Abstract
Vaccinia virus assembles two distinct lipoprotein membranes. The primary membrane contains nonglycosylated proteins, appears as crescents in the cytoplasm, and delimits immature and mature intracellular virions. The secondary or wrapping membrane contains glycoproteins, is derived from virus-modified trans-Golgi or endosomal cisternae, forms a loose coat around some intracellular mature virions, and becomes the envelope of extracellular virions. Although the mode of formation of the wrapping membrane is partially understood, we know less about the primary membrane. Recent reports posit that the primary membrane originates from the endoplasmic reticulum-Golgi intermediate compartment (ERGIC). According to this model, viral primary membrane proteins are cotranslationally inserted into the ER and accumulate in the ERGIC. To test the ERGIC model, we employed Sar1H79G, a dominant negative form of the Sar1 protein, which is an essential component of coatomer protein II (COPII)-mediated cargo transport from the ER to the ERGIC and other post-ER compartments. Overexpression of Sar1H79G by transfection or by a novel recombinant vaccinia virus with an inducible Sar1H79G gene resulted in retention of ERGIC 53 in the ER but did not interfere with localization of viral primary membrane proteins in factory regions or with formation of viral crescent membranes and infectious intracellular mature virions. Wrapping of intracellular mature virions and formation of extracellular virions did not occur, however, because some proteins that are essential for the secondary membrane were retained in the ER as a consequence of Sar1H79G overexpression. Our data argue against an essential role of COPII-mediated cargo transport and the ERGIC in the formation of the viral primary membrane. Instead, viral membranes may be derived directly from the ER or by a novel mechanism.
Vaccinia virus, the best-studied member of the family Poxviridae, has a double-stranded DNA genome of approximately 190 kbp that encodes nearly 200 polypeptides (22). Poxviruses differ from most other DNA viruses in that genome replication and virion assembly occur entirely in the cytoplasm, within discrete areas known as viral factories. Vaccinia virus morphogenesis occurs through a series of intermediate structures that have been visualized by electron microscopy (35). The first discernible structures appear as rigid, crescent-shaped membranes that evolve into spherical immature virions (IV) that enclose a nucleoprotein mass. A series of dramatic maturational events transform the IV into infectious, dense, brick-shaped intracellular mature virions (IMV). A double membrane derived from trans-Golgi or endosomal cisternae wraps some IMV, thus forming intracellular enveloped virions (IEV). The IEV are transported on microtubules to the cell periphery, where their outermost membrane fuses with the plasma membrane, leaving extracellular cell-associated virions (CEV) attached to the cell surface as well as free extracellular enveloped virions in the medium (24, 33).
To avoid ambiguity in terminology, we refer to the viral membrane that forms crescents and delimits IV and IMV as a primary membrane, to the viral membrane that delimits IEV as a secondary or wrapping membrane, and to the outer membrane of extracellular viral forms as an envelope. The origin of the primary viral membrane is not yet understood. Dales and coworkers (7) considered a de novo origin of the crescent membrane, whereas others (17, 26, 28, 34) thought this conclusion was unprecedented and suggested that the first viral membrane was derived from the ER-Golgi intermediate compartment (ERGIC). The latter suggestion was based mainly on evidence that viral membrane proteins, including the ones encoded by the A17L, A14L, and A13L genes, are cotranslationally inserted into the ER and later transported to and retained in the ERGIC, and it was also based on evidence that ERGIC markers are in membranes that are apparently continuous with the viral crescents. This topic is controversial, however, as physical connections between viral crescents and cellular membranes were not found when serial sections of infected cells were examined previously (13).
The ERGIC comprises pre-Golgi vesicular-tubular clusters that contain cargo which exits the ER via coatomer protein II (COPII) coat machinery, which includes the small GTPase Sar1 and the Sec23/24 and Sec13/31 complexes (3, 4, 12, 18, 20). A dominant negative mutated form of Sar1, specifically blocks protein exit from the ER, thus demonstrating that this pathway is essential for cargo transport to the ERGIC and other post-ER compartments (2, 18, 25, 32, 36). In the present study, we used a dominant negative form of Sar1 to determine the requirement for ER-to-ERGIC transport on the formation of the viral primary and wrapping membranes. Our results confirm the mandatory trafficking of some proteins that constitute the wrapping membrane through the secretory pathway, but they argue against an essential role of COPII-mediated cargo transport and the ERGIC in formation of the primary membrane.
MATERIALS AND METHODS
Cells and viruses.
BS-C-1, RK13, and HeLa cells were grown and maintained in Eagle's modified essential medium (EMEM) or Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). The following were all propagated in HeLa cells and titrated on BS-C-1 cells: recombinant vaccinia virus vA9L-HA, which expresses the influenza virus hemagglutinin (HA) epitope-tagged A9L protein (48); vB5R-GFP, which expresses enhanced green fluorescent protein (GFP)-tagged B5R protein (43); vT7LacOI, which expresses bacteriophage T7 RNA polymerase (44); and vGFP-Sar1H79Gi, which expresses a GFP-Sar1H79G fusion protein under an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible T7 promoter. Unless otherwise mentioned, cells were incubated at 37°C in a 5% CO2 atmosphere.
Plasmids.
Plasmid pHA-Sar1H79G, carrying an N-terminal HA-tagged Sar1H79G gene, was generously provided by Jennifer Lippincott-Schwartz, and pGFP-Sar1H79G was described previously (14). To construct pVGFP-Sar1H79G, the GFP-Sar1H79G coding sequence was amplified by PCR from pGFP-Sar1H79G and ligated to pVOTE.1 (44).
Transfection and infection. HeLa cells were grown on glass coverslips until they reached 80 to 90% confluence. One to two micrograms of Lipofectamine 2000 (Invitrogen) and 0.5 to 1 μg of DNA were diluted separately in Opti-MEM I medium (Invitrogen), mixed, incubated at room temperature for 20 min, and added to the cells for 4 to 5 h at 37°C. The Lipofectamine-DNA complex was replaced with DMEM supplemented with 10% FBS, and the incubation continued for a total of 24 h.
Virus diluted in DMEM or EMEM supplemented with 2.5% FBS was added to the cell monolayers on coverslips or in wells. After 1 h, the virus inoculum was replaced with fresh medium containing 2.5% FBS and incubated for a further 8 to 24 h.
Construction of vGFP-Sar1H79Gi.
