Abstract
To characterize epitopes on human papillomavirus (HPV) virus-like particles (VLPs), a panel of mutated HPV-16 VLPs was created. Each mutated VLP had residues substituted from HPV-31 or HPV-52 L1 sequences to the HPV-16 L1 backbone. Mutations were created on the HPV-31 and −52 L1 proteins to determine if HPV-16 type-specific recognition could be transferred. Correct folding of the mutated proteins was verified by resistance to trypsin digestion and by binding to one or more conformation-dependent monoclonal antibodies. Several of the antibodies tested were found to bind to regions already identified as being important for HPV VLP recognition (loops DE, EF, FG, and HI). Sequences at both ends of the long FG loop (amino acids 260 to 290) were required for both H16.V5 and H16.E70 reactivity. A new antibody-binding site was discovered on the C-terminal arm of L1 between positions 427 and 445. Recognition of these residues by the H16.U4 antibody suggests that this region is surface exposed and supports a recently proposed molecular model of HPV VLPs.
The human papillomavirus (HPV) virion is composed of major (L1) and minor (L2) capsid proteins. The L1 protein self-assembles into virus-like particles (VLPs) that appear identical to infectious virus both by electron microscopy and immunologically (8, 9, 11). The L2 protein is important for assembly of infectious virus (20) but does not contain the conformation-dependent and type-specific epitopes most frequently recognized by anti-HPV sera (2, 9).
The VLP is a T = 7 icosahedron composed of 72 pentamers of L1, termed capsomers. Sixty of the capsomers subunits are at hexavalent positions, interacting with six neighboring capsomers with the remaining 12 capsomers at pentavalent positions (1). The only HPV L1 crystal structure solved to date is of T1 particles (3), in which all capsomers are at pentavalent positions. In the T1 particle, capsomers interact through a portion of the C-terminal tail. This flexible arm extends away from the capsomer of origin, interacts with similar regions from neighboring capsomers and returns to the capsomer from which it came. The remainder of this C-terminal region extends around the circumference of the capsomer, participating in the core β-sheet structure, forming a short helix (h5) and finally reinserting into the core of the pentamer from which it came. Missing from this model is an intercapsomer disulfide bond, shown by others to help stabilize the VLP structure (12, 21).
A revised model for HPV VLPs was proposed by Modis et al. (18). In this model, the C-terminal extension adopts a conformation similar to its conformation in the T1 structure, but instead of returning to the capsomer of origin, the arm is displaced onto, and ultimately invades, a neighboring capsomer. The C-terminal arm is anchored by the previously described disulfide bond between the cysteine from this region (amino acids 428) and cysteine 175 of a neighboring capsomer. The new model was termed the “invading arm” model because of its similarity to the simian virus 40 and mouse polyomavirus VP1 atomic structures. A consequence of the invading arm model is that residues on the C-terminal arm, not predicted to be exposed on the surface of T1 particles, would be surface assessable. The authors noted that several amino acids in this C-terminal region are divergent among HPV types and, thus, may be important for recognition by type-specific antibodies.
All conformation-dependent type-specific monoclonal antibody (MAb) epitopes identified to date have been found to reside on one or more hypervariable loops, on the VLP surface (3, 4, 13-16, 19, 23). Despite many studies, the binding site of one MAb (H16.U4) has not been identified. The H16.U4 MAb is a type-specific antibody that was shown to neutralize pseudotype HPV-16 viral infection (20). Thus, one region and perhaps others, important for antibody recognition and viral neutralization on HPV-16 VLPs remain uncharacterized.
The purpose of this study was to identify regions of the L1 proteins important for binding by HPV-16 and HPV-31 type-specific MAbs. Hypervariable loops were found to be essential for binding by most of the MAbs tested; however, mutations on the C-terminal arm disrupted H16.U4 antibody recognition, suggesting that residues on this region are surface exposed. The data here support the invading arm model of Modis et al.(18).
MATERIALS AND METHODS
MAbs.
The production, screening, and initial characterization of HPV-16 MAbs were previously described (5). Production of HPV-52 antibodies was performed as described previously (6, 7). Briefly, HPV L1 VLPs were produced by recombinant baculovirus, purified from infected Sf9 cells (as described by Hagensee [8]) and used as immunogen. VLPs (100 μg/mouse) were mixed with complete Freund's adjuvant and injected subcutaneously into BALB/c mice. Two weeks after immunizations, the mice were sacrificed, and draining lymph nodes and spleen cells removed for fusion with the mouse myeloma fusion partner P3X63-Ag8.653. Supernatants from growing hybridomas were screened for reactivity to intact and denatured HPV-52 L1 VLPs, and positive wells were cloned and retested.
The CAMVIR-117 MAb was a gift from C. McLean (Department for Pathology, University of Cambridge).
Preparation of vectors and mutagenesis.
