Abstract
The V protein of the recently emerged paramyxovirus, Nipah virus, has been shown to inhibit interferon (IFN) signal transduction through cytoplasmic sequestration of cellular STAT1 and STAT2 in high-molecular-weight complexes. Here we demonstrate that the closely related Hendra virus V protein also inhibits cellular responses to IFN through binding and cytoplasmic sequestration of both STAT1 and STAT2, but not STAT3. These findings demonstrate a V protein-mediated IFN signal evasion mechanism that is a general property of the known Henipavirus species.
The Paramyxoviridae comprise a group of enveloped, negative-strand RNA viruses that are capable of causing human and animal disease (6). Most paramyxoviruses have the ability to inhibit interferon (IFN)-induced innate antiviral responses through direct inhibition of cellular STAT proteins that are responsible for IFN signal transduction. Within the family and also among species in a particular genus, diverse molecular mechanisms account for STAT inhibition, ranging from cytoplasmic sequestration to proteasomal degradation. These STAT inhibition properties have often been linked to a virus-encoded protein named ‘V.’ The paramyxovirus V protein is characterized by a highly conserved C-terminal domain (CTD); CTDs are approximately 50% identical within the family. V proteins are readily identifiable by their CTDs, and particularly notable is the conservation of seven cysteine residues (11, 13). This domain enables the V protein to bind two atoms of zinc, a stoichiometry similar to that found in cellular RING finger domains (11). Aside from this apparent resemblance, V proteins have no cellular homologues and the spacing of CTD cysteine residues is not in accordance with those in known cellular zinc-binding proteins (1, 2), suggesting a virus origin for this domain.
A 1994 outbreak of acute respiratory disease at an Australian horse stable resulted in the deaths of 12 horses and one horse trainer. The causative agent was identified as a paramyxovirus initially termed “equine morbillivirus” and subsequently renamed Hendra virus after the location of the initial outbreak (5, 7, 18, 19). A similar outbreak of zoonotic disease in both the animal and human populations of peninsular Malaysia was caused by Nipah virus, which is closely related to Hendra virus (3). These two species are the founding members of a new Paramyxovirus genus called Henipavirus (16, 17).
One of the features identifying these viruses as paramyxoviruses is the presence of a polycistronic gene with overlapping reading frames coding for the highly conserved V protein CTD. Despite ∼50% sequence identity within the CTD, Henipavirus V proteins have N termini that are longer and dissimilar from those of other paramyxovirus genera. Recently it was demonstrated that expression of the V protein of Nipah virus interferes with IFN signaling in avian and human systems (10, 12). Available evidence indicates that the Nipah virus V protein induces cytoplasmic sequestration of both STAT1 and STAT2 in high-molecular-weight complexes (12). To determine if this mechanism of STAT inhibition is a general property of the Henipavirus genus, the Hendra virus V protein was similarly tested for IFN signaling interference.
The V proteins of Hendra virus and Nipah virus are 58% identical in overall amino acid sequence, with 81% identity between amino acids 1 to 140, 44% identity between amino acids 141 to 405, and 83% identity within amino acids 406 to 457 (Fig. 1A). In view of these sequence similarities, it was hypothesized that Hendra virus V protein might share with Nipah virus V protein the ability to antagonize STAT protein function. To express the Hendra virus V protein in mammalian cells, cDNA expression vectors that contained Hendra virus V cDNA fused to N-terminal hemagglutinin (HA) or FLAG epitope tags were constructed. Transfection of mammalian cells with these vectors produced a single Hendra virus V protein species with an apparent molecular mass of approximately 75 kDa, identifiable by immunoblotting with tag-specific antisera (Fig. 2). To determine the ability of Hendra virus V protein to interfere with IFN signal transduction, human fibrosarcoma 2fTGH cells were transfected with IFN-inducible luciferase reporter genes along with epitope-tagged V protein expression vectors or expression vector with no insert. Stimulation with either alpha IFN (IFN-α) or IFN-γ potently induced the activity of their respective reporter gene, but expression of the Hendra virus V protein inhibited IFN signaling (Fig. 1B). Identical results for IFN signaling inhibition by Hendra virus V protein were obtained with 293T cells (data not shown). This result indicates that Hendra virus V protein is an efficient inhibitor of IFN signal transduction, similar to the Nipah virus V protein.
