0.6 | 0.0 | 153 | 0.2 | 0.0 | 155 | 0.2 | 0.0 | 155 | ||||
| Avg GAPdH | 0.1 | 0.0 | 156 | 0.0 | 0.0 | 156 | 0.0 | 0.0 | 156 | 0.1 | 0.0 | 156 | ||||
VZV open reading frame region [5′, 3′, or entire (all)] was PCR amplified and inserted into pGEM. Target DNA array coordinates (column and row) are given. rel exp, relative expression of the individual target region. std, standard deviation of mean relative expression. order, order (in decreasing magnitude) of the individual target relative expression. dpi, day postinfection.
RNA samples harvested at 1 day postinfection detected 21 (14%) of the total 147 VZV-specific array targets. Similarly, VZV-infected samples harvested at 2 days postinfection detected 136 (92%), samples harvested 3 days postinfection detected 143 (97%), and samples harvested 4 days postinfection detected 137 (93%) of the available VZV-specific array targets. Considering that 64 of the 68 predicted unique VZV ORFs contained multiple targets and that targets corresponding to ORF 14 were not present on the arrays, samples harvested 1 day postinfection detected 18 of the possible 67 VZV ORFs. Similarly, samples harvested 2 and 3 days postinfection detect 100% of the 67 VZV ORFs. Samples from day postinfection 4, which lacked detectable ORF 5 transcripts (clones 8 and 9), detected 66 of the possible 67 VZV ORFs.
A bias was detected when multiple targets were associated with a specific VZV gene, especially evident in the 1 day postinfection sample, 12 of 14 times towards the 3′ end compared to 2 of 14 times towards the 5′ end, most likely reflecting incomplete oligo(dT)-primed, first-strand cDNA synthesis resulting in underrepresentation of the 5′ start of the ORF.
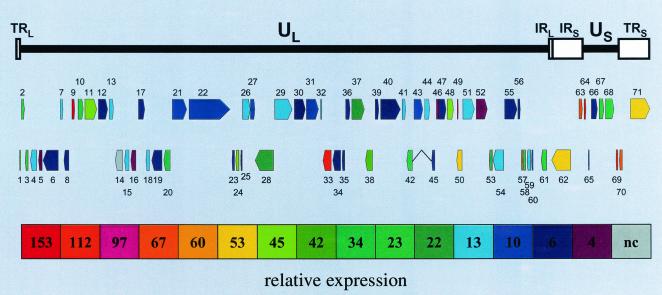
Table 3 summarizes the relative expression of VZV ORFs, given in decreasing order of abundance and as the average relative expression for both the 3′ and 5′ targets for each ORF from 2 to 4 days postinfection. The function assigned to the individual virus genes (21, 42) is also listed. Figure 3, . visually representing the transcription expression data in Table 3, shows that the four most abundant (relative expression ≈100 and greater) VZV transcripts detected during virus growth mapping to ORFs 9, 64, 33, and 49 are located throughout the virus genome.
TABLE 3.
Relative expression of VZV ORFs during lytic infection
| ORF | Relative expression | Gene function |
|---|---|---|
| 9 | 153.3 | Abundant tegument phosphoprotein |
| 64 | 121.4 | |
| 33 | 112.0 | Protease |
| 49 | 97.4 | |
| 63 | 67.7 | Tegument (transactivator/transrepressor) |
| 57 | 60.8 | |
| 62 | 53.7 | Transactivator, tegument protein |
| 50 | 53.3 | |
| 48 | 48.1 | |
| 11 | 47.9 | |
| 24 | 45.6 | |
| 38 | 42.9 | |
| 1 | 39.3 | Membrane protein |
| 58 | 38.5 | |
| 20 | 34.6 | |
| 2 | 31.8 | |
| 10 | 30.1 | Transactivator, tegument protein |
| 61 | 29.1 | Transactivator, transrepressor |
| 68 | 27.4 | Glycoprotein (gE) |
| 3 | 27.3 | |
| 42 | 23.6 | |
| 67 | 23.5 | Glycoprotein (gl) |
| 53 | 22.7 | |
| 28 | 22.6 | DNA polymerase |
| 37 | 22.4 | Glycoprotein (gH) |
| 44 | 21.9 | |
| 15 | 20.6 | |
| 54 | 20.0 | |
| 29 | 19.9 | Single-stranded DNA-binding protein |
| 51 | 17.9 | Origin-binding protein |
| 4 | 17.1 | Transactivator, tegument protein |
| 32 | 16.6 | Phosphoprotein |
| 41 | 15.3 | |
| 13 | 15.2 | Thymidylate synthetase |
| 7 | 15.2 | |
| 18 | 14.1 | Ribonucleotide reductase, small subunit |
| 59 | 13.4 | Uracil-DNA glycosylase |
| 26 | 13.3 | |
| 27 | 12.8 | |
| 22 | 12.5 | |
| 60 | 11.4 | Glycoprotein (gL) |
| 65 | 10.8 | |
| 31 | 10.7 | Glycoprotein (gB) |
| 43 | 10.7 | |
| 21 | 10.1 | |
| 36 | 9.9 | Thymidine kinase |
| 25 | 9.6 | |
| 6 | 9.3 | |
| 34 | 8.9 | |
| 39 | 8.6 | |
| 56 | 8.5 | |
| 47 | 8.5 | Protein kinase |
| 30 | 8.5 | |
| 19 | 8.4 | Ribonucleotide reductase, large subunit |
| 55 | 7.8 | |
| 17 | 7.8 | |
| 23 | 7.8 | |
| 8 | 7.5 | dUTPase |
| 66 | 6.7 | Protein kinase |
| 35 | 6.6 | |
| 40 | 6.6 | Major nucleocapsid protein |
| 45 | 6.3 | |
| 12 | 6.0 | |
| 52 | 5.9 | |
| 16 | 5.7 | |
| 46 | 5.7 | |
| 5 | 4.8 | Glycoprotein (gK) |
| 14 | nca | Glycoprotein (gC) |
ORF 14 was not cloned (nc).
