Abstract
In cultured cells, adeno-associated virus (AAV) replication requires coinfection with a helper virus, either adenovirus or herpesvirus. In the absence of helper virus coinfection AAV can integrate its genome site specifically into the AAVS1 region of chromosome 19. Upon subsequent infection with a helper virus, the AAV genome is released from chromosome 19 by a process termed rescue, and productive replication ensues. The AAV genome cloned into a plasmid vector can also serve to initiate productive AAV replication. When such constructs are transfected into cells and those cells are simultaneously or subsequently infected with a helper virus, the AAV genome is released from the plasmid. This process is thought to serve as a model for rescue from the human genomic site. In this report we present a model for rescue of AAV genomes by replication. A hallmark of this model is the production of a partially single-stranded and partially double-stranded molecule. We show that the AAV2 Rep 68 protein, together with the UL30/UL42 herpes simplex virus type 1 DNA polymerase and the UL29 single-strand DNA binding protein ICP8, is sufficient to efficiently and precisely rescue AAV from a plasmid in a way that is dependent on the AAV inverted terminal repeat sequence.
Adeno-associated virus (AAV), a member of the parvovirus family, contains a single-stranded genome of approximately 4,680 bases. The genome of AAV contains two open reading frames; one coding for the replication (Rep) proteins and the other for the structural proteins. The four Rep proteins, designated 78, 68, 52, and 40 according to their migration on sodium dodecyl sulfate (SDS)-polyacrylamide gels, are formed by utilizing different start sites and splicing patterns. Rep 68 and Rep 78 possess origin binding, DNA helicase, ATPase, and strand- as well as site-specific nicking activity. Either Rep 68 or Rep 78 is absolutely required for the replication of AAV DNA. Rep is the sole AAV protein involved in AAV DNA replication, necessitating that most replication functions are provided by non-AAV proteins. The ends of the AAV genome contain identical origins of DNA replication. Each origin consists of an inverted terminal repeat (ITR) capable of intramolecular base pairing to form a hairpin loop serving as a primer for DNA synthesis at its 3′ end. The hairpin primers can be recreated indefinitely by the process designated terminal resolution, which consists of site-specific nicking at the terminal resolution site (TRS) followed by extension from the nick to the end of the genome. The duplex ITR can then fold over and serve as a primer for full-length replication (reviewed in reference 1). The AAV Rep protein is required for each of these functions (3, 8, 16, 25).
Productive AAV replication in vivo, however, is only observed when AAV infects a cell that is simultaneously infected by a helper virus (1). Both adenovirus and herpesviruses may serve as helper viruses. In the absence of coinfection by a helper virus, AAV enters a latent state by integration of its genome into a specific region of chromosome 19 designated the AAVS1 locus. If the latently infected cell is subsequently infected by a helper virus, a copy of the latent AAV genome is released from its chromosomal context and proceeds to replicate (7a). This escape from the chromosome is termed rescue. It has also been shown that a productive infection can be established in cells infected with a helper virus if a cloned copy of the AAV genome is provided in a circular plasmid (11). The release of the AAV genome from the plasmid is thought to be analogous to the release occurring from chromosome 19 in the latently infected cell and has served as an experimental model for rescue. While the ITR is the only cis requirement for rescue, it has been noted that in the absence of the left ITR, rescue from a plasmid vector can occur at the p5 promoter region (18).
Two sorts of mechanism have been proposed to explain rescue: rescue may be carried out by cellular nucleases (7, 11, 13) or it may be coupled to DNA replication (13, 23). It has been suggested that a cellular nuclease of unknown function may catalyze rescue of the AAV genome from a circular plasmid (7). It has been observed that rescue of the AAV genome from a plasmid may be carried out by a Holliday structure-resolving activity in uninfected cellular extracts (19) and in cells (24). It has also been observed that rescue of the AAV genome in a HeLa cell extract is more efficient if the Rep 68 protein is added. Since Rep protein supported replication of AAV DNA in the same extract, this observation lent credibility to the notion that rescue could occur by a replicative mechanism (23). (It has also been suggested that Rep might have the ability to resolve Holliday structures [17].)
The testing of a replication model of rescue is difficult. Full-length AAV genomes will be the product of rescue, independent of which mechanism is used. These genomes will be amplified by DNA replication; one rescue event may, in fact, give rise to thousands of replicating AAV genomes. Additionally, in cells and cellular extracts the presence of recombinases and nucleases makes it difficult to interpret the origin of products and reaction intermediates. However, the development of a biochemically well-defined system for initiation of replication from the AAV origin (21) makes it possible to develop an assay that allows us to measure the number of rescue events and to control the subsequent amplification of the products. In addition, this assay allows the replication-based rescue mechanism to be examined in the complete absence of cellular nucleases. In this report we employ a minimal herpes simplex virus (HSV) initiation system composed of AAV Rep 68, the HSV type 1 (HSV-1) DNA polymerase UL30/UL42, and the HSV-1 single-strand DNA binding protein ICP8 (21) to test for the appearance of DNA molecules which would specifically indicate that rescue dependent on DNA replication has taken place. We show that the initiation of replication can efficiently rescue AAV from a plasmid vector.
MATERIALS AND METHODS
Plasmids and substrates.
Plasmid pL-2 was constructed by cloning the small BamHI fragment of pAV2 (9) (i.e., from outside the left ITR up to AAV base 1045) into BamHI-digested pBluescript KS. Plasmid pL0 was constructed by inserting a paired set of annealed synthetic oligonucleotides (5′GATCTGAGTCGTCTCTTGGCCAGTCCCTCTCTG3′ and 5′CGCGCAGAGAGGGAGTGGCCAAGAGACGACTCA3′) into BglII-digested, partially BssH2-digested pL-2 (partial digestion retained all but the outermost section of the ITR). Plasmid pR-8 was constructed by inserting the EcoRI-SalI fragment of pAV2 (which goes from AAV base 1985 through the right ITR to pBR base 651) into EcoRI- and SalI-digested pBluescript KS.
pR0 was constructed by inserting the BglII-MscI fragment of pL0 (partially digested to contain the whole ITR) into BglII-Msc1-digested pR-8.
pL-5 was constructed from pL0 by BsmB1 digestion followed by mung bean nuclease treatment and religation. pL-22 was constructed from pL0 by BsmB1 and BssH2 digestion (partially digested to retain the major component of the ITR) followed by treatment with the polymerase I Klenow fragment and religation.
Proteins.
HisRep 68 contains six histidine residues fused to the amino-terminal end of the full-length Rep 68 protein (15). It was produced in Escherichia coli from a pET 16b vector (New England Biolabs) and purified according to the manufacturer's instructions. The proteins encoded by the herpes simplex virus UL29, UL30, and UL42 genes were produced from stocks of recombinant Autographa californica nuclear polyhedrosis virus and purified as described previously (4, 6). The purity of each protein was ≥ 95%, as judged by SDS-polyacrylamide gel electrophoresis followed by Coomassie blue staining.