HeLa cells were infected with recombinant vaccinia virus vT7LacOI at a multiplicity of 0.1 PFU per cell for 2 h. Cells were then transfected with plasmid pVGFP-Sar1H79G by using Lipofectamine 2000. After 5 h, the Lipofectamine-containing suspension was replaced with DMEM, and the incubation was continued for 3 days. Cells were harvested and lysed, and the diluted lysates were used to infect BS-C-1 monolayers that had been pretreated and maintained in medium containing mycophenolic acid, xanthine, and hypoxanthine to select for virus expressing xanthine-guanine phosphoribosyltransferase (10). After three rounds of plaque purification, the viral DNA was screened for the presence of the inserted DNA by PCR. The recombinant virus was propagated and titrated as described previously (9).
One-step growth curve.
BS-C-1 cells were infected with 5 PFU of virus per cell in the absence or presence of 100 μM IPTG for 1 h. The virus inoculum was removed, and the cells were washed twice with fresh medium, overlaid with fresh medium with or without 100 μM IPTG, and returned to the incubator. At intervals, the medium was collected and the cells were harvested. The medium was cleared of cell debris by centrifugation at 16,000 × g for 30 s, and the virus was titrated on BS-C-1 cells in the absence of IPTG. Infected cells were washed once with fresh medium, lysed, and titrated as described above.
Antibodies.
Rabbit polyclonal anti-A36R antibody (Ab) recognizes a peptide sequence at the C terminus of the A36R protein (47), and anti-A17LN (5) and anti-A17LC (46) recognize N- and C-terminal peptides of the A17L protein, respectively. Mouse anti-A33R and anti-L1R monoclonal Abs (MAbs) were gifts of A. Schmaljohn; rat MAb 192C to the B5R protein and polyclonal serum against F13L protein were received from G. Hiller; and mouse MAb G1/93 to human ERGIC 53 protein was from H. P. Hauri and provided by J. Yewdell. Mouse anti-GFP MAb, mouse anti-HA.11 MAb, and rabbit polyclonal Ab to the HA epitope were purchased from Covance. Rhodamine red-conjugated and fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G (IgG), tetramethyl rhodamine isothiocyanate-conjugated anti-rat IgG, and anti-rabbit IgG were purchased from Jackson ImmunoResearch Laboratories. Horseradish peroxidase-conjugated anti-mouse IgG and anti-rabbit IgG were from ICNBiomedicals and Amersham, respectively.
Western blotting, metabolic labeling, and immunoprecipitation.
Western blotting was carried out essentially as described previously (14). For metabolic labeling, infected cells were incubated for 30 min in methionine- and cysteine-free EMEM without serum and then pulse-labeled with 100 μCi of [35S]methionine and [35S]cysteine per ml of the above medium for 5 to 10 min. The labeling medium was removed, and the cells were washed and harvested in cold phosphate-buffered saline or were chased for timed intervals in complete DMEM supplemented with 2 mM cysteine and 0.2 mM methionine. At each time point, cells were harvested and lysed in ice-cold radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 0.5% sodium deoxycholate) supplemented with protease inhibitor cocktail (Sigma). The lysates were incubated on ice for 10 min and centrifuged at 20,000 × g for 10 min at 4°C, and the supernatants were incubated overnight at 4°C with Ab. On the next day, 20 μl of protein G-Sepharose (Pierce) was added to each lysate and incubated as described above for 2 h. Sepharose beads were pelleted at 20,000 × g for 30 s at 4°C, washed four times with radioimmunoprecipitation assay buffer, and finally washed with phosphate-buffered saline. SDS sample buffer was added to the beads, and proteins were resolved by electrophoresis in SDS gels of 12% or 4 to 20% polyacrylamide and then visualized by autoradiography.
For endoglycosidase H (endo H) digestion of metabolically labeled protein, 20 μl of denaturing solution (2×) supplied with endo H (New England Biolabs) was added to Sepharose beads after the final wash. Samples were kept at 100°C for 10 min, incubated on ice for 2 min, and then centrifuged at 16,000 × g for 5 min at room temperature. Supernatants were collected, and 1/10 volume of supplied G5 buffer (10×) and 1,000 U of endo H were added. The samples were incubated at 37°C for 2 h, and the proteins were resolved by electrophoresis on a 12% polyacrylamide gel and detected by autoradiography.
Microscopy.
Confocal and electron microscopy were carried out as previously described (15).
RESULTS
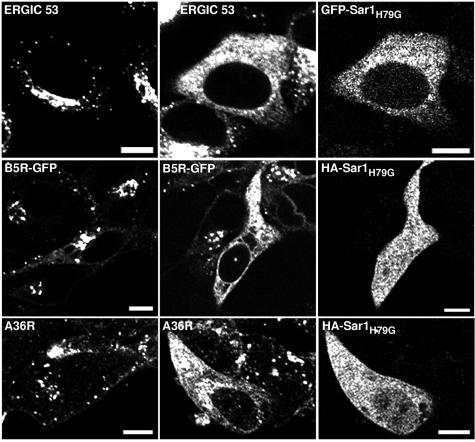
Effect of the cellular expression of Sar1H79G on the localization of viral membrane glycoproteins. Sar1H79G contains glycine substituted for the histidine at amino acid 79 of the Sar1 GTPase, localizes in the ER, and acts as a trans-dominant negative inhibitor of cargo transport from the ER to the ERGIC and other post-ER compartments (25, 36). N-terminal HA epitope-tagged and GFP-fused forms of Sar1H79G, with equivalent inhibitory activities, were used interchangeably in the present study. We examined the effect of Sar1H79G expression on the intracellular distribution of ERGIC 53, a type I membrane protein that cycles between the ER and the ERGIC and serves as a marker for the latter (11). As expected, ERGIC 53 localized primarily in the perinuclear region of mock-transfected HeLa cells (Fig. 1, row 1, column 1) but exhibited a reticular pattern and overlapped with GFP-Sar1H79G in the ER of transfected cells (Fig. 1, row 1, columns 2 and 3). An ER pattern was also noted for proteins that normally traffic through the Golgi network, including the vesicular stomatitis virus G protein and the vaccinia virus B5R and A36R proteins, when expression plasmids were cotransfected with pHA-Sar1H79G (reference 14 and data not shown). Thus, Sar1H79G expression prevented the localization of proteins in post-ER compartments.
FIG. 1.