The Invitrogen Bac-to-Bac system (Invitrogen, Carlsbad, Calif.) was used for expression of HPV L1 proteins in Spodoptera frugiperda (sf9) cells (ATCC, Manassas, Va.). To prepare vectors for use in this system the HPV-16 (114K) and HPV-31 (prototype) L1 sequences were cloned by PCR with primers including attB1 and attB2 sites (primer sequences available in supplemental materials at http://www.fhcrc.org/labs/dgalloway/virology_supplementary_data/). Primers were produced in house or purchased from Operon (Qiagen, Valencia, Calif.) or Invitrogen. Deep Vent polymerase (New England Biolabs, Beverly, Mass.) was used for high fidelity PCR, PFU turbo (Stratagene La Jolla, Calif.) was used for mutagenesis, and Taq (Invitrogen) was for analysis of recombinant bacmids. The L1 amplimers were cloned into the Gateway pDonr201 vector and from there into the pDest8 vector with Gateway technology following the manufacturer's protocol (Invitrogen). The L1 containing vectors were verified by sequencing in both directions with two primers specific for the pDest8 vector and four L1 specific primers. Sequencing was conducted with the BigDye cycle sequencing protocol on a model 377 DNA Sequencer (Applied Biosystems, Foster City, Calif.).
All mutations were made with the QuikChange mutagenesis kit (Stratagene). Mutagenesis was conducted on the L1 sequences in the pDest8 vector. When restriction enzyme sites were added, clones were identified by digestion; otherwise, clones were selected at random. All clones were sequenced as described above. Clones with the correct sequence were used for the generation of recombinant baculoviruses.
Generation of recombinant baculovirus and purification of VLPs.
Bacmids containing the L1 open reading frames were generated in DH10Bac E. coli cells with the protocol provided (Invitrogen). Recombinant bacmids containing L1 were verified by PCR with a HPV-16 or -31 L1-specific primers (forward) and the M13pUCR primer. Cells were transfected according to the method supplied with the Bac-to-Bac kit with the following modification: bacmid DNA was isolated from 10 ml of an overnight bacterial culture containing recombinant bacmid with a Qiagen tip 20 (Qiagen). Cells (sf9) were transfected with Cellfectin (Invitrogen) with 1 μg of bacmid DNA, and expression of L1 in the cells was assessed by immunoblots with CAMVIR-1 after 3 days of culture.
Production of VLPs was based on the method of Kirnbauer et al. (10). Briefly, after confirmation of high-level L1 protein expression, 100 μl of recombinant baculovirus supernatant was used to infect 150 mm plates of sf9 cells and incubated for 3 days. Cells were harvested from 20 plates and resuspended in phosphate-buffered saline (PBS, 11.69 mM phosphate, 137 mM NaCl, 2.68 mM KCl, pH 7.4) containing protease inhibitors (Complete; Roche, Indianapolis, Ind.). Cells were disrupted by douncing (50 strokes) on ice. Samples were spun at 10,000 × g for 20 min, resuspended in 3 ml of buffer (20 mM Tris, 1 M NaCl, pH 7.4) and sonicated three times for 45 s with a sonicator at 30% power. After spinning the homogenate at 10,000 × g for 20 min, supernatants were layered onto 40% sucrose cushions and spun for 2.5 h at 116,000 × g at 4°C. Pellets were resuspended in PBS with 10% CsCl (wt/wt) and layered onto 40% CsCl (in PBS) in an SW41 rotor. Samples were spun for 20 h (135,000 × g at 4°C). Bands were collected and dialyzed three times against PBS with 1 M NaCl.
Protease sensitivity assay.
A protease sensitivity assay was adapted from the method of Li et al. (12). Purified VLPs were dialyzed into digestion buffer (200 mM NaCl, 10 mM HEPES [pH 7.4], 1.0 mM CaCl2). The quantity of L1 proteins used was normalized by immunoblot analysis with CAMVIR-1. VLPs in digestion buffer were made with 10 mM dithiothreitol, by addition from a 100 mM stock solution, and incubated at 37°C for 30 min. Trypsin (sequencing grade; Roche) was serially diluted 1:3 in dilution buffer and 1 μl of diluted trypsin was added to each VLP sample (highest concentration of trypsin = 6.7 ng/ml). Samples were digested at room temperature for 1 h before stopping by addition of an equal volume of 2× sample buffer and immediately heated to 100°C for 5 min. The samples were run on 10.5% polyacrylamide gels, transferred to nitrocellulose, and immunoblotted with CAMVIR-1.
ELISAs.
The concentration of purified VLPs was normalized by denaturing enzyme-linked immunosorbent assay (ELISA) with CAMVIR-1. In these experiments VLPs were initially diluted 1:10 followed by serial dilutions (1:2) across a polypropylene plate in PBS. Diluted samples (50 μl) were transferred to an Immulon II plate (Thermo Labsystems, Franklin, Mass.) and incubated overnight at 4°C. Plates were washed four times in PBS. To denature the L1 protein, 100 μl of carbonate buffer (0.1 M, pH 9.5) containing β-mercaptoethanol (15 mM) was added to each well, and the plates were incubated at 68°C for 30 min. The plates were washed as before, and 200 μl of block (PBS with 5% goat serum and 0.05% Tween) added to each well. Following incubation at room temperature for 1 h, plates were tapped onto paper towels to remove buffer and CAMVIR-1 (1:1,000) was added in 50 μl of block. Plates were incubated for 1 h at 37°C, washed as before and 50 μl of block containing goat-anti-mouse alkaline phosphatase (1:3,000; Roche) was added to each well. Plates were incubated for 1 h at 37°C and washed as before. Phosphatase substrate (Sigma 104; Sigma Chemical, St. Louis, Mo.) was diluted in buffer (4.3 mg in 0.1 M carbonate buffer, 10 mM MgCl2, pH 9.5), 100 μl was added to each well, and the plates were incubated for 1 h at room temperature. Plates were read on a microplate reader (Elx 808; Bio-Tex Instruments Inc., Winooski, Vt.).