FIG. 3.
Relative expression of VZV genes during lytic infection. The VZV genome consists of unique long (UL) and short (US) segments, each bounded by terminal and internal inverted repeated DNA sequences (TRL/IRL and IRs/TRs). Transcription of VZV genes was determined during lytic virus growth in cell culture (Fig. 2), and relative expression of virus genes was assessed (Table 3). The relative size, direction of transcription, and expression of each of the 71 predicted virus ORFs show that the most abundant transcripts (ORFs 9, 33, 49, 63/70, and 64/69) mapped across the virus genome.
Confirmation of array results (DNase digestion).
Because array cDNA probes generated by nick translation are also expected to radiolabel residual VZV DNA, PCR was used to demonstrate the removal of residual DNA from the RNA preparations. RNA extracted at 1 to 4 days postinfection was incubated with or without reverse transcriptase (RT), and the resulting material was amplified with actin or ORF 9 primers. Resolution of RT-dependent bands at 242 bp (actin) and 452 bp (VZV ORF 9) indicated that residual DNA was eliminated from the RNA preparations, and the band intensities reflected the relative expression levels determined by array analysis (data not shown).
Confirmation of array results (Northern blot and 3′-RACE).
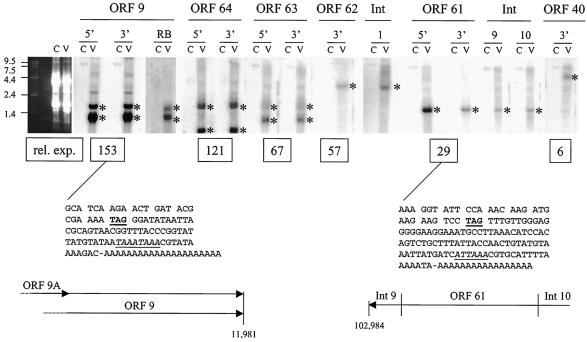
RNA from control and VZV-infected BSC-1 cells was resolved on Northern filters and hybridized with various ORF and intergenic VZV DNA probes (Fig. 4). . Both the ORF 9 probes detected two predominant transcripts at 1.68 and 1.25 kb; the ORF 64 probes detected transcripts at 1.67 and 1.01 kb; ORF 63 probes detected transcripts at 1.67 and 1.22 kb; the ORF 62 probe detected a single 3.35-kb transcript; ORF 61 probes detected a 1.54-kb transcript; and the ORF 40 probe detected a major transcript at 5.19 kb.
FIG. 4.
Northern blot analysis of VZV transcripts. Control (C) and VZV-infected (V) cell RNA was resolved on denaturing gels and hybridized to PCR-generated probes specific for the 5′ or 3′ end of ORFs 9, 64, 63, 62, 61. and 40, the antisense oligonucleotide 9RB (Table 1), or the PCR-generated intergenic (Int) fragments 1, 9, and 10. Asterisks indicate major bands detected in each lane. The relative expression (rel. exp.) of each ORF determined by array analysis (Table 3) is listed below the respective Northern blot. The DNA sequences of the ORF 9 and ORF 61 termini determined by 3′-RACE are also listed. The termination codon (TAG) is underlined, and the putative poly(A) addition signal is both italicized and underlined. The proposed ORF 9A, 9, and 61 mRNA structures are shown at the bottom of the figure.