Effect of Sar1H79G expression on the intracellular distribution of cellular ERGIC 53 and vaccinia virus IEV proteins. (Row 1) HeLa cells were either not transfected (column 1) or transfected with pGFP-Sar1H79G (columns 2 and 3) and incubated for 24 h. Cells were fixed, permeabilized, stained with an anti-ERGIC 53 MAb followed by treatment with Alexa 594-conjugated anti-mouse IgG, and viewed by confocal microscopy. (Row 2) HeLa cells were mock transfected (column 1) or transfected (columns 2 and 3) with pHA-Sar1H79G and infected 24 h later with vB5R-GFP (columns 1 to 3). At 24 h after infection, cells were fixed, permeabilized, and stained with anti-HA mouse MAb followed by rhodamine red-conjugated anti-mouse IgG (columns 2 and 3). (Row 3) HeLa cells were mock transfected (column 1) or transfected (columns 2 and 3) with pHA-Sar1H79G and infected 24 h later with vaccinia virus strain WR (columns 1 to 3). At 24 h after infection, cells were treated as described above and stained with anti-A36R rabbit Ab followed by tetramethyl rhodamine isothiocyanate-conjugated anti-rabbit IgG (column 1) or were stained with anti-A36R rabbit Ab and anti-HA mouse MAb followed by tetramethyl rhodamine isothiocyanate-conjugated anti-rabbit IgG and fluorescein isothiocyanate-conjugated anti-mouse IgG (columns 2 and 3). Bars, 10 μm.
In the next series of experiments, HeLa cells were transfected with pHA-Sar1H79G and were infected 24 h later with vaccinia virus strain WR or recombinant vaccinia virus vB5R-GFP, which expresses a functional B5R-GFP fusion protein that colocalizes with Golgi membrane markers (42, 43). In mock-transfected cells, B5R (Fig. 1, row 2, column 1) and A36R (Fig. 1, row 3, column 1) localized in the juxtanuclear Golgi region as well as in cytoplasmic punctate structures representing vesicles and IEV. In pHA-Sar1H79G-transfected cells, however, B5R (Fig. 1, row 2, columns 2 and 3) and A36R (Fig. 1, row 3, columns 2 and 3) had an ER distribution similar to that of Sar1H79G. Neighboring nontransfected cells, which were negative for Sar1H79G expression, retained the juxtanuclear distribution of B5R and A36R. Similar patterns were found in essentially all cells examined. These experiments demonstrated an inhibitory effect of Sar1H79G on the trafficking of the B5R and A36R IEV proteins.
Next, we determined the effect of Sar1H79G on the distribution of three viral primary membrane proteins: A17L, A9L, and L1R. The A17L protein was of particular interest, as its association with the ER and ERGIC of infected cells was reported previously by two groups (17, 26, 28). We mock transfected or transfected cells with Sar1H79G expression plasmids and infected the cells 24 h later with strain WR or a recombinant vaccinia virus vA9-L-HA, which expresses an epitope-tagged A9L protein (48). The cells were fixed, permeabilized, and stained with antibodies to the viral proteins and with 4′,6′-diamidino-2-phenylindole (DAPI), which stains DNA in the nucleus and cytoplasmic juxtanuclear viral factories. In the absence of Sar1H79G expression, the A9L-HA, L1R, and A17L membrane proteins localized to viral factories and to other punctate structures that could represent virions (Fig. 2, rows 1 and 2). Importantly, expression of Sar1H79G had no discernible effect on the intracellular distribution of the three primary membrane proteins (Fig. 2, rows 3 to 5). These results, which were reproduced in essentially all cells examined, suggested that viral primary membrane proteins, unlike the A36R and B5R secondary membrane proteins, are not transported from the ER through the COPII-mediated pathway, which was inhibited by the dominant negative mutated form of Sar1.
FIG.2.
Effect of Sar1H79G expression on the intracellular distribution of IMV proteins. (Rows 1 and 2) HeLa cells were infected with vA9L-HA (column 1) or WR (columns 2 and 3) and after 24 h were fixed, permeabilized, and stained with anti-HA mouse MAb (column 1), anti-L1R mouse MAb (column 2), or anti-A17LC rabbit Ab (column 3). (Rows 3 to 5) Cells were transfected with pGFP-Sar1H79G (columns 1 and 2) or pHA-Sar1H79G (column 3) and 24 h later were infected as described for rows 1 and 2. Cells were treated as described for rows 1 and 2 except that the cells in column 3 were also stained with anti-HA mouse MAb. The secondary antibodies were either rhodamine red-conjugated anti-mouse IgG or tetramethyl rhodamine isothiocyanate-conjugated anti-rabbit IgG. All cells were counterstained with the DNA binding dye DAPI. Arrows show examples of proteins colocalizing with viral DNA factories. Red fluorescence, viral proteins; blue fluorescence, DNA; green fluorescence, Sar1H79G. Bars, 10 μm.
Effect of the cellular expression of Sar1H79G on the formation of infectious virions.
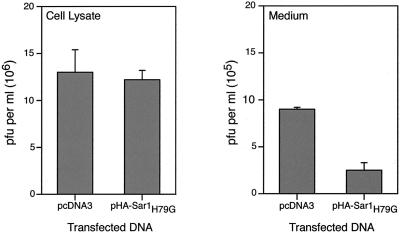
The experiments described above suggested that Sar1H79G might inhibit the production of extracellular enveloped virions but not that of IMV. To investigate this, we employed vaccinia virus strain IHD-J, which releases much more extracellular virus than the WR strain that was used in the previous experiments. After transfecting the cells with either the empty plasmid pcDNA3 or pHA-Sar1H79G for 24 h, the cells were infected with vaccinia virus strain IHD-J. After another 24 h, the medium and cells were collected separately and the infectious virus in each fraction was quantified by plaque assay. As shown in Fig. 3, Sar1H79G did not appreciably alter the amount of cell-associated infectious virus, although it decreased the infectious virus in the medium by more than threefold, despite the fact that not all cells were transfected and expressed the mutated Sar1 protein. Data showing the effects of Sar1H79G on IMV and IEV formation will be presented later.
FIG. 3.
Effect of Sar1H79G on the yields of cell-associated and released infectious virus. HeLa cells were transfected with pcDNA3 or pHA-Sar1H79G and infected 24 h later with vaccinia virus strain IHD-J. After a further 24 h, the medium was collected and cleared by high-speed centrifugation, and the cells were harvested in fresh medium, washed, and lysed. Infectious particles were determined by plaque assay on BS-C-1 cells. Data presented are the averages, with standard deviations, of three different plaque counts of two independent experiments.