DNA replication assay.
Replication rescue assays were performed essentially as described previously (21). The reaction mixture (15 μl) contained 2.7% glycerol, 40 mM HEPES (pH 7.7), 40 mM creatine phosphate (pH 7.7), 7 mM MgCl2, 4 mM ATP, 200 μM (each) CTP, GTP, and UTP, 100 μM (each) dATP, dGTP, and dTTP, 10 μM dCTP, 2 mM dithiothreitol (DTT), and 6 mM potassium glutamate. It also contained 2.0 μg of creatine phosphokinase and 10.0 μg of bovine serum albumin. In addition, 5 μCi of [α-32P]dCTP (3,000 Ci/μmol; Amersham) was added to selected reactions.
The reaction also contained 6,000 fmol of the UL29 protein, 750 fmol of the UL30/UL42 complex, and 700 fmol of HisRep 68. The reaction was incubated at 37°C for 4 h.
The Rep-only separation assay was performed as described above except that the HSV proteins were omitted and replaced by their storage buffers as follows: 10% glycerol, 20 mM HEPES, NaOH (to pH 7.6), 0.5 mM EDTA (pH 8.0), 2 mM DTT, and 300 mM NaCl for the UL29 protein and 10% glycerol, 20 mM HEPES, NaOH (to pH 7.6), 0.5 mM EDTA (pH 8.0), 2 mM DTT, and 200 mM NaCl for the UL30/UL42 complex. The reactions were incubated at 37°C for 4 h.
Both assays were terminated by the addition of 50 μl of digestion buffer (20 mM HEPES [pH 7.5], 10 mM KCl, 10 mM EDTA, 1.0% SDS, 50 mM NaCl). Products were passed through a Sephadex 50 spin column and then digested with proteinase K at 1 mg/ml for 2 h at 50°C. Aliquots of the products were separated by electrophoresis on 0.8% agarose gels with Tris-borate-EDTA buffer. Gels were dried and either stained with SYBR green or processed for autoradiography. The SYBR green data were collected by Phosphorimager (Molecular Dynamics) scanning of the dried gels with ImageQuant 1.1 software.
Two-dimensional gel electrophoresis.
For two-dimensional gel electrophoresis, nondenaturing gel electrophoresis was performed in a 0.8% agarose gel with Tris-borate-EDTA buffer; for the second, denaturing dimension, lanes were excised and transferred to a 0.8% gel containing 1 mM EDTA and 40 mm NaOH. The gels were equilibrated for 90 min in the denaturing buffer prior to electrophoresis.
Hybridization of gels.
Dried gels were rehydrated in water and incubated briefly in a prehybridization solution made up of 6× SSCP (0.72 M NaCl, 0.09 M Na citrate, 0.08 M KH2PO4, and 0.012 M EDTA [pH 6.8]), 0.3% SDS, and 0.2 mg of salmon sperm DNA per ml. A nick-translated probe of 32P-radiolabeled pLO was added for a 16-h hybridization. Hybridized gels were washed briefly in 6× SSCP with 0.5% SDS. Gels were redried and exposed to film.
RESULTS
A model for rescue by replication.
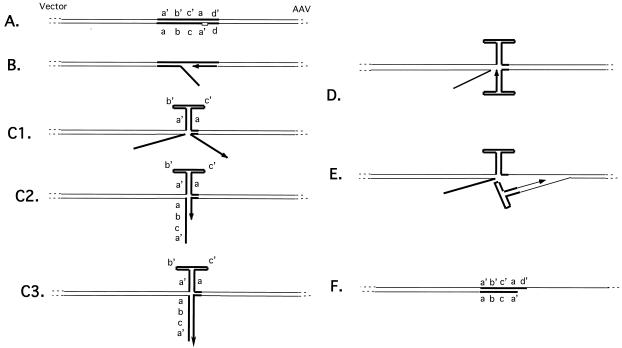
Members of our laboratory previously proposed a model for the rescue of a cloned copy of the AAV genome from a plasmid and by analogy the rescue of AAV from its site of integration into the human genome (23). According to this model, a copy of the AAV genome is replicated from the plasmid by a fold-back replication mechanism. For this model the substrate may contain only one TRS and Rep may nick the substrate only once at the TRS. We propose that replication initiates at the nick introduced by Rep at the TRS and that as replication passes through the body of the ITR, the partially single-stranded template strand will fold into a hairpin configuration (Fig. 1). This folding of the template ITR may be supported by Rep binding to the tip of the hairpin loop of the ITR (10). At this time, the replicating strand will necessarily be separated from its template (Fig. 1C, panel 1). The single-stranded newly synthesized strand can now anneal with a complementary sequence and continue extension in either of two ways. The replicating strand terminating in the a′ region can base pair with the originally displaced strand (Fig. 1C, panel 2) and resume replication until DNA synthesis reaches the end of the displaced strand (Fig. 1C, panel 3). At this point Rep may unwind the short piece of duplex DNA, allowing it to fold upon itself (Fig. 1D). In this way a standard hairpin primer is created and the replicating strand can resume replication into the AAV genome (Fig. 1E). Alternatively, if displacement occurred while replication was in the a region, i.e., just before the ITR has been completely copied and before replication has extended into non-AAV sequences, then the replicating strand can immediately fold upon itself to create a hairpin primer followed similarly by replication of AAV sequences (Fig. 1E). (Displacement occurring in either the b or c palindrome would not permit formation of a complete hairpin primer.) The result of either mechanism is rescued duplex AAV with one strand from the plasmid substrate and one newly synthesized strand. A hallmark of this model is that a hybrid molecule, i.e., double stranded in its vector part but single stranded for most of its AAV sequences, will be left behind (Fig. 1F). The point of transition between double- and single-stranded components should be at the TRS.
FIG. 1.
Diagram of model showing rescue by fold-back replication. (A) The substrate is a linearized AAV genome-carrying plasmid such that wild-type AAV sequences are juxtaposed to vector sequences immediately adjacent to the AAV ITR. Rep nicks at the TRS (small box). A replication complex is assembled and replication commences through the ITR towards the vector (B). The template ITR sequences fold into a hairpin conformation, displacing the replicating strand (C1). The replicating strand may base pair with the displaced strand (C2) and then replicate to its end (C3), at which point helicase activity can dissociate this duplex, allowing the folding up of the replicating strand and replication back into AAV sequences (D and E). Alternatively, if replication has progressed sufficiently, i.e., passed through the b and c palindromes, the replicating strand can fold upon itself immediately (D) and replicate back into AAV sequences. Also shown is the structure left behind after either mechanism of fold-back replication (F). ITR sequences are designated by thick lines.
An assay for rescue by replication.