To determine whether both the transcripts detected with the ORF 9 probes mapped to the same DNA strand, the Northern blot was probed with oligonucleotide 9FB (Table 1). This oligonucleotide maps antisense to the 3′ terminus of ORF 9 and detected both the 1.68- and 1.25-kb transcripts. The 3′ end of the ORF 9 transcripts was PCR amplified, and the DNA sequence located the poly(A) tract to a unique site 52 nucleotides distal to the ORF 9 termination codon, 12 nucleotides downstream from the predicted poly(A) addition signal. ORF 9A is a newly identified transcript mapping 107 nucleotides upstream of the ORF 9 initiation codon (38). Thus, Northern blot analysis with PCR-generated and strand-specific oligonucleotide probes, coupled with the 3′-RACE and DNA sequence results, are consistent with readthrough from the ORF 9A promoter to the ORF 9 termination/poly(A) addition signal, thereby generating two 3′-coterminal transcripts (1.69-kb ORF 9A and 1.25-kb ORF 9).
The major 1.01-kb ORF 64 transcript, detected with both 5′ and 3′ probes, corresponds to the predicted 539-bp ORF 64, indicating ≈470 nucleotides of untranslated RNA. The major 1.22-kb transcript detected by both ORF 63 probes corresponds to the predicted 833-nucleotide ORF 63, indicating 387 nucleotides of untranslated RNA. Comigrating bands at 1.67 kb were detected with all ORF 63 and ORF 64 probes, indicating that this transcript is the result of gene 64 readthrough to the gene 63 termination/poly(A) addition signal.
VZV DNA analysis indicates that gene 62 includes a 3,930-nucleotides ORF. Northern blots analyzed with ORF 62, 5′, and Int 1 probes detected a single VZV-specific transcript at ≈3.4 kb. This transcript most likely corresponds to the gene 62 transcript, with the Int-1 probe mapping to the 5′ untranslated region of the full-length transcript. The size discrepancy between the expected gene 62 transcript and that seen on the Northern blot rests in the inherent difficulty in determining the size of large transcripts on denaturing agarose gels.
A single 1.54-kb transcript was detected in VZV-infected BSC-1 RNA with probes generated from both ORF 61 termini, along with Int-9 and Int-10 DNA sequences. 3′-RACE used to amplify the end of VZV gene 61 from virus-infected cell RNA showed that the DNA sequence contains a poly(A) tract 99 nucleotides distal to the ORF 61 termination codon, 17 nucleotides distal to the predicted poly(A) addition signal. Primer extension has mapped the 5′ start of gene 61 transcription 69 nucleotides upstream of the initiation codon (31), and our present results map the 3′ terminus 99 nucleotides downstream of the termination codon. Int-9 and Int-10 probes detected the 5′ and 3′ untranslated regions, respectively, associated with the 1.54-kb ORF 61 transcripts.
ORF 40 encodes the major VZV nucleocapsid protein. This predicted 4,187-nucleotide ORF is contained within the ≈4.9-kb transcript detected with the 3′ ORF 40 probe. The low relative abundance of this transcript determined by array analysis was reflected in the Northern analysis, which required longer exposure times to detect ORF 40 transcripts. With the single exception of the ORF 61 transcript detected with the 5′ probe, the relative expression of each gene determined by array analysis was reflected in the band intensity on the Northern blots. The discrepancy noticed in ORF 61 between the relative abundance determined by array analysis and that inferred by Northern blot analysis is especially noteworthy (Fig. 4). For example, array analysis ranks ORF 61 expression below ORF 62 expression (29.1 and 53.7, respectively). However, Northern blot analysis suggests that ORF 61 expression is greater than ORF 62 expression. Some of the major factors which may interfere with both analyses are RNA stability, probe activity, and means of detection. It is possible that the ≈4-kb ORF 62 transcript is less stable than the ≈1.5-kb ORF 61 transcript, resulting in under representation of ORF 62 by Northern analysis. Alternatively, probe synthesis and hybridization kinetics for ORF 61, 5′ probe (G+C = 50.4%) may be more efficient that that of ORF 62, 3′ probe (G+C = 69.3%).
Confirmation of array results (RT-PCR analysis of intergenic regions).
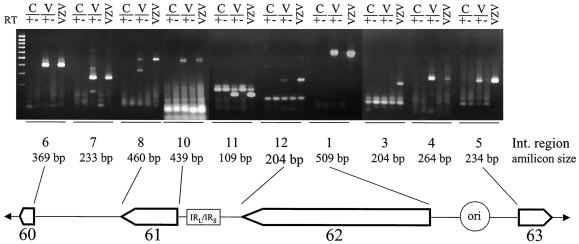
Array analysis indicated the presence of transcripts mapping to the three major intergenic regions of the VZV genome in virus-infected tissue culture cells. While Northern blot analysis mapped Int-1 and Int-9/10 probes to gene 62 and 61 transcripts, respectively, results for transcripts mapping to the remaining Int regions were not conclusive. Thus, PCR was used to confirm transcription within the intergenic regions (Fig. 5). . First-strand cDNA was synthesized from control and VZV-infected cell RNA either with or without reverse transcriptase, and PCR-amplified with primers mapping to the three major intergenic regions of the virus genome. Comparison of the PCR product obtained by amplification of VZV-infected cell cDNA with that obtained from the VZV DNA amplification showed that, with the exception of Int-3(F/R), all Int primer pairs tested yielded product transcription across these “silent” regions of the virus genome. The lack of product from control cell RNA and VZV-infected cell RNA demonstrated the specificity of amplification and removal of residual virus DNA the reactions.