When type-specific MAbs were tested, the procedure was the same as above with the elimination of the denaturing step and the use of the MAb of interest in place of CAMVIR-1. All results are presented as raw optical densities or normalized for CAMVIR-1 reactivity: [optical density of wells containing VLPi and MAb minus wells with that MAb alone/optical density of wells containing VLPi and CAMVIR-1 minus well with CAMVIR-1 alone]. The i subscript is used to emphasize that this calculation was performed separately for each VLP type used. Relative binding is defined as reactivity of an MAb to mutated VLPs divided by the reactivity of that MAb to the wild-type VLPs which that MAb specifically recognizes (with normalized values).
Graphics.
Graphs were prepared with PRIZM (GraphPad, Inc., San Diego, Calif.). The molecular model of HPV-16L1 was generated with Vector NTI (InforMax, Bethesda, Md.) based on the atomic coordinates (PDB file accession no. L1LOT). The accessibility of residues to the surface was calculated with the Deep View Swiss-PdbViewer version 3.7 (http://us.expasy.org/spdbv/).
RESULTS
Creation of mutated L1 proteins.
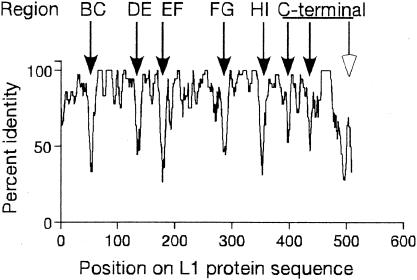
To identify regions of HPV-16 L1 important for type-specific antibody recognition, the L1 proteins from the A9 group of HPVs (types 16, 31, 33, 35, 52, 58, and 67) were aligned by Clustal W analysis (22). After alignment, the percentage of amino acid identity, within a sliding window of five amino acids in length was calculated (Fig. 1). Eight regions (indicated by arrows) were identified that were poorly conserved; this study focuses on seven of these regions (filled arrows). Five of these hypervariable regions have been shown to encode surface loops (BC, DE, EF, FG, and HI). To evaluate the importance of each of these regions in antibody binding, a series of substitution mutations was created. Each mutation substituted HPV-31 or HPV-52 residues for HPV-16 residues on the HPV-16 backbone, or conversely substituted HPV-16 residues for HPV-31 or HPV-52 sequences on the HPV-31 or HPV-52 backbones at each region of interest (Table 1). Two additional constructs, one expressing HPV-16 L1 VLPs with both the FG and HI loops substituted for the homologous HPV-31 loops (16:FG/HI), and the complementary HPV-31 L1 construct (31:FG/HI) were engineered. These chimeras were generated because substitution of the FG and HI loops from HPV-16 to the HPV 11 backbone was sufficient to transfer of HPV-16 specific binding of several MAbs (4).
FIG. 1.
Identification of poorly conserved regions of HPV L1s. The members of the A9 group of HPVs (types 16, 31, 33, 35, 52, 58, and 67) were aligned by Clustal W and the percent identity within a five-residue window between the types determined. The regions indicated on top were defined by Chen et al. (3). Arrows indicate regions of greatest variability. Filled arrows indicate the regions mutated in this study.
TABLE 1.
Residues replaced on HPV-16, HPV-31, and HPV-52 L1 proteins
| HPV backbone | Substitutiona (position) | Sequenceb | Name of constructc | Region on L1d |
|---|---|---|---|---|
| 16 | 52 (50) | FSIKNTSSGNG | 16:BC | BC loop |
| 52 | 16 (50) | *P**KP---*N | 52:BC | |
| 16 | 31 (131) | ENSNRYAGGPGVD | 16:DE | DE loop |
| 31 | 16 (130) | EN**A**ANA*** | NC | |
| 16 | 31 (176) | CSNNAITPGDC | 16:EF | EF loop |
| 31 | 16 (175) | C*N**VN**** | 31:EF | |
| 16 | 31 (261) | FFNRSGTVGESVPTDLYIKGSGSTATLANST | 16:FG | FG loop |
| 31 | 16 (260) | L***A*A***N**D*********T*N**S*N | 31:FG | |
| 16 | 31 (346) | CAAIANSDTTFKSSNFKE | 16:HI | HI loop |
| 31 | 16 (345) | ****ST*E**Y*NT**** | 31:HI | |
| 16 | 31 (396) | MNPAILEDWNFGLTTPPSGSLED | 16:410 | Hinge |
| 31 | 16 (395) | **ST*********QP**G*T*** | NC | |
| 16 | 31 (421) | FVTSQAITCQLTAPQKPKEDPFKDYVFWE | 16:430 | Carboxy-terminal arm |
| 31 | 16 (420) | *******A***HT*PA*****L*K*T*** | NC |
Amino acid position based on the HPV-16 L1 sequence.
Amino acids shown were substituted on the HPV-16, -31 or -52 backbone.
Two additional constructs, not shown here, were combinations of the FG and HI regional substitution onto HPV-16 or -31. NC, not created.