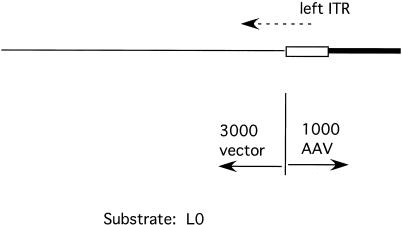
To limit the replication of the AAV genome subsequent to rescue to one cycle, we employed a substrate containing only one ITR. Figure 2 shows a schematic presentation of the basic substrates used for the assay. The plasmid pL-2 contains the left end of AAV to base 1045 (from pAV2) cloned into the polylinker of a pBluescript plasmid. In pL0 the first two bases of AAV, which are missing from pAV2, have been replaced to create a complete ITR. The plasmids were linearized to create substrates L-2 and L0, with 3,000 bases of vector DNA sequences to the outside of the ITR and the first 1,000 bases of the AAV genome to the inside of the ITR (Fig. 2).
FIG. 2.
Diagram of substrate L0 (L-2 is essentially equivalent). pL0 linearized by EcoRI digestion to form a 4,000-bp molecule is shown. The vector and AAV components are indicated. Box, AAV ITR; thick line, non-ITR AAV sequences; dashed arrow, origin and direction of replication initiation.
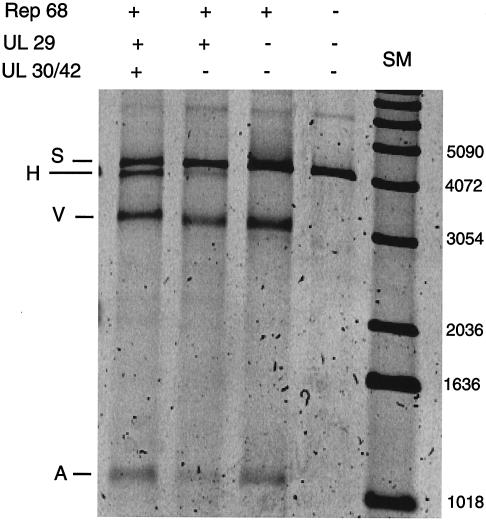
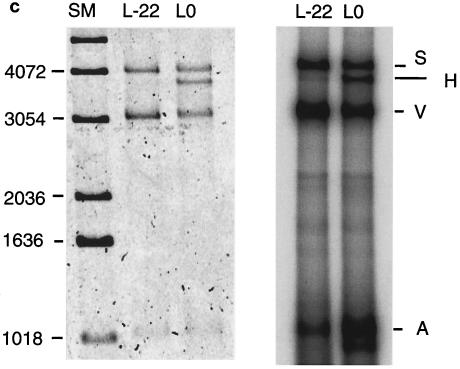
Our laboratory has previously shown that a minimal leading-strand HSV replication complex is able to initiate synthesis of DNA in a Rep-dependent manner at AAV origins of DNA replication (21). The assay used employs the AAV Rep protein, the HSV-1 DNA polymerase and its processivity factor, the UL30/UL42 complex, and the single-strand DNA binding protein ICP8 encoded by the HSV-1 UL29 gene. The substrate L0, referred to as S, was incubated with purified components of the HSV-1-AAV replication system. Figure 3 shows a SYBR green-stained gel on which products were separated. Incubation of the substrate with Rep 68 alone resulted in release of a vector, V, and an AAV component, A (Fig. 3, lane 3). The cleavage in this reaction occurs at various points in the ITR and is dependent on ATP (data not shown). The structure of the products is inconsistent with cleavage by classic Holliday structure-resolving activity, since unlike those products (19), the products of this assay are heterogeneous with respect to breakpoint. In addition, the vector products in this assay are composed of species both with and without hairpin loops (data not shown). We refer to this phenomenon as Rep-only separation. When the HSV single-stranded protein was added in addition to Rep 68, we sometimes observed a faint band migrating slightly faster than the substrate (Fig. 3, lane 2). The molecular nature of this band and its likely derivation are considered in Discussion. When the complete minimal replication complex was used in the assay, the band referred to as H was readily detected (Fig. 3, lane 1). Further experiments have demonstrated that the H product migrates faster during electrophoresis at low voltage gradients than does a fully double-stranded reference species. Furthermore, it can be stained by SYBR green, but it stains more intensely with SYBR gold than SYBR green staining of known double-stranded reference DNA (results not shown). (SYBR green stains principally double-stranded DNA, while SYBR gold stains both double- and single-stranded molecules.) Both of these observations suggest that the H product might contain double-stranded and single-stranded parts. The properties of the H product are discussed further below.
FIG. 3.
Incubation of linearized substrate with proteins from the HSV-AAV replication complex. Products were separated on a 0.8% agarose gel and visualized by SYBR green staining. Shown are the addition of Rep 68 and the products of the HSV UL29, UL 30, and UL42 genes. S, starting substrate; A, AAV component of the substrate; V, vector component of the substrate; H, putative hybrid double- and single-stranded species. Shown on the right are DNA size markers (SM).
Rescue of the AAV genome releases a product containing single-stranded and double-stranded DNA.
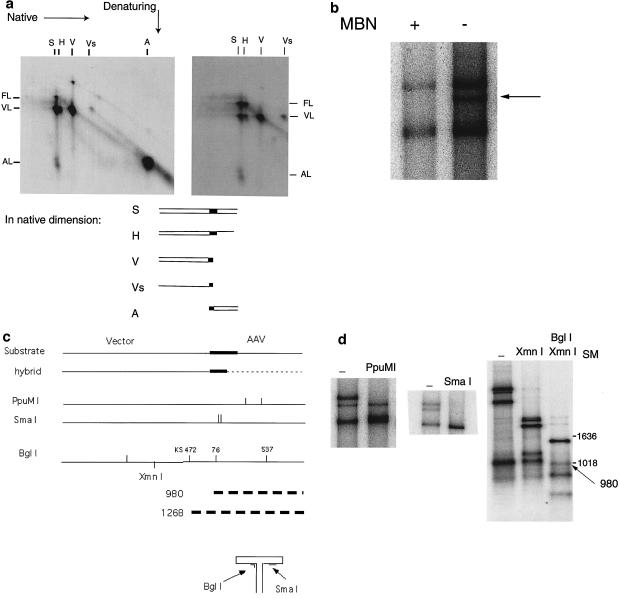
To confirm that the structure of the species designated H is that of the hybrid molecule predicted by the rescue-by-replication model, several analyses were performed. Figure 4a shows the products of an assay employing the complete four-protein replication complex separated by two-dimensional electrophoresis. Separation in the first dimension was done under neutral conditions, whereas separation in the second dimension was done under denaturing conditions. The gels were analyzed by autoradiography. Incorporation of radioactive nucleotides is shown (Fig. 4a, left panel), indicating which components of each species contained newly synthesized DNA. A hybridization of the same gel with a radioactive probe made from pL0 to detect all DNA species is also represented (Fig. 4a, right panel). Shown above the gels are the designations of each species as seen in a one-dimensional electrophoretic separation; at the sides are shown the components of each species. The substrate species shows extensive nicking by Rep at the TRS, resulting in both AAV- and vector-length components. The vector species contains molecules both with and without hairpin loops. As mentioned above, the Rep-only separation mechanism produces some vector molecules which are hairpinned. As predicted, the AAV species, which is partly the product of the fold-back rescue-by-replication mechanism, shows substantial incorporation. This product migrates in the second dimension at 1× length rather than at 2× length, indicating that it is not a hairpinned loop. This implies that Rep has nicked the hairpin loop products of rescue by replication. Also as predicted, the only labeled component of the putative hybrid molecule is of vector length. (Those replications which have passed through the ITR without rescue displace a single-stranded vector component of the substrate and replace it with a newly synthesized and therefore labeled vector sequence. The substrate molecule is recreated and capable of a second initiation event.) There is no labeled full-length or AAV-length component of the H species. Hybridization of the same gel with a probe made from the entire substrate (Fig. 4a, right panel) illustrates that the putative hybrid molecule, however, does contain a full-length component but contains no AAV-length component. The full-length strand is expected to show limited incorporation because it is predicted to be the strand that does not contain the TRS nicking site.