FIG. 5.
RT-PCR analysis of VZV intergenic DNA. Control (C) and VZV-infected (V) cell RNA was incubated with (+) or without (−) reverse transcriptase and PCR amplified with selected intergenic (Int)-specific primers. Amplified products were compared with those after PCR amplification of VZV DNA. The predicted size of the amplification product (amplicon size) for each Int region primer set is listed. Int regions 6 to 8 map between ORFs 60 and 61; Int regions 10 to 12 map between ORFs 61 and 62 and contain the IRL/IRS junction; and Int regions 1 to 5 map between ORFs 62 and 63 and contain the VZV DNA origin replication (ori).
Int-8 primers yielded two PCR products in the amplification of VZV cDNA, possibly indicating posttranscriptional processing of the full-length message. Int-11 primers resulted in two bands when VZV DNA or cDNA was amplified. The presence of the top band in all samples is the result of nonspecific amplification; however, the ≈100-bp band, present only in the VZV cDNA and in VZV DNA, approximates the 109-bp theoretical size indicating virus transcription across this region.
DISCUSSION
This study is the first to use PCR-based arrays for the simultaneous detection and quantitation of VZV transcripts. The array targets contained PCR fragments from all originally predicted ORFs (10) with the single exception of ORF 14, which contains multiple R2 reiterations that render the clones unstable. ORF S/L is the first demonstrated spliced VZV transcript and maps to the leftward end of the virus genome (19). The 21-kDa cytoplasmic ORF S/L protein is expressed during lytic virus growth (in vitro and in vivo), and null mutations of this gene yield virus with altered cell adhesion characteristics. ORF S/L-specific targets were not placed on this first-generation VZV array.
The four most abundant (relative expression > ≈100) VZV transcripts mapped to ORFs 9, 64, 33, and 49. Northern blot and DNA sequencing indicated that the ORF 9 products detected by array analysis were composed of two 3′-coterminal overlapping transcripts. ORF 9A is located 367 bp upstream from the ORF 9 initiation codon. The ORF 9-associated bands detected by Northern analysis differed by 0.34 kb. Together, the data indicate that ORF 9 and 9A are coterminal, consistent with the ORF 9 doublet band detected by Ross et al. (38), and map the 3′ terminus of these transcripts. Even though quantitation of the Northern blots indicated a ≈4-fold higher abundance of ORF 9 than ORF 9A transcripts (3.8 ± 0.4, n = 5), ORF 9 expression was greater than that of ORF 64, the second most abundant transcript detected by array analysis.
Recently, OFR 9 protein has been shown to be an abundant tegument protein phosphorylated by ORF 47-encoded protein kinase (42). Phosphorylated ORF 9 protein associates with phosphorylated IE62, and the complex is liberated during virus uncoating. Along with stabilizing the virion tegument, ORF 9 protein may also help direct IE62 to appropriate promoter sites. The VZV ORF 9 homologue in herpes simplex virus type 1 is UL49, a posttranslationally modified tegument protein which associates with cellular chromatin (1, 22). Thus, the structural ORF 9 protein may also help orchestrate virus gene expression. ORF 9A encodes a nonessential 7-kDa membrane-associated protein which is associated with syncytium formation (38).
VZV ORF 64, which encodes a predicted 19.8-kDa protein, is located within the repeat region of the unique short segment of VZV DNA and is therefore also present as ORF 69. Site-directed gene knockout studies identified a phenotype only when both ORF 64 and 69 were deleted (41). In VZV containing both ORF 64 and 69 mutations, glycoprotein E (gE) expression and syncytium formation are increased. While VZV ORF 64 and 69 proteins are dispensable for both in vitro and in vivo virus growth, they may play a role in the establishment or maintenance of VZV latency (41). Herpes simplex virus type 1 US10, the homologue to VZV ORF 64, is a capsid/tegument-associated phosphoprotein that associates with the nuclear matrix of infected cells (46).