Construction of a recombinant vaccinia virus expressing an IPTG-inducible GFP-Sar1H79G fusion protein.
We were constrained by the transfection protocol used in the initial experiments and therefore constructed a recombinant vaccinia virus with an inducible Sar1H79G gene in order to confirm and extrapolate from the above results. We used the stringently regulated VOTE system, in which the bacteriophage T7 RNA polymerase gene and E. coli lac repressor are integrated into the vaccinia virus genome (44). One lac operator regulates the expression of the T7 RNA polymerase, and another regulates the T7 promoter adjacent to the gene of interest—in this case, Sar1H79G fused at its N terminus to the GFP coding sequence (Fig. 4A). The recombinant virus, called vGFP-Sar1H79Gi, was isolated by using mycophenolic acid selection, plaque purified, analyzed by PCR, and then propagated in HeLa cells. All steps were carried out in the absence of IPTG to prevent the induction of Sar1H79G. Induction of the 48-kDa GFP-Sar1H79G was demonstrated by Western blotting that was carried out with an anti-GFP MAb (Fig. 4B). No expression was detected in the absence of inducer. The time required for Sar1H79G synthesis was compared with that for the A17L membrane protein; both were detected at 8 h after infection (data not shown). The recombinant virus made normal-sized or tiny plaques in the absence or presence of inducer, respectively (Fig. 4C). In addition, the small plaques made in the presence of inducer exhibited green fluorescence, whereas the large plaques made in the absence of inducer did not (Fig. 4D).
FIG. 4.
Characterization of a recombinant vaccinia virus expressing an inducible GFP-Sar1H79G fusion protein. (A) Diagram of portions of recombinant vaccinia virus vGFP-Sar1H79Gi. Abbreviations: Tk, thymidine kinase gene; T7 pol, bacteriophage T7 polymerase gene; lacO, E. coli lac operator; PL, vaccinia virus P11 late promoter; PE/L, vaccinia virus P7.5 early/late promoter; lacI, E. coli lac repressor gene; HA, vaccinia virus hemagglutinin gene; gpt, E. coli guanine phosphotransferase gene; SLO, stem loop lacO; EMC, encephalomyocarditis virus untranslated leader sequence; GFP-Sar1H79G, ORF encoding mutated GFP-Sar1H79G fusion protein; TT, transcription termination sequences. (B) Immunoblot showing inducible expression of GFP-Sar1H79G. HeLa cells were infected with vGFP-Sar1H79Gi in the absence and presence of 100 μM IPTG for 20 h. Proteins from the cell lysates were resolved by SDS-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and detected with anti-GFP mouse MAb. (C) Effect of IPTG on plaque size of vGFP-Sar1H79Gi. BS-C-1 cells were infected with vGFP-Sar1H79Gi and overlaid with medium containing methylcellulose and 0 or 100 μM IPTG. After 3 days, plaques were visualized with crystal violet in 20% ethanol. (D) Images of vGFP-Sar1H79Gi plaques with or without IPTG under visible (left) or fluorescent (right) light.
Effect of IPTG on vaccinia virus replication in cells infected with vGFP-Sar1H79Gi.
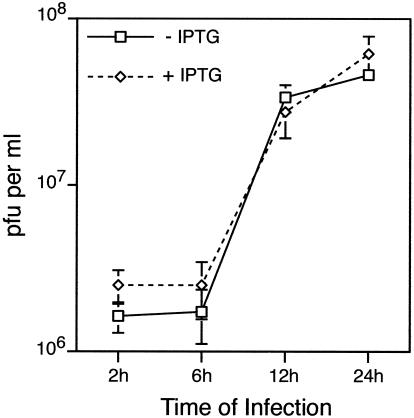
The small plaque size may result from a decrease in the production of infectious virus or from a defect in the cell-to-cell spread of virus, depending on whether Sar1H79G inhibited the formation of the viral primary membrane or that of the wrapping membrane. The presence or absence of IPTG had no effect on the production kinetics or the yield of cell-associated virus in a one-step growth experiment (Fig. 5), suggesting that the small plaque size resulted from a defect in virus spread.
FIG. 5.
One-step growth of vGFP-Sar1H79Gi in the presence or absence of IPTG. BS-C-1 cells were infected with 5 PFU of vGFP-Sar1H79Gi per cell in the absence or presence of 100 μM IPTG. Cells were harvested from triplicate cultures at 2, 6, 12, and 24 h after infection, and the infectivity of each was determined by plaque assay. Standard deviations are shown by error bars.
Effect of IPTG on distribution and processing of IEV membrane proteins in cells infected with vGFP-Sar1H79Gi.
Cells infected with vGFP-Sar1H79Gi in the presence of IPTG exhibited reticular cytoplasmic GFP fluorescence that was not seen in the absence of inducer (Fig. 6). The A33R, A36R, and B5R proteins were detected with specific antibodies and exhibited the expected Golgi membrane and peripheral punctate localization in the absence of IPTG (Fig. 6). In the presence of IPTG, these three proteins had a typical ER distribution in essentially all cells examined (Fig. 6) and extensively overlapped with Sar1H79G (data not shown). We had previously shown by cotransfection experiments that Sar1H79G did not perturb the Golgi membrane localization of the nonglycosylated palmitoylated F13L protein (14). Similar results were obtained here with vGFP-Sar1H79Gi (Fig. 6), supporting the direct association of the F13L protein with Golgi membranes.
FIG. 6.
Effect of IPTG on the intracellular distribution of GFP-Sar1H79G and IEV membrane proteins in vGFP-Sar1H79Gi-infected cells. HeLa cells were infected with vGFP-Sar1H79Gi in the absence (left column) or presence (right column) of IPTG for 20 h. Cells were fixed, permeabilized, and stained with anti-A33R mouse MAb, anti-A36R rabbit Ab, anti-F13L rabbit Ab, and anti-B5R rat MAb, followed by Alexa 594-conjugated anti-mouse IgG, tetramethyl rhodamine isothiocyanate-conjugated anti-rabbit IgG, and anti-rat IgG, respectively. Bars, 10 μm.