FIG. 4.
Analysis of putative hybrid molecule. (a) Shown in the left panel is an autoradiogram of an assay with L0 substrate and all four proteins. The horizontal dimension shows separation in a 0.8% agarose gel under native conditions. The vertical dimension shows subsequent separation under denaturing conditions. On the horizontal axis, the species seen on a native gel are indicated. S, substrate; H, hybrid; V, vector; Vs, single-stranded vector; A, AAV. On the vertical axis, the three denatured components are indicated. FL, full-length substrate; VL, vector length; AL, AAV length. The right panel is an autoradiogram made by hybridizing the same gel with a probe synthesized from L0. (b) Autoradiogram showing mung bean nuclease (MBN) digestion of the products of an assay separated on a 0.8% agarose gel. (c) Schematic showing substrate and hybrid molecules, with the ITR designated by a thick line and the putative single-stranded region of the hybrid designated by a dashed line. Below is a diagram showing the locations of the PpuMI, SmaI, and BglI restriction sites in the substrate molecule and the hybrid molecule. Also diagrammed is a hairpin loop ITR showing the recognition sites of BglI and SmaI in the ITR. The thick dashed lines indicate the derivation of the potential 980- and 1,268-bp products from BglI digestion of the hybrid species. KS472 indicates the BglI site at nucleotide 472 in the vector backbone, pBluescript. (d) Autoradiogram showing PpuMI, SmaI, and BglI restriction digestion products separated on a 0.8% agarose gel. In this gel system the double- and single-stranded species between 800 and 1,500 bases migrate almost indistinguishably. The arrow indicates the BglI-digested hybrid species at 980 bp. SM, size marker.
These observations support the following argument. The H species, the putative hybrid molecule, migrates differently than the starting substrate. Therefore, it cannot be composed of two full-length strands. The hybridization experiment shows that the H species contains a full-length DNA strand and a DNA strand of vector length. The absence of an AAV-length component means that the vector-length species cannot be derived from a full-length strand nicked at the ITR. Therefore, the H species must contain one full-length strand and one strand of vector length.
A question raised with respect to rescue by replication is the relative likelihood of fold-back replication at the ITR versus the replication complex passing through the ITR without rescue. When DNA replication passes through the ITR without rescue, a displaced single-stranded vector-length species, Vs, is produced. Each fold-back replication, on the other hand, will produce one hybrid molecule including a vector-length component (VL component of the H species). Our results from the hybridization experiment (Fig. 4a, right panel) show that the amounts of these two products are approximately equivalent, suggesting that the two reactions occur with equal probability under our experimental conditions. This implies that the rescued AAV (A in Fig. 3, for example) is about equally the product of rescue by replication and of the Rep-only separation mechanism. Since the amounts of the A component in lanes 1 and 3 (Fig. 3) are similar, a further implication is that as rescue by replication has increased, Rep-only separation has decreased.
To further analyze the structure of the H component, we used mung bean nuclease to digest single- but not double-stranded DNA. As expected, the hybrid molecule disappears but the double-stranded starting substrate and vector species do not (Fig. 4b). (We reproducibly also lost some of the substrate and vector material in our mung bean nuclease digestion reactions, but after overexposure [not shown] hybrid molecules remained undetectable in the digested lane.) This suggests that mung bean nuclease digestion removes the single-stranded AAV component, producing a DNA species migrating as the vector component. We also used restriction enzyme analyses to locate the transition between double-stranded and single-stranded DNA. Our model for rescue by replication suggests that it occurs at the TRS. Enzymes with restriction sites in the vector, i.e., double-stranded, portion of the hybrid digest both the substrate and the hybrid species (data not shown). On the other hand, enzymes, e.g., PpuMI, whose restriction sites are located in the AAV portion of the hybrid digest, as expected, the substrate but not the hybrid species (Fig. 4c and d). The model predicts that the ITR, except for the D region, should be in the double-stranded portion of the hybrid molecule. SmaI, whose recognition site is in the ITR, does digest the hybrid species (Fig. 4d). However, an SmaI site might be created when the single-stranded form of the ITR is in the hairpin conformation. In this case, the SmaI site will be in a short double-stranded region (Fig. 4c). Control experiments did not reveal SmaI cleavage of single-stranded substrates (results not shown). Thus, SmaI digestion suggests that the ITR in the H component exists as fully double-stranded DNA rather than as one single-stranded hairpin loop.
For an additional demonstration that the ITR is indeed double stranded in the hybrid molecules, we employed the restriction enzyme BglI. This enzyme has several sites in the substrate, including one site in the ITR (Fig. 4c). The BglI recognition site in the ITR is, however, not encompassed by one linear segment in the hairpin conformation of the ITR (Fig. 4c). Therefore, an ITR hairpin loop cannot be cleaved by BglI. BglI will digest the double-stranded substrate species at position 537 in the AAV part of L0 but leave the hybrid molecule undigested. If this hybrid molecule can be digested by BglI at position 76 in the ITR, a 980-bp fragment will be produced, but if the enzyme fails to digest the molecule at position 76 because the ITR is single stranded, a fragment of 1,268 bp will be formed due to digestion at position 472 in the pBluescript vector. As seen in Fig. 4d, a fragment of approximately 980 bp is present while there is no fragment of 1,268 bp. XmnI digestion was included to eliminate another BglI fragment in this size range. This result also demonstrates that in the hybrid molecule the ITR segment is double stranded rather than single stranded.
Generation of the hybrid molecule H requires DNA synthesis.