All intertypic hybrids were HPV-16/31 substitutions, with the exception of the BC loop constructs. The rationale for with HPV-52 sequences for the BC loop substitutions stems from the finding that a naturally occurring Phe to Leu mutation at position 50 (F50L) of HPV-16 disrupts H16.V5 and H16.E70 binding (23). However, residue 50 is not predicted to be surface assessable, suggesting that this mutation conferred conformational changes to other regions of the particle. The BC loop was of primary interest in this regard because it begins at position 51. Therefore, to evaluate the importance of the BC loop independent from changes induced by mutation of residue 50, we exchanged the BC loops of HPV-52 (which has a Phe at position 50) with the HPV-16 BC loop on the HPV-16 backbone and created a complementary substitution on the HPV-52 backbone (Table 1). HPV-16 VLPs with a Phe to Leu point mutation at residue 50 (16:F50L) were also created.
Conformation of mutated VLPs.
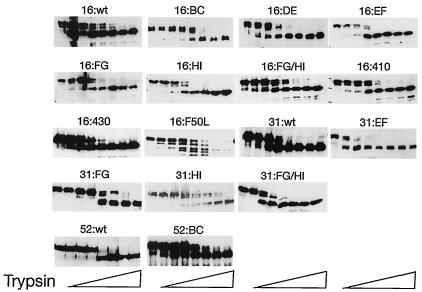
After expression and purification of HPV VLPs from sf9 cells, sensitivity to protease digestion was used to assess proper folding of VLPs. It has been shown that trypsin digestion removes a C-terminal peptide and reduces the trypsin-resistant N-terminal portion of L1 from 57 kDa to approximately 42 kDa (12). All of the mutated and wild-type VLPs tested, with the exception of 16:F50L, were reduced in size from 57 kDa to 42 kDa by treatment with trypsin suggesting that these VLPs were correctly folded (Fig. 2). VLPs tested but not shown (16:260-273 and 16:285-290) gave similar results. Very little of the 16:F50L protein was visible by immunoblotting after treatment with trypsin, indicating that 16:F50L VLPs were not in a native conformation.
FIG. 2.
Stability of VLPs in the presence of trypsin. VLPs were treated with increasing concentrations of trypsin and incubated for 1 h. Following trypsin treatment the proteins were run on polyacrylamide gels and immunoblotted with an anti-L1 antibody (CAMVIR-1). The left hand lane of each panel corresponds to samples that received no trypsin.
Binding of HPV-16 and HPV-52 MAbs to HPV-16/52 hybrid VLPs.
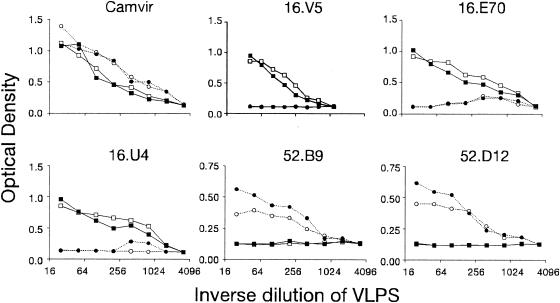
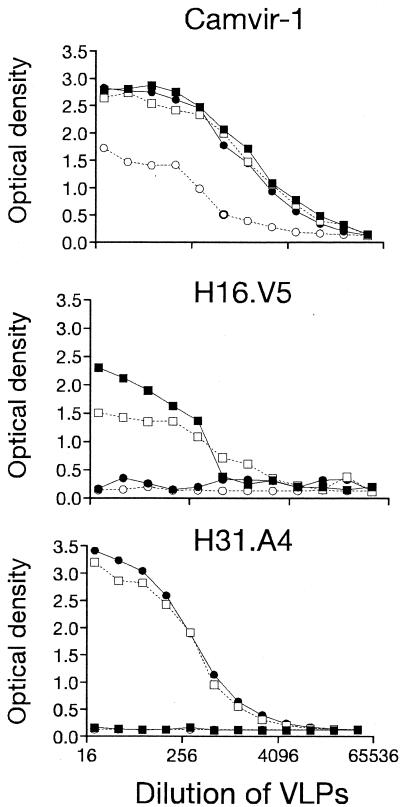
To determine if the BC loop was important for antibody recognition, HPV-16 and HPV-52 specific MAbs were used to test for loss, or transfer, of reactivity to VLPs with intertypic substitutions of this loop. The concentration of VLPs was normalized in binding assays with CAMVIR-1 (Fig. 3), an antibody that recognizes a conserved region (residues 204 to 210) of HPV L1 proteins. The three HPV-16 specific MAbs tested (H16.V5, H16.E70, H16.U4) reacted similarly to wild-type HPV-16 (16:wt) and 16:BC VLPs (Fig. 3). The two HPV-52 specific MAbs reacted somewhat more strongly to 52:wt VLPs compared with 52:BC VLPs (Fig. 3). The overall lower levels of binding by HPV-52 MAbs may be attributed to the use of tissue culture supernatants which have 10- to 100-fold lower concentrations of IgG than ascites (all HPV-16 MAbs were ascites). Binding to 52:BC VLPs by H52.B9 and H52.D12 was reduced to 63.5% and 84.2% of 52.wt, respectively. Similar results were obtained for two other HPV-52-specific MAbs, H52.F1 and H52.G5 (data not shown). No transfer of type-specific reactivity was observed for any of the HPV-16 or HPV-52 specific antibodies. Thus, the BC loop of HPV-16 was not necessary or sufficient for type specific binding by any HPV-16 specific MAb and the BC loop of HPV-52 was only partially necessary and not sufficient for any of the HPV-52 specific MAb tested. 16:F50L VLPs were tested in similar assays and several HPV-16 specific MAbs were found to have reduced binding (not shown); however, this may have resulted from the protein not folding into a native conformation required for recognition (see above).