Our Northern blot analysis suggests low-level ORF 63 readthrough to the ORF 64 termination signal. Multiple termination signals may be more common than previously thought, particularly since ORFs 18 and 19 have been shown to be coterminal (unpublished data) in cDNA libraries constructed from VZV-infected cells (8). An additional example of readthrough may be seen in the relative abundance of ORFs 42 and 45, which encode putative exons from a single processed transcript. The relative expression of ORF 42 is greater than that of ORF 45. A possible explanation is that transcription originating from upstream genes such as ORFs 40 and 41 with the termination signal supplied by ORF 43 or 44 would result in greater expression of ORF 42 than of 45. Further studies are needed to confirm such a hypothesis. Meanwhile, even the prediction that both ORFs (45 and 42) are spliced has not been confirmed.
ORFs 33 and 33.5 encode highly immunogenic proteins associated with the assembly of virus nucleocapsids (12, 16, 27, 33). The ORF 33 transcript encodes the full-length precursor protein, which is posttranslationally cleaved to yield the assembly protein protease (ORF 33) and the structural protein substrate (ORF 33.5). Thus, the nested genes are in-frame, 3′-coterminal encoding proteins with identical C termini.
The protein encoded by VZV ORF 49 has not been identified. The herpes simplex virus type 1 homologue UL11 is a posttranslationally modified, Golgi-associated, viral tegument protein that functions in nucleocapsid envelopment and egress. UL11 protein may be involved in membrane structure, since constitutive cellular expression of UL11 blocks herpes simplex virus type 1 gD-mediated virus attachment (23-25, 36).
The most abundant VZV transcripts expressed during virus propagation are associated with membrane structure (ORF 9/9A, ORF 64, and ORF 49) or capsid assembly (ORF 33/33.5). Contrary to previous assumptions that ORF 40 transcripts, encoding the major VZV nucleocapsid protein, were highly expressed during lytic virus infection (3, 6-8, 20), our array analysis suggests and Northern blot analysis confirms that ORF 40 transcripts in productively infected cells are low abundance (23-fold less abundant than ORF 9/9A).
Unlike experiments involving high-titer cell-free virus or metabolic inhibitors of protein and DNA synthesis (5), this experiment was designed to determine the relative expression of VZV genes as virus infection progresses through the culture and was not designed to determine the traditional one-step growth kinetics. As such, it might be expected that the virus genes expressed are predominantly late (i.e., virus structural genes); however, immediate-early VZV genes are also detected. Of the 67 VZV genes detected by array analysis, immediate-early genes 63 and 62 are ranked 5 and 7, immediate-early genes 61 is ranked 18, and immediate-early gene 4 is ranked 31 (Table 3). Thus, propagation of the virus by cocultivation resulting in large multinucleated syncytia is accompanied by high levels of expression of membrane-associated proteins, immediate-early proteins 63 and 62, and the expression of numerous genes whose function has not been identified (for example, ORF 57).
Oligonucleotide-based arrays have been used to determine the time-course of herpes simplex virus type 1 transcription (43). Comparison of the relative abundance of herpes simplex virus type 1 versus VZV transcripts reveals striking differences in the pattern of gene expression. For example, only the homologue of VZV ORF 64 is included among the five most abundant herpes simplex virus type 1 genes transcribed at early times after infection. Transcription of this herpes simplex virus type 1 gene diminishes late in infection and is superseded by transcription of the gene encoding the herpes simplex virus major capsid protein. Both early and late in herpes simplex virus infection, transcription of the VZV ORF 9A/9 homologue approximates the mean herpes simplex virus transcriptional level. Just as DNA sequence analysis demonstrates genetic differences between the two human neurotropic alphaherpesviruses (28), transcriptional array analysis indicates how these genetic differences are reflected functionally.
A caveat in interpreting transcriptional data arises from the ability of array and Northern blot analysis to provide only snapshot of the cell at time of harvest. Thus, high abundance may result from low-level transcription of stable RNA, while low abundance may rest in rapid degradation of an actively transcribed gene. Our studies addressed relative abundance of VZV gene transcripts, not RNA stability. On the other hand, the global approach of array analysis, besides providing a temporal description of virus gene expression, enabled discovery of novel transcripts, such as those mapping to the three major intergenic regions. Northern blot analysis confirmed three of the Int-target results and demonstrated an association with untranslated regions outside ORFs 61 and 62. PCR evidence confirmed the array analysis by demonstrating the RT-dependent amplification of cDNA within the intergenic regions. Future research is focused on mapping these transcripts, but preliminary evidence suggests posttranscriptional processing of transcripts within these regions (unpublished data).
Global transcriptional analysis is potentially valuable in several areas. For example, wild-type and VZV gene knock-out constructs can be compared to determine how the virus compensates for the lack of specific genes. Comparison of virus gene transcription in various cell lines or in human biopsy and explanted tissue in SCID-hu mice (29) might elucidate cellular effects on virus gene expression. Combining VZV transcriptional array data with wild-type and specific gene knockout mutant virus with the global cellular transcription analysis afforded by cellular gene chip experiments (18) will demonstrate the cellular response to specific virus genes. Transcriptional array analysis of parental and vaccine OKA VZV along with the recently available complete virus genetic information (14) will help to determine the molecular effects of virus attenuation.