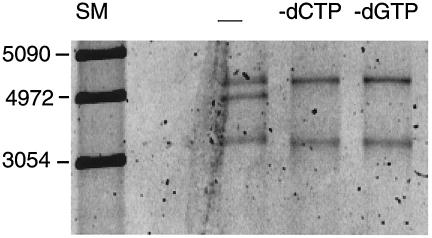
The model for rescue by replication states that the H component, the hybrid molecule, is not comprised of DNA synthesized during the rescue reaction but that its formation is due to DNA synthesis. To test this prediction, we performed three parallel assays. In each case, the L0 substrate and all four proteins, Rep 68, UL29, and the UL30/UL42 complex, were present. For one assay, a complete set of four deoxynucleoside triphosphates was included, and for two other assays, one deoxynucleoside triphosphate, either dCTP or dGTP, was omitted. As shown in Fig. 5, the hybrid molecule H was produced only when all four deoxynucleoside triphosphates were supplied. (It should be pointed out that while the vector component of the hybrid molecule does not become labeled during the rescue-by-replication reaction, it can become labeled prior to this reaction. If replication initiates but passes through the ITR without rescue, an identical substrate molecule will be recreated except that the vector sequences on the nickable strand will now be labeled.)
FIG. 5.
SYBR green-stained 0.8% agarose gel of reaction products demonstrating that generation of the hybrid molecule requires DNA synthesis. Lane SM, size marker; lane —, standard assay with all four deoxynucleoside triphosphates; lane −dCTP, standard assay with three deoxynucleoside triphosphates, with dCTP omitted; lane −dGTP, standard assay with three deoxynucleoside triphosphates, with dGTP omitted.
Rescue by replication yields AAV with an intact ITR.
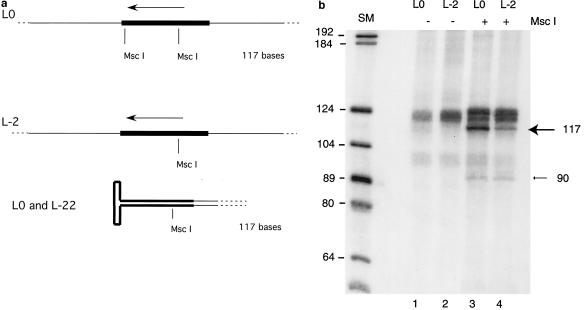
An important question is whether the AAV product of the rescue mechanism has an intact ITR. Our model predicts that the rescued AAV will contain one complete copy of the ITR in a hairpin conformation. The model also predicts that the part of the hairpin loop synthesized after separation from the template will be synthesized using the inner region of the ITR as a template (Fig. 1). To test these predictions, we compared two assays performed in parallel. In the first, the substrate was the standard L0. When the products of a rescue assay with L0 are digested with MscI, four products should result; fragments of approximately 1,000 and 3,000 bp, representing the distance between the MscI sites in the ITR, and the ends of the substrate molecule should be present. In addition, there should be a fragment of 117 bp corresponding to the distance separating the two restriction sites in the ITR (Fig. 6a). As shown in Fig. 6a, the hairpin end of the rescued AAV species will also be removed by MscI digestion. If the rescued species contains a complete hairpin loop, it will consist of one strand of DNA of 117 bases in a hairpin conformation and will have the same electrophoretic mobility under denaturing conditions as the fragment produced by cleavage of the unmodified substrate L0. The products of a replication rescue experiment with L0 as substrate were digested by MscI and analyzed by electrophoresis on a formamide-urea gel. We observed the expected product of 117 nucleotides (nt) (Fig. 6b). (Our laboratory showed previously that an ITR structure is sufficiently denatured on a formamide-urea gel to electrophorese at approximately the rate corresponding to its length in bases [19]). In the second experiment L-2 was used as substrate. L-2 lacks the outer MscI site in the ITR (Fig. 6a). Therefore, there should be no small MscI digestion product without first creating a second MscI restriction site. The fragments from an MscI digestion of a rescue assay using the L-2 substrate are shown in Fig. 6b. The presence of a small MscI digestion product indicates that fold-over occurred and that the region of the ITR containing the inner MscI site served as a template for synthesis of a second MscI site. Together, the presence of only one MscI band in the L0 experiment and the fact that the band in the L-2 assay is of equivalent length suggest that the fold-over forms an MscI ITR fragment of the same length as the MscI fragment in the original substrate, i.e., 117 bases. This is exactly as expected if a perfect ITR is created during the rescue-by-replication mechanism.
FIG. 6.
Rescued product is of the expected structure. (a) Diagram showing MscI restriction sites in L0 and L-2 as well as in the rescued hairpin loop. The arrow indicates the origin and direction of DNA synthesis. The thick line designates the ITR. (b) Autoradiogram of products separated on a formamide-urea-polyacrylamide gel. Shown are products of assays with L0 and L-2 either undigested or digested with MscI just prior to gel electrophoresis. The arrow indicates the 117-bp band. Also seen is a faint band at 90 bases in lanes 3 and 4. SM, size marker.
A close examination also reveals the presence of a fragment of 90 nt (Fig. 6b). If fold-over occurs prematurely, the result will be DNA fragments shorter than 117 nt. It is therefore possible that the 90-nt fragment represents an infrequent premature fold-over event. Our conclusion is that Rep 68 and the HSV replication enzymes can cooperate to produce a terminal ITR from a substrate with a non-terminal ITR and that it can create a complete ITR from a substrate with small deletions at the AAV-non-AAV junction.
Partially deleted ITR results in reduced amounts of hybrid molecule.
As shown in Fig. 1, it is possible that efficient rescue is dependent upon the length of the stem formed by annealing of the a and a′ regions. In wild-type AAV, this region is 42 nt long. According to our model, displacement of the replicating strand should occur while replication is copying either the a or the a′ region, i.e., the components of the stem, in order to produce a complete ITR. A reduction in the length of the stem would reduce the regions over which this transition can productively occur. In addition, a shorter a region may lessen the tendency of the single-stranded template to refold into a hairpin structure.
