Abstract
The rat zinc-finger antiviral protein (ZAP) was recently identified as a host protein conferring resistance to retroviral infection. We analyzed ZAP's ability to inhibit viruses from other families and found that ZAP potently inhibits the replication of multiple members of the Alphavirus genus within the Togaviridae, including Sindbis virus, Semliki Forest virus, Ross River virus, and Venezuelan equine encephalitis virus. However, expression of ZAP did not induce a broad-spectrum antiviral state as some viruses, including vesicular stomatitis virus, poliovirus, yellow fever virus, and herpes simplex virus type 1, replicated to normal levels in ZAP-expressing cells. We determined that ZAP expression inhibits Sindbis virus replication after virus penetration and entry, but before the amplification of newly synthesized plus strand genomic RNA. Using a temperature-sensitive Sindbis virus mutant expressing luciferase, we further showed that translation of incoming viral RNA is blocked by ZAP expression. Elucidation of the antiviral mechanism by which ZAP inhibits Sindbis virus translation may lead to the development of agents with broad activity against alphaviruses.
A previously unknown rat protein, designated zinc-finger antiviral protein (ZAP), was recently found to exhibit antiviral activity against Moloney murine leukemia virus (MMLV), a member of the Retroviridae. When challenged with an ecotropic MMLV carrying a luciferase reporter, cells expressing ZAP expressed 30 times less luciferase than did control cells. Expression of either the full-length rat ZAP or the amino-terminal one-third fused to the product of the zeocin resistance gene (NZAP-Zeo) was inhibitory, and the mechanism of the block was found to be a dramatic and specific loss of viral mRNAs from the cytoplasm, but not the nuclei, of cells (5). In our effort to better understand virus-host interactions, we tested ZAP's ability to inhibit infection by other viruses. Our studies indicate that, in addition to inhibiting MMLV replication, ZAP's range of targets also includes multiple members of the Alphavirus genus of the Togaviridae.
Alphaviruses cause significant morbidity and mortality worldwide (reviewed in reference 7). The broad host range for these viruses includes vertebrates and invertebrates, with arthropods being the usual vectors of transmission to mammals. Infection with Sindbis virus (SIN), the type alphavirus, can lead to a painful polyarthritis, while disease caused by Venezuelan equine encephalitis virus (VEE) ranges from a mild influenza-type illness to fatal encephalitis. Alphaviruses are small, enveloped RNA viruses with an icosahedral nucleocapsid (reviewed in reference 22). The SIN genome consists of a single, capped, positive-sense RNA molecule of approximately 11.7 kb and contains a 5′ untranslated region (UTR) as well as a 3′ UTR and a poly(A) tail. The 5′-terminal two-thirds of the genomic 49S RNA is directly translated to produce the four nonstructural proteins (nsPs), while the structural proteins are encoded by a subgenomic 26S RNA derived from the 3′ one-third of the genome.
The binding of alphaviruses to the cell surface is possible through a number of receptors in different species and is mediated by the viral glycoprotein E2 (22). SIN enters the cell via clathrin-coated vesicles (3), where an E1 conformational change in the acidic environment of the endosome results in membrane fusion and release of the nucleocapsid into the cytosol. After interaction of the nucleocapsids with the 60S ribosomal subunit in the cytosol, the genomic RNA is released and translated (22, 23).
Initial translation and processing of the alphavirus plus strand RNA produces the four nsPs, nsP1 to nsP4 (10, 22). Following translation of the nsPs, replication complexes are established on cellular membranes, where the genomic RNA is transcribed to the complementary minus strand. The minus strand in turn functions as a template both for the production of additional plus strand RNA and for the transcription of the 26S subgenomic molecules.
Glycoproteins PE2 and E1, a capsid protein, and a small (6-kDa) membrane-embedded protein are the final products of subgenomic translation. Assembly of nucleocapsids occurs by oligomerization of the capsid protein and selection of the genomic RNA through recognition of an encapsidation signal (4, 22, 24). Completed nucleocapsids are then transported to the plasma membrane, where they interact with viral glycoproteins and enveloped virus buds from the cell (6).
Theoretically, a protein that inhibits alphavirus replication could affect any number of these steps in the virus life cycle. In the present study, we demonstrate that cells expressing NZAP-Zeo are dramatically resistant to infection by alphaviruses. Using SIN as our alphavirus model, we show that NZAP-Zeo targets translation of the incoming viral RNA to block alphavirus replication.
MATERIALS AND METHODS
Plasmids.
Plasmid pMC-Cre, expressing the Cre recombinase, the retroviral vector pBabe-HAZ, and pBabe-NZAP-Zeo, containing a sequence which expresses the amino-terminal 254 amino acids of rat ZAP fused to the zeocin resistance gene, have been previously described (5). Plasmid pMAMNeo (Clontech) expresses the Escherichia coli neo gene, which confers resistance to G418. Plasmids pToto1101 (19) and pRR64 (11) contain the entire SIN and Ross River virus (RRV) genomes, respectively, downstream of the SP6 bacteriophage promoter sequence. Plasmid pToto1101/Luc was generated by cloning the open reading frame for firefly luciferase into the SpeI site in the coding sequence for the carboxyl-terminal portion of nsP3 in pToto1101. SIN has previously been shown to tolerate insertions in this site (G. Li and C. M. Rice, unpublished data). Plasmid pToto1101/Luc/Pol−, encoding a polymerase-defective version of the luciferase-expressing SIN, was generated from pToto1101/Luc by deletion of a 36-nucleotide (nt) KpnI fragment in the DNA encoding the nsP4 RNA-dependent RNA polymerase. Plasmids pToto1101/Luc:ts6 and pToto1101/Luc:ts110, containing single nucleotide mutations that result in amino acid changes of glycine to glutamic acid at positions 153 and 324 of Toto1101 nsP4, respectively, were generated by cloning fragments from pToto1101 constructs that contained the appropriate mutations (8) into pToto1101/Luc. At the nonpermissive temperature (40°C) RNA replication is blocked, while at the permissive temperature (28°C) SIN is able to replicate (data not shown). Plasmid pPolioRep/GFP (A. A. Kolykhalov and C. M. Rice, unpublished data) contains the T7 bacteriophage promoter followed by the poliovirus genome, with sequences encoding the enhanced green fluorescent protein replacing the majority of the poliovirus structural protein coding region. Linearization with MluI allows for the generation of RNA encoding the poliovirus replicon. Plasmid pSP64-Ren Luc-Poly(A) was generated by cloning the open reading frame for Renilla luciferase from pRL null (Promega) into the XbaI site of the pSP64 Poly(A) vector (Promega) between the SP6 promoter and 3′ poly(A) sequences. Linearization with EcoRI allows for the production of transcripts encoding Renilla luciferase.