Various methods to obtain cell-free VZV, albeit at low titers (14, 15), together with methods of probe signal enhancement (37, 45) will allow synchronized virus infection and temporal classification of VZV genes, information required for a better understanding of the virus's growth cycle and especially important when comparing VZV to the prototype neurotropic alphaherpesvirus herpes simplex virus type 1. Herpes simplex virus type 1 infection is characterized by the ordered sequential expression of immediate-early, early, and late genes (43). In this report, at the earliest time examined (1 day postinfection), little transcription (cellular or virus) was detected. The differences in gene expression between VZV (this report) and herpes simplex virus type 1 (43), especially early in virus infection, most likely reflect the mode of infection (cocultivation and unsynchronized versus synchronized high-multiplicity infection). Nevertheless, even with unsynchronized infection, we detected some VZV-specific transcripts within 24 h after infection, followed by increasing numbers and abundance of VZV-specific transcripts corresponding to all putative kinetic classes during the first 3 days (Fig. 2).
Finally, application of high-throughput technology with global VZV transcriptional array analysis to the study of human ganglia will increase our understanding of virus latency to an unprecedented level. Whereas these studies were previously limited to PCR or in situ results for selected virus genes, the technology is now available to search the entire VZV genome for transcription in a single experiment. Thus, a most vexing and basic question in the field of VZV research, i.e., which VZV genes are transcribed in latently infected human ganglia, can now be addressed.
Acknowledgments
This work was supported in part by Public Health Service grants AG 06127 and NS 32623 from the National Institutes of Health.
Expert technical assistance was provided by Carrie Essman and Jeanne Wischer. We thank Ed Wagner for insightful comments, Robert Harding for bioinformatics assistance, and Aaron Cohrs for graphic design. We also thank Marina Hoffman for editorial help and Cathy Allen for manuscript preparation.
REFERENCES
- 1.Blaho, J. A., C. Mitchell, and B. Roizman. 1994. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47, and UL49 gene products. J. Biol. Chem. 269:17401-17410. [PubMed] [Google Scholar]
- 2.Bresnahan, W. A., and T. Shenk. 2000. A subset of viral transcripts packaged within human cytomegalovirus particles. Science 288:2373-2376. [DOI] [PubMed] [Google Scholar]
- 3.Brunell, P. A., L. C. Ren, J. I. Cohen, and S. E. Straus. 1999. Viral gene expression in rat trigeminal ganglia following neonatal infection with varicella-zoster virus. J. Med. Virol. 58:286-290. [DOI] [PubMed] [Google Scholar]
- 4.Cohen, J. I., and H. Nguyen. 1998. Varicella-zoster virus ORF61 deletion mutants replicate in cell culture, but a mutant with stop codons in ORF61 reverts to wild-type virus. Virology 246:306-316. [DOI] [PubMed] [Google Scholar]
- 5.Cohrs, R., and H. Rouhandeh. 1987. Analysis of Herpesvirus sylvilagus-induced proteins in infected rabbit kidney cells by two-dimensional gel electrophoresis. Intervirology 28:206-220. [DOI] [PubMed] [Google Scholar]
- 6.Cohrs, R. J., K. Srock, M. B. Barbour, G. Owens, R. Mahalingam, M. E. Devlin, M. Wellish, and D. H. Gilden. 1994. Varicella-zoster virus (VZV) transcription during latency in human ganglia: construction of a cDNA library from latently infected human trigeminal ganglia and detection of a VZV transcript. J. Virol. 68:7900-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohrs, R. J., M. B. Barbour, R. Mahalingam, M. Wellish, and D. H. Gilden. 1995. Varicella-zoster virus (VZV) transcription during latency in human ganglia: prevalence of VZV gene 21 transcripts in latently infected human ganglia. J. Virol. 69:2674-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohrs, R. J., M. Barbour, and D. H. Gilden. 1996. Varicella-zoster virus (VZV) transcription during latency in human ganglia: detection of transcripts mapping to genes 21, 29, 62, and 63 in a cDNA library enriched for VZV RNA. J. Virol. 70:2789-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, E., S. Reddy, I. Iofin, and J. I. Cohen. 1998. Varicella-zoster virus ORF57, unlike its pseudorabies virus UL3.5 homolog, is dispensable for viral replication in cell culture. Virology 250:205-209. [DOI] [PubMed] [Google Scholar]