FIG. 3.
Binding of 16 and 52 specific MAbs to VLPs with mutations of the BC loop. VLPs 16:wt (▪), 16:BC (□), 52:wt (•), and 52:BC (○) were titrated across a plate and reacted with CAMVIR-1, H16.V5, H16.E70, H52.B9, and H52.D12 in an ELISA.
Identification of regions important for MAb binding with HPV-16/31 hybrid VLPs.
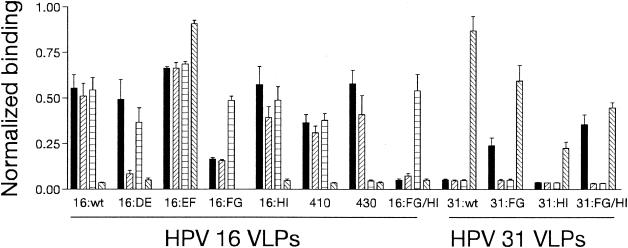
The ability of HPV-16 and HPV-31 type-specific MAbs to bind to VLPs with one or more intertypic substitutions of hypervariable loops was assessed by ELISA. As before CAMVIR-1 reactivity was used to normalize the data (not shown). 16:430 VLPs were not recognized by H16.U4 (Fig. 4), as binding was 8.6% compared with 16:wt binding. H16.U4 bound to all other mutated HPV-16 VLPs. The FG loop was required for recognition by H16.V5, with reactivity to 16:FG VLPs 29.7% relative to 16:wt VLPs (a reduction in binding of 70.3%). H16.V5 binding to VLPs with the reciprocal mutation (31:FG) was 43.4% compared with 16:wt. H16.V5 reactivity increased when both the FG and HI loops were substituted (64.1% of 16:wt binding). Recognition of VLPs by H16.E70 required loops DE (16.6% of 16:wt) and FG (21.2% of 16:wt) but binding was not transferred to HPV-31 by the FG loop alone or loops FG and HI (both < 10% of 16:wt).
FIG. 4.
Binding of MAbs to HPV-16/31 hybrid VLPs. MAbs were tested by ELISA for reactivity to HPV-16 and HPV-31 wild-type VLPs and mutated VLPs that are indicated on the x-Axis. The y-axis are normalized optical density values (see text) for H16.V5 (solid), H16.E70 (striped), H16.U4 (cross-hatched), and H31.A6 (opposite striped) binding. The error bars show standard deviations.
Transfer of type-specific recognition from HPV-31 to HPV-16 VLPs by H31.A6 was achieved by substitution of the EF loop alone (104.4% compared with 31:wt, Fig. 4). When the EF loop of HPV-31 was exchanged for HPV-16 sequences (31:EF), H31.A6 binding was abolished (Fig. 5) indicating that this region was necessary for binding. H31.A6 reactivity with other HPV-31 hybrid VLPs was 68.5% and 26.0% (compared with 31:wt binding) for the 31:FG and 31:HI hybrids, respectively (Fig. 4). Reduction of H31.A6 binding to 31:HI was partially relieved when this substitution was combined with the FG loop (51.6% of 31:wt), indicating that the reduction in binding to 31:HI was not due solely to interactions between MAb and residues on the HI loop.
FIG. 5.
EF loop is necessary and sufficient for H31.A6 binding. VLPs were serially diluted across microtiter plates and ELISAs performed with CAMVIR-1, H16.V5 or H31.A6 as indicated. The VLPs were HPV-16:wt (▪), HPV-31:wt (•), 16:EF (□), and 31:EF (○).
Identification of residues on the FG loop important H16.V5 and H16.E70 binding.
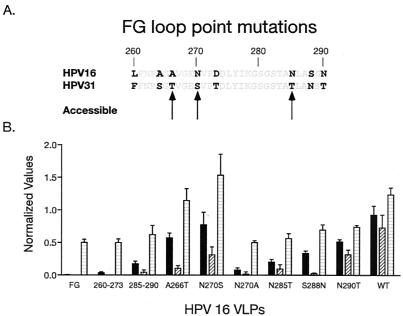
To identify residues important for recognition of the FG loop by H16.V5 and H16.E70, a series of point mutations were created. Each mutation on the FG loop substituted one or more HPV-31 residue(s) for HPV-16 residue(s) at the appropriate position with the same methodology as for the regional substitutions. The variable residues predicted to be most accessible were substituted (residues 266, 270, and 285), as well as two other variable residues close to the C terminus of the loop (residues 288 and 290). In addition to the N270S mutation, Ala was substituted at position 270 (16:N270A) to determine whether a hydrogen bond involving this residue was important for binding. The level of L1 protein bound to plates was again normalized to CAMVIR-1 reactivity (not shown). Figure 6 shows that H16.U4 bound to all mutated VLPs at levels between 40.5% (16:260-273) and 124.3% (16:N270S) compared with 16:wt. H16.V5 exhibited markedly reduced binding to three VLP preparations tested; 16:FG (0.4% of 16:wt), 16:260-273 (3.8%) and 16:N270A (8.4%). Reactivity of H16.V5 was also reduced to 16:285-290 (18.8%), 16:N285T (21.9%) and 16:S288N (36.2%) relative to 16:wt. There was no detectable H16.E70 binding above background to either the 16:FG hybrid or the 16:260-273 VLPs and minimal binding to 16:N270A, 16:S288N, 16:285-290, 16:N285T, and 16:A266T (2.2%, 3.4%, 5.8%, 13.2%, and 14.4%, respectively). These data indicated that binding of H16.V5 and H16.E70 was dependent on residues on both ends of the FG loop and, whereas, several 16-to-31 point mutations abolished H16.E70 binding none of single HPV-16 to HPV-31 point mutations tested totally disrupted H16.V5 binding.
