Abstract
Infection of Escherichia coli containing the type I restriction enzyme EcoKI by bacteriophage T7 0.3 mutants leads to restriction during the late stages of genome entry and during DNA replication. Patterns of cleavage in vivo suggest that some cutting occurs near the midpoint of two recognition sites, consistent with the idea that EcoKI translocates DNA bidirectionally through itself and cuts when two enzyme molecules collide. Rapid ejection of a 0.3+ T7 genome from a bacteriophage λ particle results in degradation of the infecting DNA by EcoKI, showing that the normal T7 DNA translocation process delays restriction. A unique recognition site inserted at the genomic left end allows EcoKI to function as a molecular motor and to translocate the remaining 39 kilobases of T7 DNA into the cell.
Type I restriction-modification systems of Escherichia coli serve to protect the bacterium from many phages (1). E. coli K-12 strains harboring the hsd locus contain the enzyme EcoKI, a multisubunit enzyme consisting of a specificity subunit and two each of restriction and modification subunits (2). EcoKI binds S-adenosylmethionine forming an activated enzyme (3), which binds the DNA sequence 5′-AACN6GTGC (4) and determines its methylation state. If the recognition sequence is nonmethylated, EcoKI hydrolyzes ATP and spools flanking DNA through itself (5); because it remains bound to its recognition sequence, highly twisted DNA loops are formed (6). A current model for cleavage site selection, determined from in vitro experiments, posits that cutting occurs when two translocating EcoKI molecules collide (7). The observation that λ mutants containing two recognition sites for EcoKI are restricted 15-fold more efficiently than expected if two enzyme molecules cut independently provides in vivo support for the model (8).
Many phages and conjugal plasmids escape restriction by type I restriction-modification systems (1, 9). Some phage genomes contain exotically modified bases (10); some plasmids and the phages T7 and T3 synthesize a direct inhibitor of type I restriction-modification systems (11–13). Inhibition of type I restriction by T7 requires stoichiometric amounts of the early protein gp0.3, which binds to EcoKI and prevents it from binding target DNA (14); gp0.3 also obstructs DNA cleavage and methylation after EcoKI has interacted with its recognition sequence (15). Escape of T7 DNA from EcoKI restriction in vivo depends on the synthesis of sufficient gp0.3 before the viral genome becomes cleaved. T7 0.3 mutants containing unmodified DNA plate on hosts harboring EcoKI at an efficiency of 10−2 to 10−4 (12), somewhat higher but similar to the efficiency of unmodified λ. Nevertheless, stocks of T7 0.3 mutants can be made on restricting cells, although lysates have low titers and progeny genomes are modified only partially. It was suggested that the incomplete restriction and modification of T7 0.3 DNA was caused by the rapid replication phase and short latent period of the phage.
Cellular internalization of T7 DNA is coupled to transcription (16–18). About 850 bp of the genetic left end are ejected efficiently from the phage particle into the cell (19). The E. coli promoters A1, A2, and A3 lie on this 850 bp (20), allowing RNA polymerase to initiate transcription and pull about 20% of the genome into the cell. After T7 RNA polymerase is synthesized, internalization of the remaining phage DNA is coupled to transcription by this enzyme. At 30°C, 9 or 10 min are required for internalization of the whole genome, but the first gene product synthesized (gp0.3) can be detected within 2 min (18, 21).
Within the 39,937-bp wild-type T7 genome lie four EcoKI recognition sites located 15,173, 26,603, 32,632, and 37,459 bp from the genetic left end (20). During infection, the first EcoKI site enters the cell after about 7 min (18); by this time, sufficient gp0.3 has accumulated to inactivate cellular pools of EcoKI. However, artificial EcoKI sites placed immediately upstream and downstream of gene 0.3 do not affect phage growth on hosts containing EcoKI (17). It was concluded that the lack of EcoKI sites in the early region of wild-type T7 DNA is not crucial in avoiding restriction and that some aspect of the T7 DNA ejection process temporarily sequesters the leading end from the nuclease.
In this work, the role of the T7 DNA entry process in enhancing escape from EcoKI restriction is investigated further. We show that transcription-coupled entry delays EcoKI restriction of T7 gene 0.3 mutants and that one-step ejection of 0.3+ T7 DNA leads to restriction. We show that EcoKI cleavage of T7 DNA in vivo results in discrete fragments whose sizes are consistent with the model positing that cutting occurs midway between two recognition sites and whose appearance suggests that translocation of two EcoKI molecules from widely separated sites may be a concerted or cooperative reaction. Finally, we show that EcoKI can act as a motor to translocate the entire T7 genome into the cell.