FIG. 1.
Hendra virus V protein inhibits IFN signaling. (A) Amino acid sequence alignment of Nipah and Hendra virus V proteins. (B) Human fibrosarcoma 2fTGH cells were transfected with expression vectors for HA-tagged Nipah virus V (NiV), Hendra virus V (HeV), or HA epitope tag expression vector with no cDNA insert. Cells were also cotransfected with either the IFN-α-inducible ISRE-luciferase reporter gene or an IFN-γ-inducible GAS-luciferase reporter gene. Cells were treated with (+) or without (−) 1,000 U of IFN-α per ml or 5 ng of IFN-γ per ml 12 h prior to lysis and luciferase assays. All bars represent average values from triplicate samples normalized to those obtained with cotransfected renilla luciferase ± standard deviations and are expressed as percentages of values for IFN-stimulated controls.
FIG. 2.
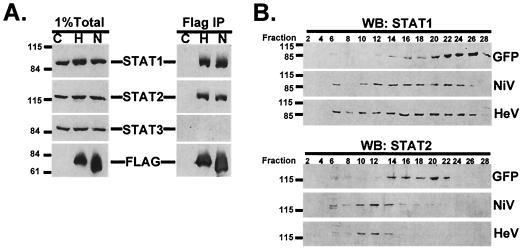
Hendra virus V forms STAT1- and STAT2-containing complexes. (A, left panel) 293T cells were transfected with either FLAG epitope expression vector with no cDNA insert (lane C), FLAG-tagged Hendra virus V expression vector (lane H), or FLAG-tagged Nipah virus V expression vector (lane N). Whole-cell extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and processed for immunoblotting with antiserum to STAT1, STAT2, STAT3 (all from Santa Cruz Biotechnology), or FLAG (Sigma). Positions of prestained molecular weight standards and STAT1, STAT2, STAT3, and FLAG Henipavirus V proteins are indicated. (A, right panel) Extracts from the left panel were immunoprecipitated (IP) by using an M2 FLAG affinity gel (Sigma) and analyzed by immunoblotting. (B) Cell extracts of 293T cells transfected with expression vectors for FLAG-tagged Nipah virus V (NiV), Hendra virus V (HeV), or green fluorescent protein (GFP) were separated by chromatography on a Superdex-200 column (Pharmacia). Positions of STAT1 and STAT2 were determined by Western blotting (WB) every other fraction. The column was calibrated with high- and low-molecular-weight calibration kits (Pharmacia).
The inhibition of IFN signal transduction by Nipah virus involves interactions between the V protein and the IFN-responsive STATs, STAT1 and STAT2 (12). To determine if the Hendra virus V protein shares the ability to bind STATs, 293T cells were transfected with expression vectors for FLAG-tagged Hendra virus V or Nipah virus V or with vector containing no cDNA insert. Cell extracts were immunoprecipitated with FLAG epitope-specific antibody, and immune complexes were processed for immunoblotting with antiserum for STAT1, STAT2, or STAT3. No STAT proteins were detected in the precipitated material from cells transfected with empty vector, but both STAT1 and STAT2 were found in the Hendra virus V immune complexes, similar to what was found in the precipitate from cells transfected with the control Nipah virus V (Fig. 2A). Neither the Hendra virus V protein nor the Nipah virus V protein could coprecipitate STAT3, despite its amino acid sequence similarity to STAT1. These data indicate that the Hendra virus V protein is similar to the Nipah virus V protein in its ability to associate specifically with STAT1 and STAT2, but not STAT3. Gel filtration of efficiently transfected 293T cell extracts expressing FLAG-tagged green fluorescent protein, Nipah virus V protein, or Hendra V protein confirms the ability of Hendra virus V to convert STAT1 and STAT2 from their latent cellular reservoirs into high-molecular-weight fractions that eluted between the calibration standards thyroglobulin (660 kDa) and catalase (232 kDa), suggesting that complexes of approximately 500 kDa are induced by Hendra virus V protein (Fig. 2B).