- 10.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 11.Fourney, R. M., J. Mlyakoshi, R. S. Day III, and M. C. Paterson. 1988. Northern blotting: efficient RNA staining and transfer. Focus 10:5-7. [Google Scholar]
- 12.Garcia-Valcarcel, M., W. J. Fowler, D. R. Harper, D. J. Jeffries, and G. T. Layton. 1997. Cloning, expression, and immunogenicity of the assembly protein of varicella-zoster virus and detection of the products of open reading frame 33. J. Med. Virol. 53:332-339. [PubMed] [Google Scholar]
- 13.Gilden, D. H., Y. Shtram, A. Friedmann, M. Wellish, M. Devlin, A. Cohen, N. Fraser, and Y. Becker. 1982. Extraction of cell-associated varicella-zoster virus DNA with Triton X-100-NaCl. J. Virol. Methods 4:263-275. [DOI] [PubMed] [Google Scholar]
- 14.Gomi, Y., H. Sunamachi, Y. Mori, K. Nagaike, M. Takahashi, and K. Yamanishi. 2002. Comparison of the complete DNA sequences of the Oka varicella vaccine and its parental virus. J. Virol. 76:11447-11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grose, C., D. M. Perrotta, P. A. Brunell, and G. C. Smith. 1979. Cell-free varicella-zoster virus in cultured human melanoma cells. J. Gen. Virol. 43:15-27. [DOI] [PubMed] [Google Scholar]
- 16.Harper, D. R., E. A. Sanders, and M. A. Ashcroft. 1995. Varicella-zoster virus assembly protein p32/p36 is present in DNA-containing as well as immature capsids. J. Med. Virol. 46:144-147. [DOI] [PubMed] [Google Scholar]
- 17.Higaki, S., B. M. Gebhardt, W. J. Lukiw, H. W. Thompson, and J. M. Hill. 2002. Effect of immunosuppression on gene expression in the herpes simplex virus type 1 latently infected mouse trigeminal ganglion. Investig. Ophthalmol. Vis. Sci. 43:1862-1869. [PubMed] [Google Scholar]
- 18.Jones, J. O., and A. M. Arvin. 2003. Microarray analysis of host cell gene transcription in response to varicella-zoster virus infection of human T cells and fibroblasts in vitro and SCIDhu skin xenografts in vivo. J. Virol. 77:1268-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kemble, G. W., P. Annunziato, O. Lungu, R. E. Winter, T. A. Cha, S. J. Silverstein, and R. R. Spaete. 2000. Open reading frame S/L of varicella-zoster virus encodes a cytoplasmic protein expressed in infected cells. J. Virol. 74:11311-11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennedy, P. G., E. Grinfeld, and J. W. Gow. 1999. Latent Varicella-zoster virus in human dorsal root ganglia. Virology 258:451-454. [DOI] [PubMed] [Google Scholar]
- 21.Kinchington, P. R., and J. I. Cohen. 2000. Viral proteins, p. 74-104. In A. M. Arvin and A. A. Gershon (ed.), Varicella-zoster virus. Cambridge University Press, Cambridge, United Kingdom.
- 22.Leslie, J., F. J. Rixon, and J. McLauchlan. 1996. Overexpression of the herpes simplex virus type 1 tegument protein VP22 increases its incorporation into virus particles. Virology 220:60-68. [DOI] [PubMed] [Google Scholar]
- 23.Loomis, J. S., J. B. Bowzard, R. J. Courtney, and J. W. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacLean, C. A., B. Clark, and D. J. McGeoch. 1989. Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J. Gen. Virol. 70:3147-3157. [DOI] [PubMed] [Google Scholar]
- 25.MacLean, C. A., A. Dolan, F. E. Jamieson, and D. J. McGeoch. 1992. The myristylated virion proteins of herpes simplex virus type 1: investigation of their role in the virus life cycle. J. Gen. Virol. 73:539-547. [DOI] [PubMed] [Google Scholar]
- 26.Maguire, H. F., and R. W. Hyman. 1986. Polyadenylated, cytoplasmic transcripts of varicella-zoster virus. Intervirology 26:181-191. [DOI] [PubMed] [Google Scholar]
- 27.McMillan, D. J., J. Kay, and J. S. Mills. 1997. Characterization of the proteinase specified by varicella-zoster virus gene 33. J. Gen. Virol. 78:2153-2157. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell, B. M., D. C. Bloom, R. J. Cohrs, D. H. Gilden, and P. G. Kennedy. 2003. Herpes simplex virus type 1 and varicella-zoster virus latency in ganglia. J. Neurovirol. 9:194-204. [DOI] [PubMed] [Google Scholar]