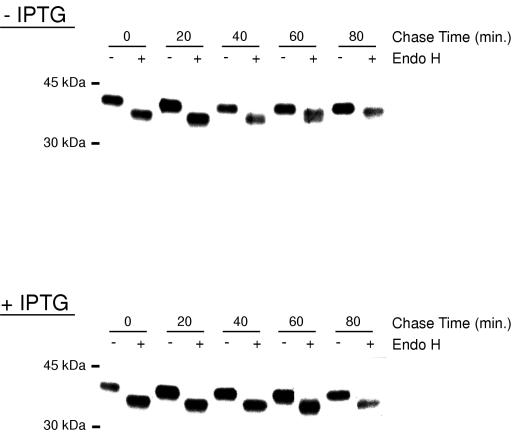
To confirm the block in ER cargo transport by biochemical means, we analyzed the endo H sensitivity of the B5R glycoprotein (21). Endo H can digest the high-mannose carbohydrate side chains of glycoproteins that have been synthesized in the ER but can no longer do so after the glycoproteins have been transported to the Golgi network, where trimming of the mannose side chains and addition of complex sugars occurs. The acquisition of endo H resistance, although often incomplete, is a generally accepted sign of ER-to-Golgi trafficking. Cells were infected with vGFP-Sar1H79Gi in the absence or presence of IPTG for 8 h and pulse-labeled with [35S]methionine and [35S]cysteine for 5 min. Cells were then incubated in medium supplemented with cold methionine, cysteine, and IPTG as required. At intervals, the cells were lysed and the B5R protein was immunoprecipitated from the lysate and tested for endo H sensitivity. In the absence of IPTG, some resistance to endo H occurred between 40 and 80 min (Fig. 7). At 60 min, a barely resolved doublet was detected. In the presence of IPTG, the B5R glycoprotein remained sensitive to endo H during the entire chase period (Fig. 7). Thus, the biochemical experiments also indicated that Sar1H79G inhibited trafficking of the B5R glycoprotein from the ER.
FIG. 7.
Endo H sensitivity of the B5R glycoprotein synthesized in the absence or presence of IPTG. BS-C-1 cells were infected with 5 PFU of vGFP-Sar1H79Gi per cell in the absence or presence of IPTG. After 8 h, cells were pulse-labeled with [35S]methionine and [35S]cysteine for 5 min and then washed and chased for 0, 20, 40, 60, and 80 min in medium supplemented with unlabeled cysteine, methionine, and IPTG as required. Cells were lysed immediately after the pulse or chase, and the B5R glycoprotein was captured with Ab, subjected to Endo H digestion, resolved by SDS-polyacrylamide gel electrophoresis, and visualized by autoradiography.
Effect of IPTG on the distribution of IMV membrane proteins in cells infected with vGFP-Sar1H79Gi.
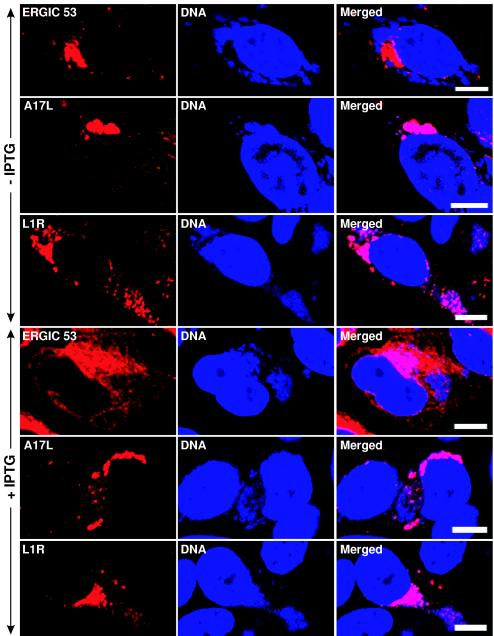
Next, we determined the effect of induced Sar1H79G expression on the intracellular distribution of the A17L and L1R viral membrane proteins. HeLa cells infected with vGFP-Sar1H79Gi were first stained with anti-ERGIC 53, anti-A17L, or anti-L1R Abs, and then with DAPI to visualize the cytoplasmic viral DNA factories. In the absence of IPTG, ERGIC 53 localized to the juxtanuclear Golgi area and appeared to be largely excluded from the DAPI-staining cytoplasmic viral DNA factories (Fig. 8, row 1). In contrast, the A17L and L1R proteins overlapped with the viral DNA factories in the perinuclear areas (Fig. 8, rows 2 and 3). In the presence of IPTG, ERGIC 53 was retained in the ER, giving a reticular pattern, whereas A17L and L1R still overlapped with the viral DNA factories and did not exhibit a reticular pattern (Fig. 8, rows 4 to 6). This result was found in essentially all examined infected cells. In addition, the studies were repeated with a BS-C-1 cell line that constitutively expresses the T7 RNA polymerase (45). Under these conditions, expression of Sar1H79G was detected 1 h before A17L, but the localization of the A17L and L1R proteins was still unaffected (data not shown). We also determined that the A17L protein was proteolytically processed (as previously described [5]) in the presence of Sar1H79G (data not shown), a finding consistent with the absence of any effect of this inhibitor on viral membrane formation. Thus, the induction of Sar1H79G failed to perturb the distribution of two IMV membrane proteins, while effectively blocking trafficking of IEV proteins and ERGIC 53 from the ER.
FIG. 8.
Effect of IPTG on the intracellular location of viral membrane proteins in cells infected with vGFP-Sar1H79Gi. HeLa cells were infected with vGFP-Sar1H79Gi in the absence (rows 1 to 3) or presence (rows 4 to 6) of IPTG for 20 h. The cells were fixed, permeabilized, and stained with anti-ERGIC 53 MAb, anti-L1R mouse MAb, and anti-A17LC rabbit Ab, followed by Alexa 594-conjugated anti-mouse IgG and tetramethyl rhodamine isothiocyanate-conjugated anti-rabbit IgG, respectively. Cells were then stained with DAPI. Red, secondary antibodies; blue, DAPI. Bars, 10 μm.
Vaccinia virus morphogenesis in cells infected with vGFP-Sar1H79Gi.
Electron microscopy was used to visualize the effects of Sar1H79G on the formation of virus structures. In cells infected for 20 h in the absence of IPTG, all forms of vaccinia virus, including IMV, IEV, and CEV, were visualized (Fig. 9A to C). When Sar1H79G was induced, IV and IMV appeared normal and abundant (Fig. 9D to F). However, the cell surface appeared devoid of virus particles, and neither IEV nor CEV was detected in numerous cell sections examined (Fig. 9D).
FIG.9.