FIG. 6.
Identification of residues on the FG loop important for MAb binding. The sequence of HPV-16 and HPV-31 L1 are compared (A). Residues that are predicted to be >45% surface accessible are indicated with arrows and bold type is used to indicate nonconserved residues. CAMVIR-1, H16.V5, H16.E70, and H16.U4 were tested for binding to VLPs that were serially diluted prior to testing. The normalized binding values are presented (B), with each MAb represented by bars, as described in the legend to Fig. 4 (error bars show standard deviations).
DISCUSSION
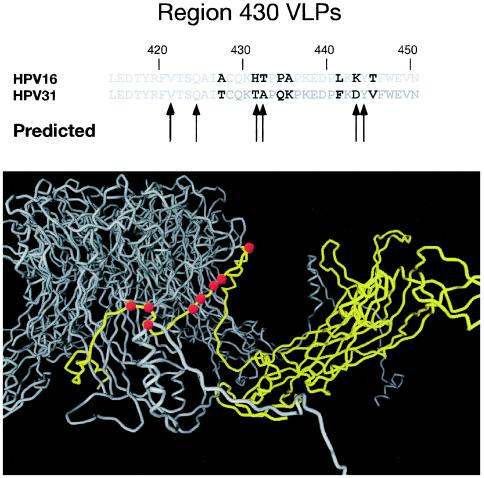
The HPV L1 model proposed by Modis et al. (18) postulated that the C-terminal arm of the molecule, important for intercapsomer interactions, would be exposed to the surface and possibly express type-specific epitopes. In support of this model, we show evidence that the C-terminal region around residue 430 was important for recognition by the type-specific MAb H16.U4. Six residues between position 421 and 444 were proposed by Modis et al. to be hypervariable and surface accessible and, therefore, of potential immunologic importance. Three of these residues (421, 424 and 444) were conserved between types HPV-31 and HPV-16 and were therefore unchanged in the 16:430 construct. Four residues in this region (435, 436, 442 and 446) were mutated in these studies but were not predicted by Modis et al. to be potential antibody binding sites.
Figure 7 highlights the residues that were changed on the C-terminal arm on the Modis molecular model. Although the data presented here support a model in which the C-terminal region is at the virus surface, the possibility exists that mutations in this region conferred a conformational change to an adjacent region on the VLP surface that was required for antibody binding. This possibility is unlikely, however, because all of the other HPV-16 type-specific antibodies tested recognized 16:430 VLPs indicating that there were no major conformational changes. Trypsin digestion experiments also suggested that 16:430 VLPs folded into a conformation similar to wild-type particles. H16.U4 has exhibited neutralizing activity in some studies (19) but not others (23), and thus antibody responses to the H16.U4 epitopes as defined here may be important for protective immune responses. There was one other residue on the C-terminal tail proposed to be of potential immunologic importance, residue 413. Substitution of HPV-31 residues in this region had no effect on type-specific binding as 16:410 VLPs (that include a mutation of residue 413) had a binding profile similar to 16:wt for all HPV-16 MAbs tested.
FIG. 7.
Position of mutations made on the C-terminal arm of HPV-16 L1. The upper panel compares the sequence of HPV-16 and HPV-31 with the differences emphasized with bold type. The arrows indicate residues predicted by Modis et al. 18 to be potentially immunogenic. In the lower panel the carbon backbone of HPV-16 is shown based on the Modis model showing the C-terminal arm from a neighboring capsomer (in yellow) as it wraps around and inserts into a capsomer (gray). The mutations in this region are indicated as larger red balls. The view is from the side.
The epitope recognized by the only HPV-31 specific antibody tested here was found to be a simple epitope comprised of a single loop. The EF loop of HPV-31 was shown to be necessary for H31.A6 binding as the antibody did not recognize HPV-31 VLPs with a HPV-16 EF loop. The EF loop was also demonstrated to be sufficient for transfer of H31.A6 type-specific binding to the HPV-16 VLPs. The EF loop, in combination with the BC loop, was previously identified as part of an HPV 6 epitope (16). A potential criticism of these observations is that because binding was ablated by the transfer of the HPV-16 EF loop on to the HPV-31 backbone, there were no conformation-dependent MAbs that recognized these particles and thus we could not be certain that these particles had folded correctly. However, 31:EF VLPs that were resistant to trypsin digestion (Fig. 2) were recognized by HPV-31 immune human sera (supplementary Fig. 1 may be found at http://www.fhcrc.org/labs/dgalloway/virology_supplementary_data/) and produced particles that looked similar to wild-type particles by electron microscopy (supplemental material Fig. 2), suggesting that the particles had folded correctly.