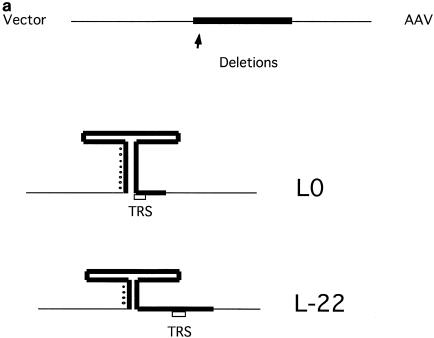
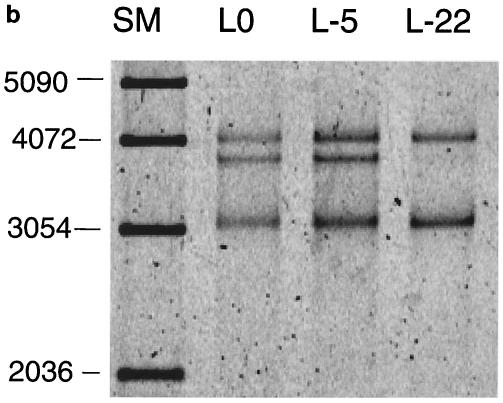
To test this prediction, we compared three substrates with different terminal deletions of AAV (Fig. 7a). L0 contains the full ITR. L-5 and L-22 are missing the outermost 5 and 22 bases from the ITR (i.e., the outer component of the stem-forming region), respectively. They were constructed by deleting these regions from the parental plasmid pL0. Rescue experiments were performed to examine how the ITR deletions affect rescue by replication. The fraction of substrate converted to the hybrid species was only marginally reduced when L-5 was used as substrate, but it was reduced by more than 80% when using the L-22 substrate (Fig. 7b). (An alternative explanation to this result might be that binding of Rep 68 to the newly synthesized and folded-over hairpin loop may be required before synthesis is resumed. This binding might be reduced since the 22-nt deletion eliminates a part of the Rep binding site.) In contrast, the amount of the V species, the vector part, was not reduced by these deletions in the ITR. The vector species is only produced by the Rep-only rescue mechanism. It seems that the Rep-only mechanism is not dependent on the missing sequences.
FIG. 7.
Terminally deleted ITRs result in less production of hybrid species and fewer replicated AAV species. (a) Schematic of potential hairpin structures of L0 and L-22 substrates. The thick line denotes the ITR sequence. (b) SYBR green staining of products from an assay with L0, L-5, and L-22 substrates which were separated on a 0.8% agarose gel, showing reduced hybrid formation with terminally deleted ITRs. (c) Left panel, SYBR green staining of products of an assay with L0 and L-22 substrates. Right panel, an autoradiograph of the same gel showing relatively greater incorporation into the AAV species in the L0 substrate. S, full-length substrate; H, hybrid species; V, vector component; A, AAV component. SM, size marker.
A comparison of the Rep-only mechanism and the rescue-by-replication mechanism for generating AAV under competitive conditions is presented in Fig. 7c. The L0 and L-22 substrates were used in rescue experiments and the products were analyzed by gel electrophoresis (Fig. 7c). Though each assay has the four proteins required for rescue by replication, since the L-22 substrate has little capacity to be rescued by the replication mechanism, its rescue will be done almost entirely by the Rep-only mechanism. The L0 substrate can be rescued by both mechanisms. The left panel shows SYBR green staining, while the right panel shows new DNA synthesis by autoradiography of the same gel. SYBR green staining demonstrates that similar amounts of vector (V) and AAV (A) products were produced from the L-22 and L0 substrates. However, autoradiography revealed that the A product derived from L0 was more highly radiolabeled, indicating that it was in part the product of new DNA synthesis (Fig. 7c, right panel). This confirms that when L-22 is used as substrate, the replication complex is much more likely to pass through the ITR rather than generating the fold-back ITR that results in rescue by replication. Replication through the ITR radiolabels the vector component as demonstrated. The vector component and the AAV component are ultimately separated by Rep 68 cleavage of the double-stranded product, i.e., the Rep-only separation mechanism, but in this case only the vector component is heavily labeled.
Partially deleted ITRs result in greater amounts of Rep-only separation.
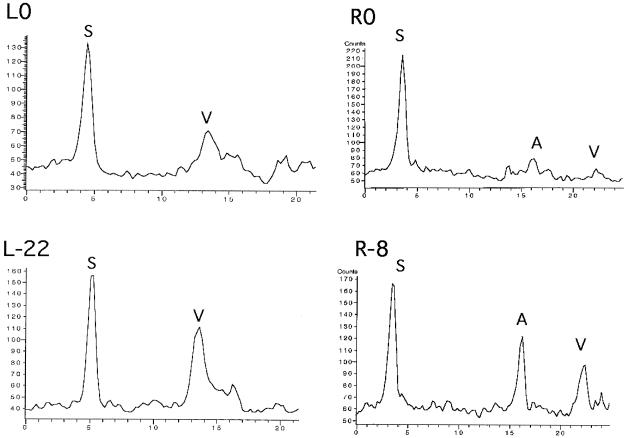
We have examined directly how the efficiency of the Rep-only mechanism is affected by deletions in the AAV ITR. We compared L0 and L-22 as well as R0 and R-8. The latter pair was created by cloning the right side of pAV2, including the right ITR, into pBluescript plasmids. The linearized substrate R0 contains a complete ITR, while R-8 is missing the outermost 8 bases of the ITR. The plasmid sequences at the vector-ITR junction are identical in these two substrates. We performed assays in which the substrates were incubated with only the Rep protein, i.e., the three HSV-1 replication proteins were omitted, as in the assay shown in Fig. 3, lane 3. Products of these assays were separated on agarose gels that were subsequently stained with SYBR green. Shown in Fig. 8 are graphs of these gels. (In the R0 and R-8 clones the AAV component is much larger and so is present on the gel as a larger fragment than the vector component. In L0 and L-22, the AAV component, being only 1,000 bp long, has migrated too far to the right and so is off the graph). In the constructs from both the left and the right ITRs, the partially deleted ITR shows increased amounts of Rep-only separation, as measured by the relative amounts of vector component, in comparison to the full-length ITR. This is precisely the reverse of the replication rescue assay, in which the deleted ITR demonstrates greatly reduced amounts of rescue.
FIG. 8.
Deleted ITRs result in greater amounts of Rep-only rescue. Shown are graphs from SYBR green staining of 0.8% agarose gels in which assay products were separated. In each case, higher-molecular-weight material is to the left and lower-molecular-weight material is to the right. Substrates were incubated with Rep for 4 h (HSV proteins omitted) and then separated by electrophoresis on agarose gels. L0, left ITR with no missing bases; L-22, left ITR with terminal 22 bases missing; R0, right ITR with no bases missing; R-8, right ITR with 8 terminal bases missing. S, full-length substrate; V, vector component; A, AAV component.