Effect of Sar1H79G expression on the maturation and production of IMV and IEV. RK13 cells were infected with vGFP-SarH79Gi at 5 PFU per cell in the absence or presence of 100 μM IPTG. At 20 after infection, cells were harvested and processed for electron microscopy. All forms of vaccinia virus, including CEV (A), IMV (B), and IEV (C), were visualized in the cells infected without IPTG, whereas only IV (E) and IMV (F) but no CEV or IEV (D) were detected in the cells infected in the presence of IPTG.
Localization of ERGIC 53 in vaccinia virus-infected cells.
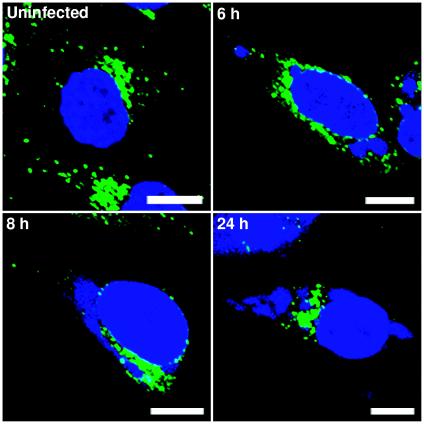
Although ERGIC 53 was previously reported to concentrate and localize in viral factories (26), we did not find this distribution in cells infected with vGFP-Sar1H79Gi in the absence of IPTG (Fig. 8). There was a possibility that this discrepancy might somehow be connected with the recombinant virus or linked to the time at which the infected cells were examined in our study. Therefore, we repeated the experiment by staining cells with Ab to ERGIC 53 and DAPI at 6, 8, and 24 h after infection with WR. At each time, however, the areas that stained intensely with ERGIC 53 Ab appeared to be excluded from DNA-staining factories (Fig. 10).
FIG. 10.
Intracellular localization of ERGIC 53 in cells infected with vaccinia virus strain WR. HeLa cells were infected with WR and at 6, 8, and 24 h after infection were fixed, permeabilized, and stained with mouse anti-ERGIC 53 MAb, followed by Alexa 488-conjugated anti-mouse IgG. Cells were counterstained with DAPI. Green, Alexa 488; blue, DAPI. Note that ERGIC 53 is excluded from viral DNA factories at all time points. Bars, 10 μm.
DISCUSSION
The mechanism by which poxvirus membranes form in the cytoplasm of infected cells is the most intriguing and least understood of all steps in virion morphogenesis. The idea that the primary viral membrane forms de novo is generally disfavored, partly on conceptual grounds. In several recent papers, an alternative model has been formulated in which viral proteins are inserted cotranslationally into the ER and accumulate in the ERGIC, the tubules of which form the viral crescent membranes (17, 26, 28, 34). The evidence supporting the model consists largely of electron microscopic images showing Ab to A17L and other viral membrane proteins in association with the ERGIC and ERGIC markers in tubules that are near or associated with viral crescents. However, one cannot conclude from static images that the viral proteins associated with the ERGIC are destined to form the viral membrane or whether they have been diverted from the viral membrane pathway. A difficulty in finding ERGIC or other cellular membrane markers associated with crescents and the membranes of immature or mature virions (16) can be plausibly explained by viral protein displacement, but such an explanation leaves some uncertainty as to the validity of the ERGIC model. We considered that a test of the ERGIC model of viral membrane origin could be performed by blocking cargo transport between the ER and ERGIC. The ERGIC model would receive strong support if the A17L and other membrane proteins accumulated in the ER and viral crescent membranes then failed to form under such conditions. On the other hand, the ERGIC model's plausibility would be weakened if viral proteins still localized in viral factories and viral membranes then formed normally. Fortunately for our purposes, cell biologists have studied the ER exit mechanism intensively, and trans-dominant negative inhibitors of ER protein export have been developed. The most widely used inhibitors have point mutations in the gene encoding Sar1 GTPase, which is an essential component in COPII-mediated transport from the ER (25, 36).
We overexpressed the dominant negative inhibitor Sar1H79G by using two different but complementary methods. First, we transfected a eukaryotic expression plasmid 24 h before infection so that inhibition of cargo transport would be established prior to the time of virus addition. The use of epitope-tagged or GFP-fused forms of Sar1H79G allowed us to specifically examine, by using confocal microscopy, those cells that expressed the inhibitor. For the second approach, we constructed a novel recombinant vaccinia virus that expresses Sar1H79G regulated by a strong, inducible T7 promoter that is turned on at the time of viral membrane protein synthesis or before. Importantly, identical results were obtained with both approaches. Sar1H79G expression had no effect on the localization of the A17L and other viral membrane proteins in the viral factories, and it had no effect on the formation of viral membranes or viral morphogenesis stages up to and including the stage of IMV formation. Moreover, Sar1H79G did not reduce the yield of infectious cell-associated virus. Nevertheless, the cellular protein ERGIC 53 and the viral glycoproteins that form the wrapping membranes were retained in the ER as determined by confocal microscopy as well as by sensitivity to endo H digestion. Furthermore, the formation of IEV and extracellular virions was completely blocked, demonstrating the stringency of the effects of Sar1H79G. We concluded that COPII-mediated cargo transport from the ER to the ERGIC is not required for the formation of the primary viral membranes but is necessary for the formation of the wrapping membranes.
Some of the proteins that constitute the wrapping membrane are typical integral membrane proteins, and their retention in the ER upon overexpression of Sar1H79G was therefore anticipated. In previous cotransfection experiments, we showed that Sar1H79G did not prevent the association of the nonglycosylated F13L protein with Golgi membranes (14), and a similar result was obtained here in the context of a viral infection. The F13L protein is palmitoylated and appears to associate directly with Golgi membranes. Golgi membrane association of F13L, however, is insufficient for wrapping of the IMV.
The effect of brefeldin A on vaccinia virus assembly superficially resembles that of Sar1H79G; e.g., IMV form but vaccinia virus envelopment is inhibited (40). However, unlike Sar1H79G, brefeldin A does not prevent ER export but deregulates membrane traffic, resulting in Golgi proteins entering a tubule pathway back to the ER (19). Therefore, the effect of brefeldin A cannot be used to argue for or against the ERGIC model.
During our study with vGFP-Sar1H79Gi, we noted that the A17L and L1R primary viral membrane proteins localized in DNA-containing viral factories, whereas ERGIC 53, a marker for the ERGIC, did not do so, even in the absence of inducer. Because this result conflicted with a confocal microscopic image in a recent paper by Risco and collaborators (26), we repeated the experiment with cells that were infected with the WR strain of vaccinia virus. Again, intense ERGIC 53-staining areas were excluded from the factories at all time points examined. Our data do not, however, rule out the presence of individual ERGIC tubules extending into factory regions, since these might not be detected by confocal microscopy. Immunoelectron microscopy would be necessary to resolve these structures, and Risco et al. (26) have obtained electron microscopic images of tubules staining with Ab to ERGIC 53 near viral membranes.