Cell lines.
All cell lines were maintained at 37°C in humidified chambers containing 5% CO2. Vero cells (American Type Culture Collection [ATCC] CCL-81; African green monkey) were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Invitrogen). BHK-J cells, a derivative of BHK-21 cells (ATCC CCL-10; hamster kidney fibroblasts), were cultured in minimum essential medium (MEM; Sigma) supplemented with 7.5% FBS and have been described previously (15). Rat2 (ATCC CRL-1764; rat thymidine kinase-negative fibroblast) cells transduced with the retroviral vector pBabe-HAZ (Rat2-HA-Zeo) or pBabe-NZAP-Zeo (Rat2-NZAP-Zeo) were maintained in DMEM supplemented with 10% FBS and 100 μg of zeocin (Invitrogen)/ml.
Viruses, replicons, and viral infection.
SIN and RRV stocks were produced by electroporation of BHK-J cells (BTX Electro Square Porator, EMC 830; 5 pulses at 99 μs and 960 V) with viral RNA generated by transcription in vitro, in the presence of a cap analog, with XhoI-linearized pToto1101 (19) or derivatives and SacI-linearized pRR64 (11), respectively, as the template. Virus present in the medium 24 to 48 h later was harvested and stored at −80°C. Semliki Forest virus (SFV) (14) and vesicular stomatitis virus (VSV), San Juan strain, stocks were a gift from Milton Schlesinger. Herpes simplex virus type 1 (HSV-1) strain KOS (ATCC VR-1493) stocks were produced by low-multiplicity infection of Vero cells as described previously (18). Yellow Fever virus (YF) strain 17D stocks were generated by electroporation of in vitro-transcribed viral RNA (1) as previously described (12). Packaged SIN replicons derived from the S.A.AR86 isolate and expressing the green fluorescent protein (GFP) under the control of the viral subgenomic promoter were generated as described previously (9) and were a gift from Mark T. Heise. Packaged VEE replicons expressing GFP, a gift from Bob Johnston and Nancy Davis, were generated as described previously (16).
Viral infections, unless indicated otherwise, were carried out in six-well culture dishes, with 0.2 ml of inoculum per well. SIN, RRV, SFV, and VSV, as well as packaged SIN and VEE replicons, were diluted in Dulbecco's phosphate-buffered saline (DPBS; Gibco) supplemented with 1% FBS prior to infection. HSV-1 was diluted in DMEM containing 10% FBS; YF was diluted in DMEM containing 2% FBS. After removal of the medium, the inoculum was applied and cells were incubated for 1 h at 37°C with occasional rocking. For temperature-sensitive mutants, infection and subsequent incubation were carried out at the permissive (28°C) or nonpermissive (40°C) temperature.
Virus titration.
Virus stocks and growth curve samples were titered in duplicate by infection of permissive cells with 10-fold serial dilutions of sample, followed by visual enumeration of plaques. For alphavirus and YF stocks, titers were determined with either BHK-J or Rat2-HA-Zeo cells and an MEM overlay containing 1.2% agarose and supplemented with 2% FBS, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Plaques were enumerated by crystal violet staining after ∼48 (alphaviruses) or ∼72 h (YF). Titers of temperature-sensitive SIN stocks were determined at 28°C, with staining and enumeration after 4 days. The titer of the VSV stock was similarly determined by using an MEM overlay containing 0.6% agarose, with plaque enumeration after ∼18 h. For alphavirus, YF, and VSV growth curve samples, titers were determined on BHK-J cells. HSV-1 stocks and growth curve samples were titered on Vero cells as described previously (18).
Growth curves.
Rat2-HA-Zeo and Rat2-NZAP-Zeo cells were seeded at 7 × 105 per 9.6-cm2 dish in 3 ml of medium. The following day the cells were infected at the multiplicities of infection (MOI) indicated in the figure legends. For alphaviruses, YF, and VSV, following the 1 h of infection the inoculum was removed and the cells were washed twice with DMEM, supplemented with 10% FBS, or DPBS (YF). Three (alphaviruses and YF) or 2 ml (VSV) of fresh medium was then added to each dish. Medium overlying the cells was frozen (−80°C) at various times after infection and titered on BHK-J cells as described above. For HSV-1 growth curves, following infection the cells were washed twice with DPBS and 3 ml of fresh medium was added to each dish. Dishes were frozen at −80°C at various times after infection. After thawing, cells and medium were scraped off the plate, vortexed, and subjected to titration on Vero cells.
Analysis of SIN RNA replication.
Dishes (55 cm2) seeded with either Rat2-HA-Zeo or Rat2-NZAP-Zeo cells at 2.8 × 106 cells/plate 24 h previously were either mock infected or infected with SIN (MOI of 5) in DMEM supplemented with 2% FBS and l μg of actinomycin D (Sigma)/ml. Following the 1-h infection, 7 ml of medium containing l μg of actinomycin D/ml and 10 μCi of [5,6-3H]uridine (ICN; 41 Ci/mmol)/ml was added to each dish. Total RNA was isolated from the cells with Trizol reagent (Gibco) at various times after infection and was size separated by 1.2% formaldehyde denaturing agarose gel electrophoresis. Newly synthesized viral RNA was detected by fluorography.
Preparation of 32P-radiolabeled SIN.