DISCUSSION
In this report we describe an assay which combines the initiation of DNA replication with rescue of the AAV genome from a linearized plasmid molecule. Members of our laboratory have previously shown that a minimal set of purified HSV-1 replication proteins, the HSV-1 polymerase UL30, the polymerase accessory protein UL42, and the DNA single-strand DNA binding protein UL29, could cooperate with Rep to initiate replication of the AAV genome (21). Initiation was specific, since it was dependent on both the AAV Rep 68 protein and the AAV origin of replication. The same experimental strategy has now been used to examine release of the AAV genome from a plasmid vector and a model for rescue by replication is presented. According to this model, a partially single-stranded and partially double-stranded molecule referred to as the H component should be produced when DNA replication, starting at a nick induced by Rep in the AAV ITR, folds back on itself while replicating through the ITR and copies the AAV part of the plasmid. The molecular nature of the H component was examined by gel electrophoresis experiments under native and denaturing conditions as well as by nuclease digestion experiments. In summary, these experiments show that the H component consists of one full-length DNA strand and a complementary strand corresponding to the vector component of the plasmid and part of the AAV ITR. (In Fig. 3, lane 2, there is a faint band which may have a structure similar to the H band. The particular assay shown in Fig. 3 was the only case in which this species was detectable. We have observed that the Rep protein in the presence of a single-strand binding protein is able to nick and unwind substantial segments of DNA [data not shown], and we think that occurs here. The point of the assay in Fig. 3 is that replication can greatly enhance the production of this species and that, if replication is promoting the production of this species, it is indicative that replication is of the sort that simultaneously accomplishes rescue.) The released AAV product terminates in a full-length ITR in a hairpin conformation.
The observation of an efficient Rep-only mechanism leading to separation of vector from AAV sequences is intriguing and deserves further study. At present we do not know how the Rep-only separation (which requires that both strands be cut) is accomplished, and we cannot rule out the possibility that a factor copurifying with Rep plays a role. We have no evidence that the AAV genomes released by this mechanism have their ITRs sufficiently intact to serve as substrates for replication initiation. The observation that Rep-only separation is enhanced with defective ITRs, i.e., ITRs which we have shown will not undergo efficient rescue by replication (Fig. 7), suggests a possible role for Rep-only separation that is complementary to that for rescue by replication.
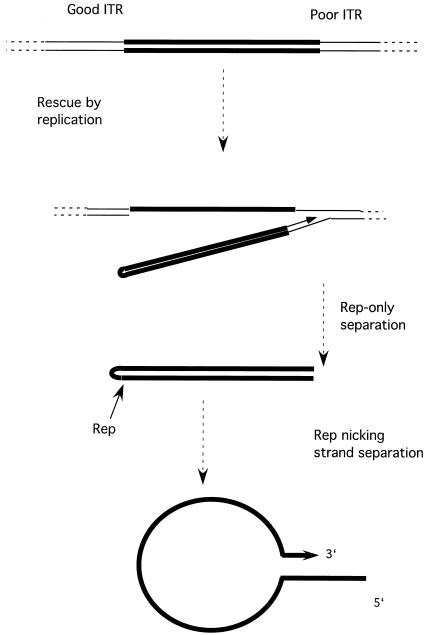
These results suggest the following model for rescue of the AAV genome from a plasmid vector and by analogy from its integrated state in the human genome (Fig. 9). DNA replication initiates at a nick introduced by Rep in the ITR of an integrated copy of the AAV genome. As replication passes through the ITR, the newly replicated strand is displaced due to self-annealing by the now single-stranded template strand. Replication will subsequently turn back by a fold-back mechanism, producing a hairpin loop with a complete ITR. If the ITR at the other end of the genome is intact, it can lead to rescue by the same mechanism. In cases in which the distal ITR is defective (which is apparently frequently the case in latently infected cells), the AAV genome can be rescued by the as yet poorly characterized Rep-only mechanism. In that instance, the product will be an AAV genome with a complete hairpin ITR at one end and a truncated ITR at the other end. Nicking of this structure at the complete ITR followed by strand separation (22) will permit the formation of a panhandle structure. The complete copy of the ITR, the product of rescue by replication, can be used as a template to synthesize a second complete copy of the ITR on the truncated strand as has been previously proposed (12).
FIG. 9.
Model of rescue of the AAV genome from plasmid or human genomic sequences. See text for explanation. AAV sequences are denoted by thick lines.
Does this model apply to the release of the AAV2 genome from its integrated site in the latently infected cell? A hallmark of the rescue-by-replication mechanism, the H component, is readily detected in vitro, but it seems unfortunately less likely that it could be detected in vivo. Therefore, the argument for the significance of this mechanism must rest on its ability to explain results from in vivo experiments with cell culture, combined with the demonstration of its feasibility using purified proteins.
It is possible that the Rep-dependent replication might serve as the predominant rescue mechanism in vivo for two reasons. Firstly, it is an efficient mechanism which has the capacity to replace missing nucleotides at the AAV-plasmid junction by copying them from the inner region of the ITR. Secondly, there is little absolute dependence upon cellular proteins such as a putative Holliday structure-resolving enzyme. The only proteins that would be strictly required would be a minimal DNA synthesis machinery and the AAV Rep protein, the very same factors needed for the subsequent productive replication.
Rescue may also occur by non-Rep-dependent mechanisms. For example, rescue by an unidentified Holliday structure-resolving activity has been observed both in cell extracts and in intact cells (19, 24). A key point about Rep-dependent rescue, however, is that it would only occur in latently infected cells when the Rep protein is expressed, i.e., when the cell is infected by a helper virus. Rescue by a non-Rep-dependent or non-helper-dependent mechanism would seem an unlikely choice for a principle rescue mechanism, since rescue could then occur when productive replication of AAV was not possible. In our experiments we have used HSV-1 replication enzymes. It would be of great interest to know if a similar reaction could be carried out by cellular replication enzymes as when adenovirus is the helper virus. We believe that this is likely. The original observation that Rep increased rescue was made in an extract in which the replication machinery was of cellular origin (23). Interestingly, it has been found that the addition of the adenovirus DNA binding protein, Ad-DBP, to a HeLa extract greatly increased the apparent fold-back replication in that system compared to the human single-stranded binding protein RPA (20). Perhaps Ad-DBP might permit refolding of the single-stranded AAV ITR and therefore be more efficient at promoting the rescue of the AAV genome. However, it cannot be stated with certainty that rescue occurred by fold-back replication in that report. Therefore, further experiments are needed to determine the mechanism by which Ad-DBP facilitates rescue of integrated AAV genomes.
Apart from the basic enzymatic requirements, there are almost certainly additional processes that play significant roles in control of rescue of AAV genomes. The replication rescue assays for this report were performed with the left ITR only. The same assays performed with the right ITR demonstrate less rescue by fold-back replication (data not shown). This result is reminiscent of results by Bohenzky and Berns in which substrates with ITR mutations demonstrated some ascendance of the left ITR over the right ITR in cell-based replication rescue assays (2) and suggests a possible involvement of internal AAV sequences. In the chromosomal context, the accessibility of the integrated AAV genome, nearby transcriptional activity, etc., would be expected to affect rescue. These processes will be determined by cellular factors and sequences proximal to the chromosome 19 integration site (5). It may be that the AAVS1 region is a site which allows easy rescue of the AAV genome. It has been noted by Gottlieb and Muzyczka (7) that production of virus from an AAV genome integrated at AAVS1 is substantially greater than that from the same copy number of transfected plasmid. An interesting suggestion by these workers is that the difference in virus production may in part reflect a difference in rescue efficiency. Understanding the rescue mechanisms of the AAV and recombinant AAV genomes from plasmids may help in recombinant AAV production.