Where does this leave us with regard to viral membrane formation? Are we back to the original de novo membrane model? Not necessarily. First, we must point out that our experiments demonstrated that COPII-mediated cargo transport from the ER to the ERGIC was not required for viral membrane formation, but we cannot rule out the possibility that the ERGIC is involved under normal conditions when cargo transport is unaffected. Nevertheless, we do not feel that the previous data supporting a role of the ERGIC are convincing for reasons already mentioned. In light of the data presented here, the possibility that the tubules which form the viral crescent membranes are derived directly from the ER, as has been proposed for the membrane that underlies the capsid of African swine fever virus (1, 6, 30), should be considered. In place of the COPII-mediated ER exit pathway, poxviruses might encode a novel mechanism for inducing tubule formation from the ER. After all, poxviruses encode their own transcription system (23), their own DNA replication system (37), and even their own cytoplasmic disulfide bond pathway (31). At present, the only vaccinia virus conditional lethal mutant that is blocked in viral membrane formation and which does not exhibit a general defect in viral protein synthesis, maps to the F10L protein kinase (38, 41). Several viral membrane proteins are substrates of the kinase and are therefore potentially involved in tubule formation. The A17L gene encodes one such membrane protein (5, 8). Although repression of A17L expression prevents the formation of viral crescent membranes, tubular-vesicular structures accumulate adjacent to dense granular viral material (27, 46). Therefore, A17L expression might be a step removed from the hypothetical inducer of tubule formation. The membrane protein A14L is also phosphorylated by the F10L kinase (5, 39), but repression of A14L expression blocks morphogenesis at a later stage than does repression of A17L (29, 39). Additional putative membrane proteins encoded by vaccinia virus remain to be characterized.
Acknowledgments
We thank Jennifer-Lippincott Schwartz for a plasmid, Brian Ward for vB5R-GFP, Andrea Weisberg for electron microscopy, Owen Schwartz for helping in confocal microscopy, and Norman Cooper for providing tissue culture cells.
REFERENCES
- 1.Andres, G., R. Garcia-Escudero, C. Simon-Mateo, and E. Vinuela. 1998. African swine fever virus is enveloped by a two-membraned collapsed cisterna derived from the endoplasmic reticulum. J. Virol. 72:8988-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aridor, M., S. I. Bannykh, T. Rowe, and W. E. Balch. 1995. Sequential coupling between COPII and COPI vesicle coats in endoplasmic reticulum to Golgi transport. J. Cell Biol. 131:875-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aridor, M., K. N. Fish, S. Bannykh, J. Weissman, T. H. Roberts, J. Lippincott-Schwartz, and W. E. Balch. 2001. The Sar1 GTPase coordinates biosynthetic cargo selection with endoplasmic reticulum export site assembly. J. Cell Biol. 152:213-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barlowe, C., L. Orci, T. Yeung, M. Hosobuchi, S. Hamamoto, N. Salama, M. F. Rexach, M. Ravazzola, M. Amherdt, and R. Schekman. 1994. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77:895-907. [DOI] [PubMed] [Google Scholar]
- 5.Betakova, T., E. J. Wolffe, and B. Moss. 1999. Regulation of vaccinia virus morphogenesis: phosphorylation of the A14L and A17L membrane proteins and C-terminal truncation of the A17L protein are dependent on the F10L protein kinase. J. Virol. 73:3534-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cobbold, C., J. T. Whittle, and T. Wileman. 1996. Involvement of the endoplasmic reticulum in the assembly of African swine fever virus. J. Virol. 70:8382-8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dales, S., and E. H. Mosbach. 1968. Vaccinia as a model for membrane biogenesis. Virology 35:564-583. [DOI] [PubMed] [Google Scholar]
- 8.Derrien, M., A. Punjabi, R. Khanna, O. Grubisha, and P. Traktman. 1999. Tyrosine phosphorylation of A17 during vaccinia virus infection: involvement of the H1 phosphatase and the F10 kinase. J. Virol. 73:7287-7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Earl, P. L., B. Moss, L. S. Wyatt, and M. W. Carroll. 1998. Generation of recombinant vaccinia viruses, p. 16.17.1-16.17.19. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. Wiley Interscience, New York, N.Y.
- 10.Falkner, F. G., and B. Moss. 1988. Escherichia coli gpt gene provides dominant selection for vaccinia virus open reading frame expression vectors. J. Virol. 62:1849-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauri, H. P., O. Nufer, L. Breuza, H. B. Tekaya, and L. Liang. 2002. Lectins and protein traffic early in the secretory pathway. Biochem. Soc. Symp. 73-82. [DOI] [PubMed]
- 12.Hauri, H. P., and A. Schweizer. 1992. The endoplasmic reticulum-Golgi intermediate compartment. Curr. Opin. Cell Biol. 4:600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollinshead, M., A. Vanderplasschen, G. L. Smith, and D. J. Vaux. 1999. Vaccinia virus intracellular mature virions contain only one lipid membrane. J. Virol. 73:1503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Husain, M., and B. Moss. 2003. Intracellular trafficking of a palmitoylated membrane-associated protein component of enveloped vaccinia virions. J. Virol. 77:9008-9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Husain, M., A. Weisberg, and B. Moss. 2003. Topology of epitope-tagged F13L protein, a major membrane component of extracellular vaccinia virions. Virology 308:233-242. [DOI] [PubMed] [Google Scholar]
- 16.Krauss, O., R. Hollinshead, M. Hollinshead, and G. L. Smith. 2002. An investigation of incorporation of cellular antigens into vaccinia virus particles. J. Gen. Virol. 83:2347-2359. [DOI] [PubMed] [Google Scholar]
- 17.Krijnse-Locker, J., S. Schleich, D. Rodriguez, B. Goud, E. J. Snijder, and G. Griffiths. 1996. The role of a 21-kDa viral membrane protein in the assembly of vaccinia virus from the intermediate compartment. J. Biol. Chem. 271:14950-14958. [DOI] [PubMed] [Google Scholar]
- 18.Kuge, O., C. Dascher, L. Orci, T. Rowe, M. Amherdt, H. Plutner, M. Ravazzola, G. Tanigawa, J. E. Rothman, and W. E. Balch. 1994. Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J. Cell Biol. 125:51-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lippincott-Schwartz, J., N. B. Cole, and J. G. Donaldson. 1998. Building a secretory apparatus: role of ARF1/COPI in Golgi biogenesis and maintenance. Histochem. Cell Biol. 109:449-462. [DOI] [PubMed] [Google Scholar]
- 20.Lippincott-Schwartz, J., T. H. Roberts, and K. Hirschberg. 2000. Secretory protein trafficking and organelle dynamics in living cells. Annu. Rev. Cell Dev. Biol. 16:557-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathew, E. C., C. M. Sanderson, R. Hollinshead, M. Hollinshead, R. Grimley, and G. L. Smith. 1999. The effects of targeting the vaccinia virus B5R protein to the endoplasmic reticulum on virus morphogenesis and dissemination. Virology 265:131-146. [DOI] [PubMed] [Google Scholar]
- 22.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 23.Moss, B. 1994. Vaccinia virus transcription, p. 185-205. In R. Conaway and J. Conaway (ed.), Transcription mechanisms and regulation. Raven Press, New York, N.Y.