To generate a high-titer radiolabeled SIN stock, we made use of the fact that SIN release is inhibited under low-salt conditions (17). Cells were seeded at 7.3 × 106 cells per 145-cm2 dish and were infected the following day with SIN (MOI of 10) diluted in DPBS supplemented with 1% FBS and l μg of actinomycin D/ml. Following the 1-h incubation, the inoculum was removed and the cells were washed extensively with low-salt, low-phosphate MEM (LSM; made with MEM containing 60 mM NaCl and 0.1 mM NaH2PO4 · H2O and supplemented with 7.5% FBS). After the last wash, LSM supplemented with 1 μg of actinomycin D/ml and 0.1 mCi of 32P/ml as orthophosphate (Amersham Pharmacia) was added. After 24 h, the radioactive medium was removed and cells were washed with LSM. SIN was harvested from each dish by two sequential 15-min incubations with high-salt MEM (MEM containing 216 mM NaCl and supplemented with 7.5% FBS), with the same wash medium for each plate. The washes were pooled (∼15 ml) and immediately loaded onto two 15 to 30% sucrose gradients made up in virus dilution buffer (VDB) containing 50 mM Tris · HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, and 200 μg of bovine serum albumin/ml (fraction V; Amersham). After centrifugation (120,000 × g) for 2 h at 4°C, fractions were collected and an aliquot of each was counted by liquid scintillation spectrophotometry. Peak fractions were pooled, diluted in VDB, and pelleted by centrifugation (100,000 × g, 4°C for 3 h) to remove the sucrose. The virus pellet was resuspended in VDB, and aliquots were frozen at −80°C. The titer was determined on Rat2-HA-Zeo cells.
Viral binding assay.
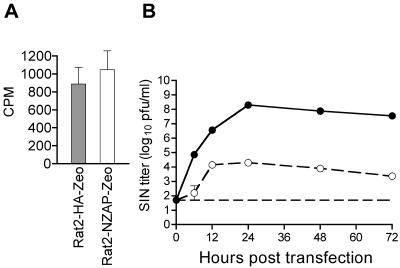
Rat2-HA-Zeo and Rat2-NZAP-Zeo cells were seeded at 3.5 × 105 cells per well in collagen (Sigma)-coated 12-well plates. The following day, 10,000 cpm (∼100 PFU per cell) of 32P-labeled SIN diluted in 0.1 ml of binding medium (BM) was added to each well. BM consisted of DPBS supplemented with 0.9 mM CaCl2 · 2H2O, 0.5 mM MgCl2 · 6H2O, and 1% FBS. Binding was carried out on a rocker at 4°C for 1 h with frequent additional manual rocking. After 1 h of binding the cells were washed four times with 2 ml of BM per well and lysed with 150 μl of 1% sodium dodecyl sulfate (Fisher), and the amount of bound virus in a 100-μl aliquot was determined by liquid scintillation spectrophotometry.
Transfection of viral RNA.
For transfections, Rat2-HA-Zeo and Rat2-NZAP-Zeo cells were seeded in six-well plates at 7 × 105 cells per well. The next day the cells were transfected with Lipofectin (Gibco) or TransMessenger transfection reagent (Qiagen). For wild-type SIN, cells were washed with Opti-MEMI (Gibco) and were transfected with in vitro-transcribed SIN Toto1101 RNA by using a 200-μl cocktail of 8 μg of SIN RNA and 8 μg of Lipofectin in Opti-MEMI. After incubation at room temperature for 10 min, the cells were washed with Opti-MEMI and 3 ml of DMEM, containing 2% FBS, was added to each well. A 200-μl aliquot of the medium was removed and replaced with fresh medium at various times after transfection. Samples were stored at −80°C and titered on BHK-J cells. PolioRep/GFP was similarly transfected into the cells with 3.5 μg of RNA per well. Toto1101/Luc/Pol− and Renilla luciferase RNAs were introduced into Rat2-HA-Zeo and Rat2-NZAP-Zeo cells according to the manufacturer's recommendations by using 1 μg of RNA with a 1:4 ratio of RNA to TransMessenger transfection reagent.
Determination of luciferase activity.
Cell lysates were harvested with 1× passive lysis buffer, and luciferase activity was determined with the Dual-Luciferase reporter assay system or luciferase assay system (Promega) according to the manufacturer's recommendations. Luciferase activity was measured on a Lumat LD 9507 (EG&G Berthold).
DNA transfections for Cre-mediated insert removal.
Cells were seeded at 1.5 × 106 cells per 55-cm2 plate and transfected the following day with 10 μg of plasmid pMAMNeo or 10 μg of pMC-Cre in combination with 1 μg of pMAMNeo by using 85 μg of SuperFect transfection reagent (Qiagen). Cells were placed under G418 (Invitrogen) selection (0.5 mg/ml) the following day. For pMC-Cre-transfected cells, individual clones were picked and expanded. Cre-mediated removal was verified by PCR as described previously (5).
Flow cytometry.
Cells were harvested, fixed with 2% paraformaldehyde in phosphate-buffered saline and analyzed for expression of GFP using a FACSCalibur cytometer and CellQuest software (Becton Dickinson), analyzing 10,000 events for each sample. Gates were set with uninfected or untransfected cells such that less than 1% of the cells fell within the positive gate. Unpaired t tests were performed using GraphPad Prism, version 3.0a, for Macintosh (GraphPad Software, San Diego, Calif).
RESULTS
Cells expressing NZAP-Zeo are highly resistant to infection with SIN.
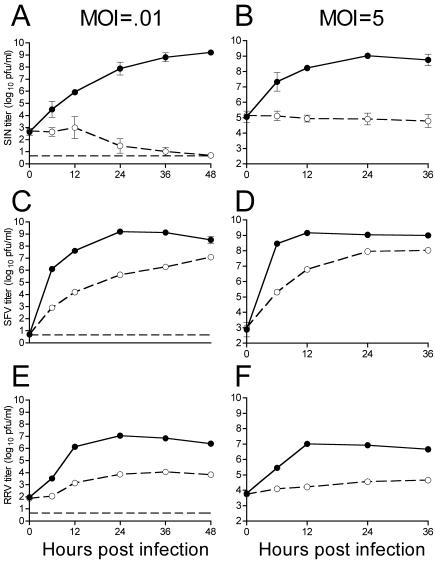
Expression of rat ZAP was previously shown to inhibit MMLV replication in Rat2 cells. The blocked infection correlated with a marked reduction in viral transcripts present in the cytoplasm without affecting nuclear viral transcript levels (5). To explore the range of viruses affected by ZAP, we tested its ability to inhibit SIN, an RNA virus whose replication is strictly cytoplasmic. Rat fibroblast cells stably transduced with vector alone (Rat2-HA-Zeo) or with a vector expressing the amino-terminal one-third of ZAP fused to the zeocin resistance gene (Rat2-NZAP-Zeo) were infected with SIN at MOIs of 0.01 and 5 (Fig. 1A and B). Expression of ZAP resulted in a dramatic inhibition of SIN replication, with little or no virus production observed in the Rat2-NZAP-Zeo cells, while control cells challenged with SIN had viral titers of ∼108 PFU/ml in both the high- and low-MOI infections. This effect was still observed when cells were challenged with SIN at an MOI of 25, calculated based on titration of the SIN stock on Rat2-HA-Zeo cells (data not shown). Additionally, full-length rat ZAP without the zeocin resistance gene fusion was inhibitory to SIN when expressed from plasmid pZAP-myc (5) in human T-REx-293 (Invitrogen) cells (not shown).