The above observation and mechanism seem consistent with previously reported observations of rescue in cellular assays using plasmid constructs. In particular, it has been observed that rescue could correct missing AAV bases at the AAV-vector junction (13, 14). In a second observation related to rescue, Samulski et al. noted an eightfold decrease in the production of virus in an ITR construct with a deletion of 13 bases (pSUB201) compared with one containing full-length ITRs (pSM620) (12). The mechanism of this report can explain why a large deletion of terminal bases would lead to reduced rescue. Finally, this assay suggests an explanation as to why previous attempts to separate rescue from replication may have been unsuccessful. The initiation of replication at an embedded ITR seems designed to automatically result in rescue.
Acknowledgments
This work was supported in part by NIH grant GM62234 (to R.M.L.).
REFERENCES
- 1.Berns, K. I., and C. Giraud. 1996. Biology of adeno-associated virus. Curr. Top. Microbiol. Immunol. 218:1-23. [DOI] [PubMed] [Google Scholar]
- 2.Bohenzky, R. A., and K. I. Berns. 1989. Interactions between the termini of adeno-associated virus DNA. J. Mol. Biol. 206:91-100. [DOI] [PubMed] [Google Scholar]
- 3.Brister, J. R., and N. Muzyczka. 1999. Rep-mediated nicking of the adeno-associated virus origin requires two biochemical activities, DNA helicase activity and transesterification. J. Virol. 73:9325-9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crute, J. J., and I. R. Lehman. 1989. Herpes simplex-1 DNA polymerase. Identification of an intrinsic 5′—-3′ exonuclease with ribonuclease H activity. J. Biol. Chem. 264:19266-19270. [PubMed] [Google Scholar]
- 5.Dutheil, N., F. Shi, T. Dupressoir, and R. M. Linden. 2000. Adeno-associated virus site-specifically integrates into a muscle-specific DNA region. Proc. Natl. Acad. Sci. USA 97:4862-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falkenberg, M., D. A. Bushnell, P. Elias, and I. R. Lehman. 1997. The UL8 subunit of the heterotrimeric herpes simplex virus type 1 helicase-primase is required for the unwinding of single strand DNA-binding protein (ICP8)-coated DNA substrates. J. Biol. Chem. 272:22766-22770. [DOI] [PubMed] [Google Scholar]
- 7.Gottlieb, J., and N. Muzyczka. 1988. In vitro excision of adeno-associated virus DNA from recombinant plasmids: isolation of an enzyme fraction from HeLa cells that cleaves DNA at poly(G) sequences. Mol. Cell. Biol. 8:2513-2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Hoggan, M. D., G. F. Thomas, F. B. Thomas, and F. B. Johnson. 1972. Continuous “carriage” of adenovirus-associated virus genome in cell cultures in the absence of helper adenoviruses, p. 243-249. In Proceedings of the Fourth Lepetit Colloquium, Cocoyac, Mexico. North Holland Publishing Co., Amsterdam, The Netherlands.
- 8.Im, D. S., and N. Muzyczka. 1990. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell 61:447-457. [DOI] [PubMed] [Google Scholar]
- 9.Laughlin, C. A., J. D. Tratschin, H. Coon, and B. J. Carter. 1983. Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene 23:65-73. [DOI] [PubMed] [Google Scholar]
- 10.Ryan, J. H., S. Zolotukhin, and N. Muzyczka. 1996. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J. Virol. 70:1542-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samulski, R. J., K. I. Berns, M. Tan, and N. Muzyczka. 1982. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc. Natl. Acad. Sci. USA 79:2077-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samulski, R. J., L. S. Chang, and T. Shenk. 1987. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J. Virol. 61:3096-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samulski, R. J., A. Srivastava, K. I. Berns, and N. Muzyczka. 1983. Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell 33:135-143. [DOI] [PubMed] [Google Scholar]
- 14.Senapathy, P., J. D. Tratschin, and B. J. Carter. 1984. Replication of adeno-associated virus DNA. Complementation of naturally occurring rep− mutants by a wild-type genome or an ori− mutant and correction of terminal palindrome deletions. J. Mol. Biol. 179:1-20. [DOI] [PubMed] [Google Scholar]
- 15.Smith, D. H., P. Ward, and R. M. Linden. 1999. Comparative characterization of Rep proteins from the helper-dependent adeno-associated virus type 2 and the autonomous goose parvovirus. J. Virol. 73:2930-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snyder, R. O., R. J. Samulski, and N. Muzyczka. 1990. In vitro resolution of covalently joined AAV chromosome ends. Cell 60:105-113. [DOI] [PubMed] [Google Scholar]
- 17.Wang, X. S., S. Ponnazhagan, and A. Srivastava. 1996. Rescue and replication of adeno-associated virus type 2 as well as vector DNA sequences from recombinant plasmids containing deletions in the viral inverted terminal repeats: selective encapsidation of viral genomes in progeny virions. J. Virol. 70:1668-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, X. S., and A. Srivastava. 1997. A novel terminal resolution-like site in the adeno-associated virus type 2 genome. J. Virol. 71:1140-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward, P., and K. I. Berns. 1991. In vitro rescue of an integrated hybrid adeno-associated virus/simian virus 40 genome. J. Mol. Biol. 218:791-804. [DOI] [PubMed] [Google Scholar]
- 20.Ward, P., F. B. Dean, M. E. O'Donnell, and K. I. Berns. 1998. Role of the adenovirus DNA-binding protein in in vitro adeno-associated virus DNA replication. J. Virol. 72:420-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward, P., M. Falkenberg, P. Elias, M. Weitzman, and R. M. Linden. 2001. Rep-dependent initiation of adeno-associated virus type 2 DNA replication by a herpes simplex virus type 1 replication complex in a reconstituted system. J. Virol. 75:10250-10258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward, P., and R. M. Linden. 2000. A role for single-stranded templates in cell-free adeno-associated virus DNA replication. J. Virol. 74:744-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ward, P., E. Urcelay, R. Kotin, B. Safer, and K. I. Berns. 1994. Adeno-associated virus DNA replication in vitro: activation by a maltose binding protein/Rep 68 fusion protein. J. Virol. 68:6029-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao, X., W. Xiao, J. Li, and R. J. Samulski. 1997. A novel 165-base-pair terminal repeat sequence is the sole cis requirement for the adeno-associated virus life cycle. J. Virol. 71:941-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou, X., I. Zolotukhin, D. S. Im, and N. Muzyczka. 1999. Biochemical characterization of adeno-associated virus Rep68 DNA helicase and ATPase activities. J. Virol. 73:1580-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]