The H16.V5 epitope was previously characterized by Christensen et al. (4) as being a complex epitope composed of multiple regions (the FG and HI loops). In agreement with this finding, we found that the FG loop was required for recognition and both the FG and HI loops were necessary for transfer of HPV-16-specific binding onto HPV-31. The regions substituted in this study were somewhat different from those in Christensen's study. The FG and HI loops in this study were residues 260 to 290 and 349 to 358, respectively, compared with 266 to 297 and 339 to 365 in the report by Christensen et al. (4).
It has been proposed that the F50L point mutation that disrupts H16.V5 and H16.E70 binding (23) does so by altering the conformation of residues on the FG loop (3). An alternate hypothesis is that this mutation alters the conformation of the BC loop to which it is adjacent. To address this question, hybrid VLPs were created where the HPV-52 BC loop was substituted on the HPV-16 L1 backbone and the HPV-16 BC loop was substituted on the HPV-52 L1 backbone. HPV-16 VLPs with an F50L mutation were shown here to be degraded by trypsin indicating a failure to fold correctly and, therefore, no conclusions could be drawn regarding epitopes with these particles. Although the BC loop has been shown to be important for the binding of some HPV 6 and HPV-16 MAbs (4,16), this region was not required for any of the HPV-16 MAbs tested here. All of the HPV-52 MAbs tested exhibited only modest reductions in binding to HPV-52 VLPs with BC loop substitutions suggesting that the BC loop was not directly involved in binding to any of the MAbs tested.
Residues at both ends of the FG loop were shown here to be important for binding by H16.V5 and H16.E70. To characterize which residues were important for antibody binding, a series of point mutations and smaller regional mutations along the FG loop were examined. VLPs with four intertypic substitutions between amino acids 260 and 273 (16:260-273), and VLPs with three substitutions between residues 285 and 290 (16:285-290) both showed substantial loss of reactivity to H16.V5 and H16.E70. Previous studies had identified residues 266 and 282 as being important for 16.E70 binding but not for H16.V5 binding (19, 23). None of the point mutations tested in this study (A266T, N270S, N285T, S288N, and N290T) were found to be essential for H16.V5 binding. H16E.70 binding was more sensitive to point mutations on the FG loop, with the greatest loss of binding seen to VLPs with substitutions at positions 285, 288, and 266.
Having a polar residue at position 270 was important for both H16.V5 and H16.E70 binding because substitution of Asn270 with Ala greatly reduced antibody reactivity. Both H16.V5 and H16.E70 had reduced binding to 16:N270S VLPs but binding was reduced dramatically to 16:N270A VLPs. Although, Ser and Ala are similar sized residues (somewhat smaller than Asn), Ser has a polar side chain that can participate in a hydrogen bond much like Asn. Thus, our data suggest that Asn270 may participate in a hydrogen bond that is important for antibody recognition of the FG loop by both H16.V5 and H16.E70.
The H16.E70 epitope was found here to be a complex epitope because both the FG and DE loops were found to be necessary for binding. The finding that the FG loop was required for recognition confirms previous studies (4,19). This is the first study to demonstrate that the DE loop was also important for H16.E70 recognition. The DE loop has also been shown to be essential for binding to HPV 11 by several MAbs (13,14). However, Christensen et al. (4) found that H16.E70 binding could be transferred to HPV 11/16 hybrid VLPs that did not contain the HPV-16 DE loop but possessed the HPV-16 C terminus from residue 172 onward. The difference between our findings and those of Christensen's may indicate that substitution of the DE loop conferred a conformational change to another region directly involved in antibody-VLP reactivity. Only the transfer of H16.E70 recognition to other HPV types with defined variable regions will resolve which region, in addition to the FG loop is important for H16.E70 binding.
One region of the L1 protein that was highly variable between types was not tested here (Fig. 1). The extreme C terminus is highly basic and is thought to interact with viral DNA on the inside of the virion; however, the C terminus was not part of the crystal structure (3).
In summary, the MAb binding profiles characterized here defined epitopes that were either simple (binding to one loop only) or complex (binding to multiple loops). Data for one HPV-16 specific MAb (H16.U4) supported the proposed structure of VLPs in which the C-terminal arm is surface exposed. The epitope recognized by H31.A6 was identified and residues at both ends of the FG loop were found to be important for H16.V5 and H16.E70 binding. The DE loop was also found to be important for H16.E70 binding, but this finding conflicts with previous studies of this antibody. In the future, it will be important to define which epitopes are dominant in the human antibody response.
Acknowledgments
We thank Robert Garcea for helpful discussions.
This work was supported by grants CA42792 and AI38382 from the National Institutes of Health.