FIG. 1.
ZAP inhibits multiple members of the Alphavirus genus. Rat2-HA-Zeo cells expressing vector alone (filled circles) and Rat2-NZAP-Zeo cells expressing the amino-terminal portion of ZAP fused to the product of the zeocin resistance gene (open circles) were infected with SIN (A and B), SFV (C and D), or RRV (E and F) at MOIs of 0.01 and 5, as indicated. SIN MOIs were calculated based on stock titers determined on BHK-J cells, while SFV and RRV MOIs were based on stock titers determined on Rat2-HA-Zeo cells. At the indicated times after infection, medium was harvested and virus growth was determined by titration in duplicate on permissive cells. A separate well was utilized for each time point, and each experiment was done in duplicate. Dashed lines (A, C, and E), plaque assay detection limit. Data are mean log titers ± standard errors of the means; error bars for some points are obscured by the symbol.
NZAP-Zeo expression is required for the resistance of Rat2-NZAP-Zeo cells to SIN infection.
To show that expression of the cDNA insert contained in pBabe-NZAP-Zeo was necessary for the SIN inhibition seen in Rat2-NZAP-Zeo cells, we made use of the LoxP nucleotide sequence located in the U3 region of the long terminal repeat (LTR) of the pBabe-HAZ retroviral vector (5). Because the U3 sequence is duplicated upon reverse transcription of the vector, both LTRs in the resulting provirus contain LoxP sites. This allows for excision of the provirus from the genome by the Cre recombinase when a plasmid expressing Cre is introduced into cells. Rat2-NZAP-Zeo cells were cotransfected with plasmids pMC-Cre and pMAMNeo, and stably transfected cells were isolated by selection in G418. Clones from two independent transfections were isolated and analyzed by PCR for the presence of the NZAP-Zeo insert and tested for their ability to support SIN infection (Fig. 2). The ability of SIN to replicate was restored in all four clones from which the NZAP-Zeo insert had been removed, indicating that inhibition is not due to retroviral disruption of a host gene and that NZAP-Zeo expression is required for inhibition of SIN replication.
FIG. 2.
NZAP-Zeo expression is necessary for the resistance of Rat2-NZAP-Zeo cells to SIN infection. Rat2-HA-Zeo, Rat2-NZAP-Zeo, Rat2-NZAP-Zeo cells stably transfected with pMAMNeo (pMamNeo) and clones of Rat2-NZAP-Zeo cells stably transfected with pMAMNeo in combination with pMC-Cre (Cre1, -2, -3, and -4) were infected with SIN at MOIs of 0.01 and 5 as indicated, and the virus produced after 24 h was determined by titration in duplicate. Dashed line, plaque assay detection limit. Each cell line was tested in duplicate; data are mean log titers ± standard errors of the means. The retroviral insert present in each of the tested cell populations was amplified by PCR, and the ethidium-stained gel is shown at the top.
ZAP inhibits multiple members of the Alphavirus genus.
Given ZAP's impressive 3- to 8-log-unit inhibition of SIN, we tested other members of the Alphavirus genus. Although not as dramatic as the SIN inhibition, inhibition of both SFV (Fig. 1C and D) and RRV (Fig. 1E and F) by ZAP after infection at both low and high MOIs was significant. Although SFV eventually reached titers in NZAP-Zeo-expressing cells approximately 1 log unit lower than those in control cells, the inhibition was greater at earlier time points (∼3 log units). RRV showed a consistent 2- to 3-log-unit reduction in titer in ZAP-expressing cells compared to its titer in control cells.
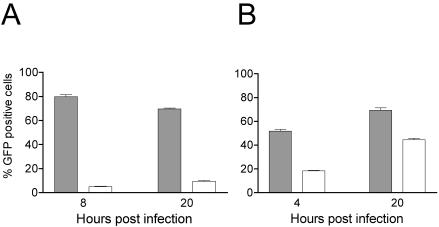
For our studies on VEE we chose to use a packaged self-replicating replicon that lacks the viral structural genes and that expresses GFP under the control of the viral subgenomic promoter. We first validated the assay by confirming that replication of a similar SIN-based replicon was inhibited by ZAP. For this we utilized a SIN replicon (9) derived from strain S.A.AR86, a strain isolated from mosquitoes and likely responsible for febrile illnesses in humans (reviewed in reference 20). The replicon expresses GFP under the control of the subgenomic promoter and lacks the viral structural genes. NZAP-Zeo-expressing and control cells were infected with SIN replicon-containing particles, and replication was monitored by flow cytometry. Expression of GFP indicates that the incoming replicon RNA was translated to generate the nonstructural proteins, that minus strand RNA was generated, and that subgenomic RNAs were transcribed and translated to produce GFP. Expression of ZAP inhibited the number of cells (<10%) expressing GFP, compared to control cells, where 70 to 80% of the cells were GFP positive (Fig. 3A). ZAP also inhibited, although less dramatically, replication of VEE (Fig. 3B). When analyzed by flow cytometry at 4 h postinfection (p.i.), 18% of the NZAP-Zeo-expressing cells infected with the VEE replicon showed GFP expression compared to 52% of control cells (a 65% reduction). When analyzed 20 h p.i., 44% of the NZAP-Zeo-expressing cells and 69% of the control cells expressed GFP (a 36% reduction). These data indicate that replication of VEE is inhibited in the NZAP-Zeo-expressing cells and that expression of the alphavirus structural genes is not required for ZAP's inhibitory activity. Taken together with the viral titer results from SIN, SFV, and RRV infections, these results demonstrate that NZAP-Zeo expression results in a broad resistance to alphaviruses, blocking replication at a step prior to translation of the viral structural genes encoded by the subgenomic RNA.