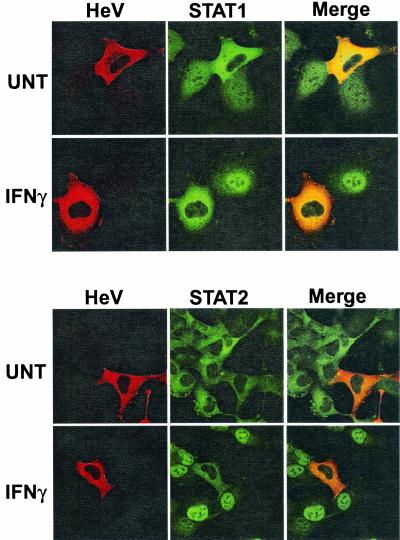
To assess the subcellular localization of Hendra virus V protein and determine its effect on STAT distribution, 2fTGH cells transfected with expression plasmid encoding HA-tagged Hendra virus V protein were either fixed directly or stimulated with IFN for 30 min prior to fixation and permeabilization. Cells were then stained sequentially, first with antiserum to the HA epitope and then with antiserum for either STAT1 or STAT2. In unstimulated cells with no Hendra virus V expression, STAT1 was detected in both the cytoplasm and the nucleus while STAT2 accumulated exclusively in the cytoplasm (Fig. 3). The Hendra virus V protein was detected only in the cytoplasm, a pattern similar to that reported for Nipah virus V protein (12). Expression of the Hendra virus V protein dramatically altered the basal subcellular localization of STAT1, resulting in a predominantly cytoplasmic staining pattern. Hendra virus V protein had no detectable effect on the distribution of STAT2 in unstimulated cells. In response to IFN stimulation, both STAT1 and STAT2 efficiently translocate to the nuclei of nontransfected cells. However, expression of the Hendra virus V protein prevented the IFN-dependent nuclear redistribution of both STAT1 and STAT2. Thus, Hendra virus V protein, like the Nipah virus V protein, antagonizes IFN signaling by preventing IFN-inducible STAT nuclear localization.
FIG. 3.
Hendra virus V protein blocks nuclear import of STAT1 and STAT2. 2fTGH cells were transfected with HA-tagged Hendra virus V expression plasmid (HeV) and were either unstimulated (UNT) or treated for 30 min with 5 ng of IFN-γ per ml (IFNγ) to visualize the nuc1ear accumulation of STAT1 or with 1,000 U of IFN-α per ml (IFNα) to visualize the nuclear accumulation of STAT2. Cells were fixed, permeabilized, and stained sequentially for HA and then STAT1 or STAT2.
In conclusion, the data presented here indicate that expression of the Hendra virus V protein causes IFN signaling inhibition for both the IFN-α and the IFN-γ pathways. The Hendra virus V protein accumulates in the cytoplasm of cells and forms molecular interactions with STAT proteins, STAT1 and STAT2, preventing their IFN-induced nuclear accumulation. All of these activities are shared with the V protein of Nipah virus, suggesting little divergence between these two viruses with respect to their innate immune evasion activities. While these findings suggest that STAT inhibition in the Henipavirus genus is uniform, a great diversity of mechanisms has been revealed for V protein-mediated IFN signaling evasion among the Paramyxovirus family. For example, measles virus, a member of the Morbillivirus genus, interferes with IFN-induced STAT nuclear transport but does not alter the basal STAT1 distribution pattern like Henipavirus V proteins (8), while the Rubulavirus species SV5, HPIV2, and mumps virus target STATs for polyubiquitylation and proteosome-dependent degradation (4, 9, 14, 15, 20, 21). Nonetheless, individual rubulaviruses differ with respect to their targeting abilities, with SV5 targeting STAT1, HPIV2 targeting STAT2, and mumps virus targeting STAT1 and STAT3. It must be noted, however, that with most, if not all, of these viruses, additional immune evasion and replication phenotypes have been revealed either by V protein expression or by observing the phenotypes of recombinant V-deficient viruses. Hence, in the lack of specific data for Nipah virus- and Hendra virus-infected cells, it remains possible that differences exist between these species. Irrespective of this possibility, the data presented here clearly demonstrate that STAT-directed IFN signaling inhibition is a property of both Nipah virus and Hendra virus V proteins.