REFERENCES
- 1.Baker, T. S., W. W. Newcomb, N. H. Olson, L. M. Cowsert, C. Olson, and J. C. Brown. 1991. Structures of bovine and human papillomaviruses. Analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys. J. 60:1445-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter, J. J., M. E. Hagensee, M. C. Taflin, S. K. Lee, L. A. Koutsky, and D. A. Galloway. 1993. HPV-1 capsids expressed in vitro detect human serum antibodies associated with foot warts. Virology 195:456-462. [DOI] [PubMed] [Google Scholar]
- 3.Chen, X. S., R. L. Garcea, I. Goldberg, G. Casini, and S. C. Harrison. 2000. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol. Cell 5:557-567. [DOI] [PubMed] [Google Scholar]
- 4.Christensen, N. D., N. M. Cladel, C. A. Reed, L. R. Budgeon, M. E. Embers, D. M. Skulsky, W. L. McClements, S. W. Ludmerer, and K. U. Jansen. 2001. Hybrid papillomavirus L1 molecules assemble into virus-like particles that reconstitute conformational epitopes and induce neutralizing antibodies to distinct HPV types. Virology 291:324-334. [DOI] [PubMed] [Google Scholar]
- 5.Christensen, N. D., J. Dillner, C. EkLund, J. J. Carter, G. C. Wipf, C. A. Reed, N. M. Cladel, and D. A. Galloway. 1996. Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus-like particles as defined by monoclonal antibodies. Virology 223:174-184. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, N. D., R. Kirnbauer, J. T. Schiller, S. J. Ghim, R. Schlegel, A. B. Jenson, and J. W. Kreider. 1994. Human papillomavirus types 6 and 11 have antigenically distinct strongly immunogenic conformationally dependent neutralizing epitopes. Virology 205:329-335. [DOI] [PubMed] [Google Scholar]
- 7.Christensen, N. D., and J. W. Kreider. 1990. Antibody-mediated neutralization in vivo of infectious papillomaviruses. J. Virol. 64:3151-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagensee, M. E., N. Yaegashi, and D. A. Galloway. 1993. Self-assembly of human papillomavirus type 1 capsids by expression of the L1 protein alone or by coexpression of the L1 and L2 capsid proteins. J. Virol. 67:315-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirnbauer, R., F. Booy, N. Cheng, D. R. Lowy, and J. T. Schiller. 1992. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc. Natl. Acad. Sci. USA 89:12180-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirnbauer, R., N. L. Hubbert, C. M. Wheeler, T. M. Becker, D. R. Lowy, and J. T. Schiller. 1994. A virus-like particle enzyme-linked immunosorbent assay detects serum antibodies in a majority of women infected with human papillomavirus type-16. J. Natl. Cancer Inst. 86:494-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirnbauer, R., J. Taub, H. Greenstone, R. Roden, M. Dürst, L. Gissmann, D. R. Lowy, and J. T. Schiller. 1993. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J. Virol. 67:6929-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, M., P. Beard, P. A. Estes, M. K. Lyon, and R. L. Garcea. 1998. Intercapsomeric disulfide bonds in papillomavirus assembly and disassembly. J. Virol. 72:2160-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludmerer, S. W., D. Benincasa, and G. E. Mark III. 1996. Two amino acid residues confer type specificity to a neutralizing, conformationally dependent epitope on human papillomavirus type 11. J. Virol. 70:4791-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludmerer, S. W., D. Benincasa, G. E. Mark III, and N. D. Christensen. 1997. A neutralizing epitope of human papillomavirus type 11 is principally described by a continuous set of residues which overlap a distinct linear, surface-exposed epitope. J. Virol. 71:3834-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludmerer, S. W., W. L. McClements, X. M. Wang, J. C. Ling, K. U. Jansen, and N. D. Christensen. 2000. HPV11 mutant virus-like particles elicit immune responses that neutralize virus and delineate a novel neutralizing domain. Virology 266:237-245. [DOI] [PubMed] [Google Scholar]
- 16.McClements, W. L., X. M. Wang, J. C. Ling, D. M. Skulsky, N. D. Christensen, K. U. Jansen, and S. W. Ludmerer. 2001. A novel human papillomavirus type 6 neutralizing domain comprising two discrete regions of the major capsid protein L1. Virology 289:262-268. [DOI] [PubMed] [Google Scholar]
- 17.McLean, C. S., M. J. Churcher, J. Meinke, G. L. Smith, G. Higgins, M. Stanley, and A. C. Minson. 1990. Production and characterisation of a monoclonal antibody to human papillomavirus type 16 with recombinant vaccinia virus. J. Clin. Pathol. 43:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Modis, Y., B. L. Trus, and S. C. Harrison. 2002. Atomic model of the papillomavirus capsid. EMBO J. 21:4754-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roden, R. B., A. Armstrong, P. Haderer, N. D. Christensen, N. L. Hubbert, D. R. Lowy, J. T. Schiller, and R. Kirnbauer. 1997. Characterization of a human papillomavirus type 16 variant-dependent neutralizing epitope. J. Virol. 71:6247-6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roden, R. B., H. L. Greenstone, R. Kirnbauer, F. P. Booy, J. Jessie, D. R. Lowy, and J. T. Schiller. 1996. In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype. J. Virol. 70:5875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sapp, M., C. Fligge, I. Petzak, J. R. Harris, and R. E. Streeck. 1998. Papillomavirus assembly requires trimerization of the major capsid protein by disulfides between two highly conserved cysteines. J. Virol. 72:6186-6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White, W. I., S. D. Wilson, F. J. Palmer-Hill, R. M. Woods, S. J. Ghim, L. A. Hewitt, D. M. Goldman, S. J. Burke, A. B. Jenson, S. Koenig, and J. A. Suzich. 1999. Characterization of a major neutralizing epitope on human papillomavirus type 16 L1. J. Virol. 73:4882-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]