- 24.Moss, B., and B. M. Ward. 2001. High-speed mass transit for poxviruses on microtubules. Nat. Cell Biol. 3:E245-E246. [DOI] [PubMed] [Google Scholar]
- 25.Pepperkok, R., M. Lowe, B. Burke, and T. E. Kreis. 1998. Three distinct steps in transport of vesicular stomatitis virus glycoprotein from the ER to the cell surface in vivo with differential sensitivities to GTPγS. J. Cell Sci. 111:1877-1888. [DOI] [PubMed] [Google Scholar]
- 26.Risco, C., J. R. Rodriguez, C. Lopez-Iglesias, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 2002. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 76:1839-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodríguez, D., C. Risco, J. R. Rodríguez, J. L. Carrascosa, and M. Esteban. 1996. Inducible expression of the vaccinia virus A17L gene provides a synchronized system to monitor sorting of viral proteins during morphogenesis. J. Virol. 70:7641-7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez, J. R., C. Risco, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 1997. Characterization of early stages in vaccinia virus membrane biogenesis: implications of the 21-kilodalton protein and a newly identified 15-kilodalton envelope protein. J. Virol. 71:1821-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez, J. R., C. Risco, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 1998. Vaccinia virus 15-kilodalton (A14L) protein is essential for assembly and attachment of viral crescents to virosomes. J. Virol. 72:1287-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rouiller, I., S. M. Brookes, A. D. Hyatt, M. Windsor, and T. Wileman. 1998. African swine fever virus is wrapped by the endoplasmic reticulum. J. Virol. 72:2373-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Senkevich, T. G., C. L. White, E. V. Koonin, and B. Moss. 2002. Complete pathway for protein disulfide bond formation encoded by poxviruses. Proc. Natl. Acad. Sci. USA 99:6667-6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shima, D. T., N. Cabrera-Poch, R. Pepperkok, and G. Warren. 1998. An ordered inheritance strategy for the Golgi apparatus: visualization of mitotic disassembly reveals a role for the mitotic spindle. J. Cell Biol. 141:955-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, G. L., A. Vanderplasschen, and M. Law. 2002. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 83:2915-2931. [DOI] [PubMed] [Google Scholar]
- 34.Sodeik, B., R. W. Doms, M. Ericsson, G. Hiller, C. E. Machamer, W. van't Hof, G. van Meer, B. Moss, and G. Griffiths. 1993. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J. Cell Biol. 121:521-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sodeik, B., and J. Krijnse-Locker. 2002. Assembly of vaccinia virus revisited: de novo membrane synthesis or acquisition from the host? Trends Microbiol. 10:15-24. [DOI] [PubMed] [Google Scholar]
- 36.Storrie, B., J. White, S. Rottger, E. H. Stelzer, T. Suganuma, and T. Nilsson. 1998. Recycling of Golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J. Cell Biol. 143:1505-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Traktman, P. 1996. Poxvirus DNA replication, p. 775-793. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Traktman, P., A. Caligiuri, S. A. Jesty, and U. Sankar. 1995. Temperature-sensitive mutants with lesions in the vaccinia virus F10 kinase undergo arrest at the earliest stage of morphogenesis. J. Virol. 69:6581-6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Traktman, P., K. Liu, J. DeMasi, R. Rollins, S. Jesty, and B. Unger. 2000. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: construction and characterization of a tetracycline-inducible recombinant. J. Virol. 74:3682-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulaeto, D., D. Grosenbach, and D. E. Hruby. 1995. Brefeldin A inhibits vaccinia virus envelopment but does not prevent normal processing and localization of the putative envelopment receptor P37. J. Gen. Virol. 76:103-111. [DOI] [PubMed] [Google Scholar]
- 41.Wang, S., and S. Shuman. 1995. Vaccinia virus morphogenesis is blocked by temperature-sensitive mutations in the F10 gene, which encodes protein kinase 2. J. Virol. 69:6376-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward, B. M., and B. Moss. 2000. Golgi network targeting and plasma membrane internalization signals in vaccinia virus B5R envelope protein. J. Virol. 74:3771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward, B. M., and B. Moss. 2001. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J. Virol. 75:4802-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward, G. A., C. K. Stover, B. Moss, and T. R. Fuerst. 1995. Stringent chemical and thermal regulation of recombinant gene expression by vaccinia virus vectors in mammalian cells. Proc. Natl. Acad. Sci. USA 92:6773-6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whetter, L. E., S. P. Day, E. A. Brown, O. Elroy-Stein, and S. M. Lemon. 1994. Analysis of hepatitis A virus translation in a T7 polymerase-expressing cell line. Arch. Virol. 9(Suppl.):291-298. [DOI] [PubMed] [Google Scholar]
- 46.Wolffe, E. J., D. M. Moore, P. J. Peters, and B. Moss. 1996. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J. Virol. 70:2797-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolffe, E. J., A. Weisberg, and B. Moss. 2001. The vaccinia virus A33R protein provides a chaperone function for viral membrane localization and tyrosine phosphorylation of the A36R protein. J. Virol. 75:303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeh, W. W., B. Moss, and E. J. Wolffe. 2000. The vaccinia virus A9L gene encodes a membrane protein required for an early step in virion morphogenesis. J. Virol. 74:9701-9711. [DOI] [PMC free article] [PubMed] [Google Scholar]