Acknowledgments
We are grateful to Pablo De Iaonnes and the members of the Cortes laboratory for use of the FPLC Superdex 200 column and to members of the Horvath laboratory for helpful discussions and comments on the manuscript.
This study was supported by research grants to C.M.H. from the American Cancer Society (research scholar grant 103079) and the NIH (AI-48722 and AI-50707). J.J.R is a trainee of the integrated training program in pharmacological sciences (GM-62754).
REFERENCES
- 1.Borden, K. L. 2000. RING domains: master builders of molecular scaffolds? J. Mol. Biol. 295:1103-1112. [DOI] [PubMed] [Google Scholar]
- 2.Capili, A. D., D. C. Schultz, I. F. Rauscher, and K. L. Borden. 2001. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 20:165-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chua, K. B., W. J. Bellini, P. A. Rota, B. H. Harcourt, A. Tamin, S. K. Lam, T. G. Ksiazek, P. E. Rollin, S. R. Zaki, W. Shieh, C. S. Goldsmith, D. J. Gubler, J. T. Roehrig, B. Eaton, A. R. Gould, J. Olson, H. Field, P. Daniels, A. E. Ling, C. J. Peters, L. J. Anderson, and B. W. Mahy. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432-1435. [DOI] [PubMed] [Google Scholar]
- 4.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Field, H., P. Young, J. M. Yob, J. Mills, L. Hall, and J. Mackenzie. 2001. The natural history of Hendra and Nipah viruses. Microbes Infect. 3:307-314. [DOI] [PubMed] [Google Scholar]
- 6.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In B. N. Fields, D. M. Knipe, P. M. Howley, and D. E. Griffin (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 7.Murray, K., P. Selleck, P. Hooper, A. Hyatt, A. Gould, L. Gleeson, H. Westbury, L. Hiley, L. Selvey, B. Rodwell, et al. 1995. A morbillivirus that caused fatal disease in horses and humans. Science 268:94-97. [DOI] [PubMed] [Google Scholar]
- 8.Palosaari, H., J. P. Parisien, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2003. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 77:7635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parisien, J.-P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 10.Park, M. S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paterson, R. G., G. P. Leser, M. A. Shaughnessy, and R. A. Lamb. 1995. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology 208:121-131. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez, J. J., J. P. Parisien, and C. M. Horvath. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 76:11476-11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas, S. M., R. A. Lamb, and R. G. Paterson. 1988. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell 54:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulane, C. M., and C. M. Horvath. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304:160-166. [DOI] [PubMed] [Google Scholar]
- 15.Ulane, C. M., J. J. Rodriguez, J.-P. Parisien, and C. M. Horvath. 2003. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J. Virol. 77:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, L., and B. T. Eaton. 2002. Henipavirus (Paramyxoviridae, Paramyxovirinae), p. 641-644. In C. A. Tidona, G. Darai, and C. Büchen-Osmond (ed.), The Springer index of viruses. Springer, Berlin, Germany.
- 17.Wang, L., B. H. Harcourt, M. Yu, A. Tamin, P. A. Rota, W. J. Bellini, and B. T. Eaton. 2001. Molecular biology of Hendra and Nipah viruses. Microbes Infect. 3:279-287. [DOI] [PubMed] [Google Scholar]
- 18.Wang, L. F., W. P. Michalski, M. Yu, L. I. Pritchard, G. Crameri, B. Shiell, and B. T. Eaton. 1998. A novel P/V/C gene in a new member of the Paramyxoviridae family, which causes lethal infection in humans, horses, and other animals. J. Virol. 72:1482-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yob, J. M., H. Field, A. M. Rashdi, C. Morrissy, B. van der Heide, P. Rota, A. bin Adzhar, J. White, P. Daniels, A. Jamaluddin, and T. Ksiazek. 2001. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 7:439-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokosawa, N., S. Yokota, T. Kubota, and N. Fujii. 2002. C-terminal region of STAT-1alpha is not necessary for its ubiquitination and degradation caused by mumps virus V protein. J. Virol. 76:12683-12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383-390. [DOI] [PubMed] [Google Scholar]