FIG. 3.
ZAP inhibits replication of a GFP-expressing VEE replicon. Rat2-HA-Zeo cells expressing vector alone (shaded bars) and Rat2-NZAP-Zeo cells expressing the amino-terminal portion of ZAP fused to the product of the zeocin resistance gene (open bars) were seeded in 12-well plates and infected with packaged SIN (A) or VEE (B) replicons expressing GFP. The percentage of cells expressing GFP was determined by flow cytometry at the indicated times after infection. Each bar represents the mean of results for three independent wells, with error bars indicating the standard deviations. The unpaired two-tailed t test indicates a significant difference between the mean values obtained after VEE infection of Rat2-HA-Zeo cells and those obtained after infection of Rat2-NZAP-Zeo cells (P < 0.0001 at 4 h, P = 0.0002 at 20 h). The results shown are similar to those from one other independent experiment.
ZAP expression does not produce a general antiviral state.
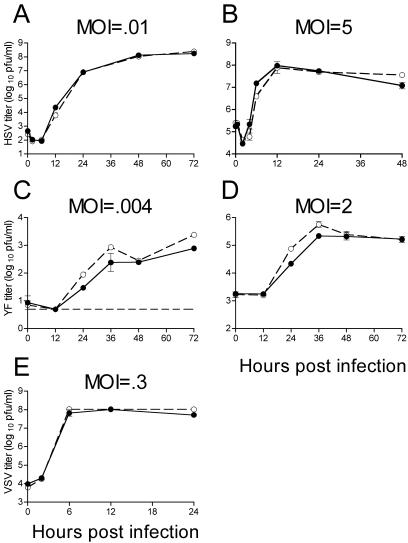
Given that NZAP-Zeo expression inhibits replication of members of two divergent virus families, we wondered whether ZAP exerts a general antiviral effect. Therefore, we challenged our cell lines with other viruses (Fig. 4). We tested a DNA virus within the Herpesviridae, HSV-1, and found that it grows to identical titers in NZAP-Zeo-expressing and control cells (Fig. 4A and B), demonstrating some specificity to ZAP's antiviral effects. We also tested several other RNA viruses. YF, the prototype virus of the Flavivirus genus within the Flaviviridae, grew to similar titers in the two cells lines (Fig. 4C and D). Similarly, VSV, a member of the Rhabdoviridae, replicated equivalently in the two cell lines (Fig. 3E). We utilized a poliovirus replicon expressing GFP to assess replication of a member of the Picornaviridae and found similar numbers of GFP-positive cells after transfection of the two cell lines with replicon RNA (Fig. 5). Although not an extensive survey, these results indicate that expression of ZAP does not render a cell resistant to all viruses. We chose SIN as a model alphavirus for further studies on the mechanism of ZAP's inhibition given the abundance of information and reagents available for this virus.
FIG. 4.
HSV-1, YF, and VSV are not inhibited by ZAP. Rat2-HA-Zeo cells expressing vector alone (filled circles) and Rat2-NZAP-Zeo cells expressing the amino-terminal portion of ZAP fused to the product of the zeocin resistance gene (open circles) were infected at the indicated MOIs with HSV-1 (A and B), YF (C and D), and VSV (E), and virus growth was determined by titration in duplicate on permissive cells. MOIs were calculated based on stock titers determined on Vero (HSV-1), BHK-J (YF), or Rat2-HA-Zeo (VSV) cells. A separate well was utilized for each time point, and the experiment was done in duplicate. Dashed line (C), plaque assay detection limit. Data are mean log titers ± standard errors of the means; error bars for some points are obscured by the symbol.
FIG. 5.
Replication of a poliovirus replicon is not inhibited by ZAP. Rat2-HA-Zeo cells expressing vector alone (shaded bars) and Rat2-NZAP-Zeo cells expressing the amino-terminal portion of ZAP fused to the product of the zeocin resistance gene (open bars) were seeded in six-well plates and transfected with 3.5 μg of PolioRep/GFP RNA encoding a GFP-expressing poliovirus replicon per well. The percentage of cells expressing GFP was determined by flow cytometry 8 h after transfection. Each bar represents the mean of results for three independent wells, with error bars indicating the standard deviations. Similar results were obtained in two independent experiments.
ZAP's block to SIN replication is likely after binding, penetration, and uncoating.
Experiments utilizing the SIN replicon (Fig. 3) indicated that the block to SIN replication was early, at a step prior to the translation of detectable levels of structural proteins derived from the viral subgenomic RNA. We tested whether ZAP might inhibit very early events in the SIN life cycle. The binding of virus at the cell surface to its corresponding receptor is the first stage in a viral infection. To examine whether ZAP expression inhibits binding, we utilized 32P-labeled SIN to measure SIN binding to Rat2-HA-Zeo and Rat2-NZAP-Zeo cells (Fig. 6A). SIN binding was unaffected by NZAP-Zeo expression. While a block to penetration and/or uncoating of SIN might be achieved by ZAP, this seems unlikely, since YF also enters the cell via the endocytic pathway and is unaffected by NZAP-Zeo expression. We chose to bypass early steps of attachment and entry by transfection of SIN RNA directly into the cytoplasm of Rat2-NZAP-Zeo cells (Fig. 6B). If a block to binding, penetration, and/or uncoating is the mechanism by which ZAP inhibits SIN replication, then direct transfection of the RNA should overcome this inhibition and restore the ability of SIN to grow. However, SIN replication after transfection of SIN RNA into NZAP-Zeo cells was significantly impaired, with a 3- to 4-log-unit decrease in viral titers in ZAP-expressing cells compared to titers in control cells. Virus production seen at the later time points represents a combination of virus generated directly from RNA and virus generated by a subsequent second-round infection of untransfected cells. However, the large decrease in titer seen at the earliest time point (6 h) suggests that ZAP is able to block SIN replication at a stage after release of the viral RNA into the cytoplasm. In addition, ZAP's block of SIN replication was also observed after transfection of a GFP-expressing SIN replicon RNA that is unable to generate virus and thus incapable of second-round infection. Notably, the two cell lines were found to be equally transfectable by using RNA derived from a GFP-expressing poliovirus replicon (Fig. 5).