- 29.Moffat, J. F., M. D. Stein, H. Kaneshima, and A. M. Arvin. 1995. Tropism of varicella-zoster virus for human CD4+ and CD8+ T lymphocytes and epidermal cells in SCID-hu mice. J. Virol. 69:5236-5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moffat, J. F., L. Zerboni, M. H. Sommer, T. C. Heineman, J. I. Cohen, H. Kaneshima, and A. M. Arvin. 1998. The ORF47 and ORF66 putative protein kinases of varicella-zoster virus determine tropism for human T cells and skin in the SCID-hu mouse. Proc. Natl. Acad. Sci. 95:11969-11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagpal, S., and J. M. Ostrove. 1991. Characterization of a potent varicella-zoster virus-encoded trans-repressor. J. Virol. 65:5289-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ostrove, J. M., W. Reinhold, C. M. Fan, S. Zorn, J. Hay, and S. E. Straus. 1985. Transcription mapping of the varicella-zoster virus genome. J. Virol. 56:600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Preston, V. G., J. Kennard, F. J. Rixon, A. J. Logan, R. W. Mansfield, and I. M. McDougall. 1997. Efficient herpes simplex virus type 1 (herpes simplex virus type 1) capsid formation directed by the varicella-zoster virus scaffolding protein requires the carboxy-terminal sequences from the herpes simplex virus type 1 homologue. J. Gen. Virol. 78:1633-1646. [DOI] [PubMed] [Google Scholar]
- 34.Reddy, S. M., M. Williams, and J. I. Cohen. 1998. Expression of a uracil DNA glycosylase (UNG) inhibitor in mammalian cells: varicella-zoster virus can replicate in vitro in the absence of detectable UNG activity. Virology 251:393-401. [DOI] [PubMed] [Google Scholar]
- 35.Reinhold, W. C., S. E. Straus, and J. M. Ostrove. 1988. Directionality and further mapping of varicella zoster virus transcripts. Virus Res. 9:249-261. [DOI] [PubMed] [Google Scholar]
- 36.Roller, R. J., and B. Roizman. 1994. A herpes simplex virus 1 US11-expressing cell line is resistant to herpes simplex virus infection at a step in viral entry mediated by glycoprotein D. J. Virol. 68:2830-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross-Macdonald, P., P. S. Coelho, T. Roemer, S. Agarwal, A. Kumar, R. Jansen, K. H. Cheung, A. Sheehan, D. Symoniatis, L. Umansky, M. Heidtman, F. K. Nelson, H. Iwasaki, K. Hager, M. Gerstein, P. Miller, G. S. Roeder, and M. Snyder. 1999. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402:413-418. [DOI] [PubMed] [Google Scholar]
- 38.Ross, J., M. Williams, and J. I. Cohen. 1997. Disruption of the varicella-zoster virus dUTPase and the adjacent ORF9A gene results in impaired growth and reduced syncytia formation in vitro. Virology 234:186-195. [DOI] [PubMed] [Google Scholar]
- 39.Sciortino, M. T., M. Suzuki, B. Taddeo, and B. Roizman. 2001. RNAs extracted from herpes simplex virus 1 virions: apparent selectivity of viral but not cellular RNAs packaged in virions. J. Virol. 75:8105-8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shenkar, R., J. P. Elliott, K. Diener, J. Gault, L. J. Hu, R. J. Cohrs, T. Phang, L. Hunter, R. E. Breeze, and I. A. Awad. 2003. Differential gene expression in human cerebrovascular malformations. Neurosurgery 52:465-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sommer, M. H., E. Zagha, O. K. Serrano, C. C. Ku, L. Zerboni, A. Baiker, R. Santos, M. Spengler, J. Lynch, C. Grose, W. Ruyechan, J. Hay, and A. M. Arvin. 2001. Mutational analysis of the repeated open reading frames ORFs 63 and 70 and ORFs 64 and 69 of varicella-zoster virus. J. Virol. 75:8224-8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spengler, M., N. Niesen, C. Grose, W. T. Ruyechan, and J. Hay. 2001. Interactions among structural proteins of varicella zoster virus. Arch. Virol. (Suppl.) 2001:71-79. [DOI] [PubMed]
- 43.Stingley, S. W., J. J. Ramirez, S. A. Aguilar, K. Simmen, R. M. Sandri-Goldin, P. Ghazal, and E. K. Wagner. 2000. Global analysis of herpes simplex virus type 1 transcription with an oligonucleotide-based DNA microarray. J. Virol. 74:9916-9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Straus, S. E., H. S. Aulakh, W. T. Ruyechan, J. Hay, T. A. Casey, G. F. Vande Woude, J. Owens, and H. A. Smith. 1981. Structure of varicella-zoster virus DNA. J. Virol. 40:516-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Gelder, R. N., M. E. von Zastrow, A. Yool, W. C. Dement, J. D. Barchas, and J. H. Eberwine. 1990. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc. Natl. Acad. Sci. 87:1663-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamada, H., T. Daikoku, Y. Yamashita, Y. M. Jiang, T. Tsurumi, and Y. Nishiyama. 1997. The product of the US10 gene of herpes simplex virus type 1 is a capsid/tegument-associated phosphoprotein which copurifies with the nuclear matrix. J. Gen. Virol. 78:2923-2931. [DOI] [PubMed] [Google Scholar]