FIG. 6.
ZAP affects SIN replication after binding, penetration, and uncoating. (A) SIN binding. Rat2-HA-Zeo (shaded bar) and Rat2-NZAP-Zeo (open bar) cells were incubated in 12-well plates as described in Materials and Methods with 10,000 cpm of radioactive SIN (∼100 PFU/cell) for 1 h at 4°C to allow binding without penetration of SIN. Cells were washed extensively to remove unbound virus, and, after lysis in sodium dodecyl sulfate, the amount of bound virus was determined by counting an aliquot of the lysate. The experiment was done in triplicate and is representative of three independent experiments. Data are means, with error bars indicating the standard deviations. (B) SIN RNA transfection. SIN RNA was introduced via cationic lipid transfection into Rat2-HA-Zeo cells expressing vector alone (filled circles) and Rat2-NZAP-Zeo cells (open circles). At the indicated times after transfection, an aliquot of the medium was harvested, and virus growth was determined by titration in duplicate on permissive cells. Dashed line, plaque assay detection limit. The experiment was done in duplicate and is representative of two independent experiments. Data are mean log titers ± standard errors of the means; error bars for some points are obscured by the symbol.
ZAP blocks the amplification of viral plus strand genomic SIN RNA.
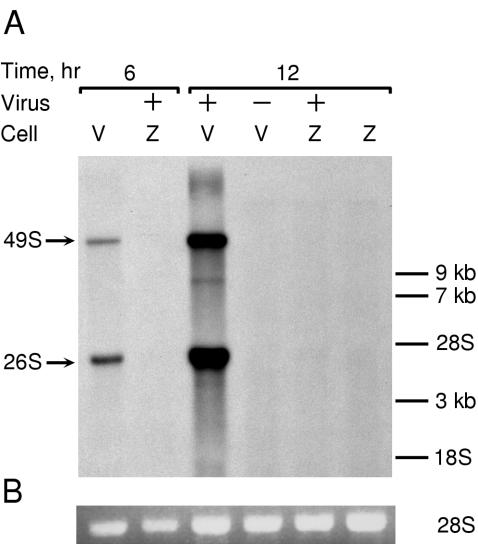
To further determine at which step in the viral life cycle NZAP-Zeo acted, we looked at the production of new genomic and subgenomic RNA after infection. Rat2-HA-Zeo and Rat2-NZAP-Zeo cells were infected with SIN in the presence of [3H]uridine and actinomycin D, and RNA was harvested at various times after infection (Fig. 7). By 6 h p.i., newly made SIN genomic 49S and subgenomic 26S RNA was observed in control cells but not in the NZAP-Zeo-expressing cells. Levels of newly synthesized SIN RNA were increased at 12 h p.i. in control cells, but SIN RNA was absent in NZAP-Zeo-expressing cells. This effect could still be seen at 36 h p.i. (not shown). Taken with the results from the binding and transfection experiments described above, these results suggest that ZAP inhibits alphavirus replication by blocking a step in the virus life cycle at some point after virus uncoating but before the efficient amplification of new plus strand genomic RNA.
FIG. 7.
ZAP blocks the production of new plus strand genomic RNA. (A) Rat2-HA-Zeo (V) and Rat2-NZAP-Zeo (Z) cells were mock-infected (−) or infected (+) with SIN (MOI = 5, as titered on BHK-J cells) in the presence of actinomycin D (1 μg/ml) as described in Materials and Methods. After infection, [3H]uridine was added, and RNA was harvested at the indicated times after infection. Total RNA (5 μg) was size separated by denaturing gel electrophoresis, and, after ethidium bromide staining, the gel was treated for fluorography and exposed to film for 28 h at −80°C. On prolonged exposure (1 week) a faint 49S signal, likely due to a small percentage of permissive cells in the culture, can be seen in infected Rat2-NZAP-Zeo cells harvested at 12 h. Migration of RNA size markers is shown on the right. Arrows, locations of the SIN genomic (49S) and subgenomic (26S) RNAs. (B) Ethidium bromide staining of ribosomal 28S RNA prior to fluorography indicates equal loading of RNA on the gel. The results are representative of two independent experiments.
ZAP blocks translation of incoming SIN RNA.
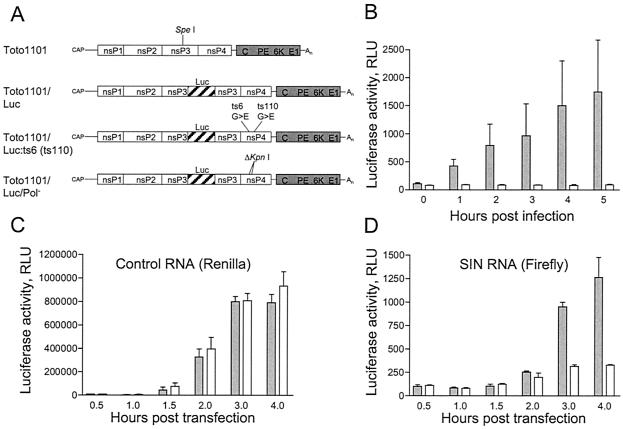
After uncoating, translation of incoming viral RNA is the first step that must occur in order to generate the proteins necessary to assemble a replication complex. Since replicated RNA is identical to incoming RNA, measurements of SIN translation include contributions made by both incoming and newly synthesized RNA. We have generated a SIN mutant, designated Toto1101/Luc, which expresses firefly luciferase as an in frame fusion within nsP3. The Toto1101/Luc virus and derivatives (described below) are shown schematically in Fig. 8A. Toto1101/Luc is viable and allows for simple quantification of the levels of viral polyprotein generated by translation of incoming as well as replicated viral RNA. To compare translation in control cells that replicate the viral RNA and in NZAP-Zeo-expressing cells that do not, we made use of temperature-sensitive SIN mutants that are defective in RNA replication at the nonpermissive temperature (21). Temperature-sensitive mutants ts6 and ts110 have mutations that have been mapped to specific residues of nsP4 (8). The temperature sensitivity mutations were therefore engineered into Toto1101/Luc to allow for the direct quantification of the translation of incoming RNA that cannot be replicated. Stocks of luciferase-expressing SIN harboring the ts6 or ts110 mutations of nsP4 were generated and titered at the permissive temperature (28°C). Infection of cells at the nonpermissive temperature (40°C) with Toto1101/Luc:ts6 indicates that translation of incoming viral RNA is markedly reduced in ZAP-expressing cells compared to that in control cells (Fig. 8B). Similar results were obtained with Toto1101/Luc:ts110 (not shown). The block to SIN translation is not due to a general translation block, since levels of translation of a capped Renilla luciferase reporter RNA in the two cell lines are identical (Fig. 8C). We generated an additional SIN mutant, designated Toto1101/Luc/Pol−, that is incapable of replication, due to a 36-nt deletion in the nsP4 region encoding the viral RNA-dependent RNA polymerase. As expected, the ZAP-mediated block to SIN RNA translation is still apparent when steps of binding, penetration, and uncoating are bypassed by directly introducing the replication-defective Toto1101/Luc/Pol− RNA into the cells (Fig. 8D).
FIG. 8.
ZAP blocks translation of incoming SIN RNA. (A) Schematic representation of Toto1101 virus and luciferase-expressing variants. Solid lines, UTRs. Open and shaded boxes, locations of the nonstructural- and structural-protein-encoding regions, respectively. The approximate locations of the nonstructural proteins nsP1, nsP2, nsP3, and nsP4 and structural protein capsid (C), glycoproteins (PE and E1), and the 6-kDa protein are shown, as is the location of the cap and poly(A) tail (An). SpeI indicates the restriction site used for cloning the luciferase (Luc) gene into pToto1101. Hatched bars within nsP3, firefly luciferase-encoding region. The 36-nt deletion resulting in a defective RNA-dependent RNA polymerase is indicated (ΔKpnI), as are the glycine (G)-to-glutamic acid (E) mutations of ts6 and ts110. (B) Rat2-HA-Zeo control cells (shaded bars) and Rat2-NZAP-Zeo cells (open bars) were infected with firefly luciferase-expressing SIN temperature-sensitive mutant Toto1101/Luc:ts6 (MOI = 0.006). Binding was carried out at 4°C. After being washed, the cells were incubated at the nonpermissive temperature (40°C). At the indicated times after infection, the cells were lysed and luciferase activity was measured with the luciferase assay system (Promega). (C and D) Rat2-HA-Zeo control cells (shaded bars) and Rat2-NZAP-Zeo cells (open bars) were transfected for 30 min with a mixture of 0.9 μg of capped Toto1101/Luc/Pol− RNA and 0.1 μg of capped control RNA encoding Renilla luciferase. Cells were lysed at the indicated times, and Renilla (C) and firefly (D) luciferase activities were measured with the Dual-Luciferase reporter assay system (Promega). Renilla luciferase activity indicates translation of the control RNA (C), while firefly luciferase activity indicates translation of SIN RNA (D). For panels B to D the data are representative of two independent experiments. Bars represent the means of triplicate samples ± standard deviations. RLU, relative light units.
DISCUSSION
ZAP was originally isolated as a protein whose expression confers resistance to MMLV replication. Retroviruses, which exist in both DNA and RNA states, have a complex life cycle and use both cellular cytoplasmic and nuclear machinery to propagate their genomes. ZAP was found to block MMLV replication by preventing the accumulation of cytoplasmic viral RNA, without affecting levels of viral transcripts in the nucleus (5). In these studies we showed that ZAP also potently inhibits multiple members of the Alphavirus genus of the Togaviridae, including SIN, RRV, SFV, and VEE. We tested both tissue culture-adapted (Fig. 1A and B) and virulent (Fig. 3A) strains of SIN and found both to be inhibited by ZAP. Of the alphaviruses tested, inhibition of SIN was most impressive, followed by RRV, with SFV and VEE showing less-dramatic, although significant, inhibition. The reason(s) for the difference in susceptibility to ZAP-mediated inhibition is unclear but may unfold once the mechanism of inhibition is determined.
Similar to its effect on MMLV, ZAP was found to block SIN replication by preventing the accumulation of new genomic viral RNA in the cytoplasm. Since alphaviruses are replicated and propagated entirely in an RNA state, utilizing only cytoplasmic machinery, ZAP-mediated viral inhibition must not directly require nuclear machinery. The mechanism(s) by which ZAP inhibits viruses from two divergent families is unknown. Our data indicate that ZAP blocks the generation of nsPs from SIN RNA, leading to a failure to replicate and accumulate viral RNA. Whether the block is a result of RNA degradation, altered RNA targeting, and/or a direct inhibition of the translation process is unknown. The block to viral translation is not due to induction of a general antiviral state and likely does not involve interferon, since several viruses, including the highly interferon-sensitive VSV, grew to equivalent titers in control and ZAP-expressing cells. No obvious similarity between MMLV and SIN replication cycles compared to those of viruses unaffected by ZAP explains the pattern of viral inhibition seen. Since MMLV genome transcription occurred normally in the presence of ZAP (5), it is tempting to consider that a common mechanism occurring in the cytoplasm underlies ZAP-mediated inhibition of the accumulation of viral genomic MMLV and SIN RNA. Interestingly, two viruses (YF and poliovirus) which, like SIN, are positive stranded and replicate entirely within the cytoplasm were unaffected by ZAP. MMLV and SIN RNA genomes both contain a 5′ cap and 3′ poly(A) tail. However, neither of these structures alone was able to confer susceptibility to ZAP-mediated inhibition. YF contains a 5′ cap structure and lacks a poly(A) tail, and poliovirus lacks a 5′ cap but contains a poly(A) tail, yet neither was inhibited by ZAP. Whether ZAP inhibits MMLV and alphaviruses by a common mechanism or by distinct mechanisms remains to be determined.
ZAP contains four potential CCCH-type zinc-fingers similar to those found in proteins known to bind RNA, including tristetraprolin, a protein that negatively regulates the stability of several RNAs (2, 13). Similarly, ZAP might directly interact with viral RNAs and affect their stability. Alternatively ZAP might bind SIN RNA and alter its intracellular trafficking or ability to interact appropriately with the translational machinery. ZAP might also interfere with a protein-protein or nucleic acid-protein interaction that is necessary for polysome association or translation of SIN RNA without directly binding to SIN RNA. Although members of the Alphavirus genus have extensive sequence homology, there is no obvious sequence homology between MMLV and SIN. Due to a lack of proofreading ability in their RNA-dependent RNA polymerases, alphaviruses replicate their genomes with relatively low fidelity, the error rate being in the range of 10−4 to 10−5 (reviewed in reference 22). As a result, one might expect escape mutants to arise. We have been unable to isolate such SIN mutants that can overcome ZAP's effect (not shown). This suggests that, if ZAP binds directly to the SIN RNA, it either targets multiple sites within the genome or interacts with a conserved sequence required for virus survival.
ZAP exhibits broad-spectrum activity against alphaviruses, which cause widespread human disease. Determining the mechanism of ZAP's inhibition of SIN translation will shed light on ZAP's normal cellular function and may lead to the development of new antiviral therapies. For example, treatments that upregulate ZAP expression might be used as therapeutic interventions. If ZAP has a direct interaction with a host or viral factor that results in viral inhibition, then small molecules that mimic this interaction could be pursued. Such approaches could lead to the development of panalphavirus therapies for preventing encephalitic disease or debilitating arthritis due to alphavirus infection.
Acknowledgments
This work was supported in part by Basil O'Connor Starter Scholar Research Award Grant no. 5-FY98-905 from the March of Dimes Birth Defects Foundation (M.R.M.) and by the Greenberg Medical Research Institute.
We thank Richard Kuhn for plasmid pRR64, Mark Heise for SIN, and Bob Johnston and Nancy Davis for VEE replicon stocks, respectively, Milton Schlesinger for the SFV and VSV stocks, Beate Kümmerer for YF stocks, and David Leib for HSV-1 strain KOS.
REFERENCES
- 1.Bredenbeek, P. J., E. A. Kooi, B. Lindenbach, N. Huijkman, C. M. Rice, and W. J. Spaan. 2003. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 84:1261-1268. [DOI] [PubMed] [Google Scholar]
- 2.Carballo, E., W. S. Lai, and P. J. Blackshear. 2000. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95:1891-1899. [PubMed] [Google Scholar]
- 3.DeTulleo, L., and T. Kirchhausen. 1998. The clathrin endocytic pathway in viral infection. EMBO J. 17:4585-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frolova, E., I. Frolov, and S. Schlesinger. 1997. Packaging signals in alphaviruses. J. Virol. 71:248-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao, G., X. Guo, and S. P. Goff. 2002. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297:1703-1706. [DOI] [PubMed] [Google Scholar]
- 6.Garoff, H., R. Hewson, and D. J. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin, D. E. 2001. Alphaviruses, p. 917-962. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 8.Hahn, Y. S., A. Grakoui, C. M. Rice, E. G. Strauss, and J. H. Strauss. 1989. Mapping of RNA− temperature-sensitive mutants of Sindbis virus: complementation group F mutants have lesions in nsP4. J. Virol. 63:1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heise, M. T., D. A. Simpson, and R. E. Johnston. 2000. Sindbis-group alphavirus replication in periosteum and endosteum of long bones in adult mice. J. Virol. 74:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kääriäinen, L., and T. Ahola. 2002. Functions of alphavirus nonstructural proteins in RNA replication. Prog. Nucleic Acid Res. Mol. Biol. 71:187-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn, R. J., H. G. Niesters, Z. Hong, and J. H. Strauss. 1991. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology 182:430-441. [DOI] [PubMed] [Google Scholar]
- 12.Kümmerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai, W. S., E. Carballo, J. M. Thorn, E. A. Kennington, and P. J. Blackshear. 2000. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to AU-rich elements and destabilization of mRNA. J. Biol. Chem. 275:17827-17837. [DOI] [PubMed] [Google Scholar]
- 14.Liljestrom, P., S. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindenbach, B. D., and C. M. Rice. 1997. trans-complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608-9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacDonald, G. H., and R. E. Johnston. 2000. Role of dendritic cell targeting in Venezuelan equine encephalitis virus pathogenesis. J. Virol. 74:914-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierce, J. S., E. G. Strauss, and J. H. Strauss. 1974. Effect of ionic strength on the binding of Sindbis virus to chick cells. J. Virol. 13:1030-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rader, K. A., C. E. Ackland-Berglund, J. K. Miller, J. S. Pepose, and D. A. Leib. 1993. In vivo characterization of site-directed mutations in the promoter of the herpes simplex virus type 1 latency-associated transcripts. J. Gen. Virol. 74:1859-1869. [DOI] [PubMed] [Google Scholar]
- 19.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simpson, D. A., N. L. Davis, S. C. Lin, D. Russell, and R. E. Johnston. 1996. Complete nucleotide sequence and full-length cDNA clone of S.A.AR86, a South African alphavirus related to Sindbis. Virology 222:464-469. [DOI] [PubMed] [Google Scholar]
- 21.Strauss, E. G., E. M. Lenches, and J. H. Strauss. 1976. Mutants of Sindbis virus. I. Isolation and partial characterization of 89 new temperature-sensitive mutants. Virology 74:154-168. [DOI] [PubMed] [Google Scholar]
- 22.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wengler, G., and C. Gros. 1996. Analyses of the role of structural changes in the regulation of uncoating and assembly of alphavirus cores. Virology 222:123-132. [DOI] [PubMed] [Google Scholar]
- 24.White, C. L., M. Thomson, and N. J. Dimmock. 1998. Deletion analysis of a defective interfering Semliki Forest virus RNA genome defines a region in the nsP2 sequence that is required for efficient packaging of the genome into virus particles. J. Virol. 72:4320-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]