Materials and Methods
Bacteriophages and Bacteria.
T7 mutants sRK836 and the 0.3 deletion mutant D364 were kindly provided by F. W. Studier (Brookhaven National Laboratory, Upton, NY). T71.7∷λ- cosΔ10-NB1 and procedures for packaging a T7 genome in λ-capsids have been described (18). sRK836sK0 was constructed as follows. D364 was alternately grown on the EcoKI-containing strain IJ891 (E. coli K-12 ΔlacX74 thi) and the isogenic nonrestricting Δhsd strain IJ1133 [IJ891 Δ(mcrC-mrr)∷Tn10] to enrich for mutants that have lost EcoKI recognition sites. After several passages, phages then were propagated alternately on IJ1133 and on IJ891 containing pSB2, a plasmid that overexpresses hsdR (8), to increase the selection pressure for loss of EcoKI sites. A two-step procedure was necessary, because D364 does not plate at detectable frequencies on IJ891(pSB2). A phage growing normally in the presence of pSB2 was selected: D364sK0 is insensitive to EcoKI but is restricted by EcoBI to the same degree as D364. IJ1178, isogenic to IJ891, except that it contains hsdB, was used as the strain containing EcoBI. The left-end 11.5-kilobase (kb) BglII fragment of sRK836 DNA, which contains a cloned EcoKI site at position 836 on the genome but no natural EcoKI site, was then ligated to the right-end 28.4-kb BglII fragment of D364sK0 DNA to create the phage sRK836sK0.
Stocks of sRK836, sRK836sK0, and D364 containing unmethylated DNA were prepared on IJ922 (E. coli K-12 ΔlacX74 supE44 galK2 galT22 mcrA dam-13∷Tn9 rfbD1 mcrB1 hsdS3). DNA translocation experiments were performed with the strains IJ891 and IJ1133 or their recD6001∷Tn10:Kn derivatives, respectively named IJ1409 and IJ1190; each strain also contained the dam plasmid pTP166 (22).
Assays of Phage Genome Entry into Cells.
The assay has been described (18). Briefly, bacteria containing pTP166 were grown in LB + 100 μg/ml ampicillin to a cell density of 2 × 108 per ml at 30°C. When rifampin was required, cells were collected and resuspended at 30°C in LB medium containing 500 μg/ml rifampin. Cells were then incubated at 30°C for 10 min before adding unmethylated phages at a multiplicity of 0.4. At various times, a portion of the infected culture was added to ice-cold phenol-ethanol [2% (vol/vol) phenol/75% (vol/vol) ethanol/8 mM EDTA/20 mM sodium acetate, pH 5.2]. DNA was extracted and digested with either DpnI or Sau3AI. DNA fragments were separated by electrophoresis on an agarose gel and transferred to a nylon membrane. Immobilized DNA was hybridized with 32P-labeled T7 DNA and visualized by autoradiography.
Results
Cleavage of T7 0.3 DNA Occurs During the Late Stages of DNA Entry and the DNA Replication Phase.
T7 0.3 mutants inefficiently lyse liquid cultures of bacteria containing EcoKI (12), suggesting that, although EcoKI reduces the fitness of the infecting population, a small number of phages escape cleavage. One-step growth experiments with the 0.3 deletion mutant D364 (previously grown on a Δhsd host) on an hsd+ strain show directly that gp0.3 is not absolutely required to escape EcoKI restriction. An average burst of nine progeny per infective center was obtained (Table 1). However, only 13% of infected cells make an infective center, a value consistent with the low efficiency of plaque formation by the phage on this host.
Table 1.
Plating efficiencies and burst sizes
| Strain | Host | Genotype | Relative efficiency of plating | Progeny per infective center |
|---|---|---|---|---|
| D364 | IJ1133 | Δhsd | 1 | 72 |
| IJ891 | hsdK+ | 3.7 × 10−3 | 9* | |
| IJ891(pSB2) | hsdK+ | <10−10 | ||
| IJ1178 | hsdB+ | 1.6 × 10−4 | ||
| D364sK0 | IJ1133 | Δhsd | 1 | 87 |
| IJ891 | hsdK+ | 1.2 | 94 | |
| IJ891(pSB2) | hsdK+ | 0.83 | ||
| IJ1178 | hsdB+ | 3.4 × 10−4 | ||
| sRK836sK0 | IJ1133 | Δhsd | 1 | |
| IJ891 | hsdK+ | 1.0 | ||
| IJ1178 | hsdB+ | 0.87 |
*The ratio of infective centers per infected cell was 0.13.
To determine more precisely when DNA cleavage occurs, Dam-overproducing hsd+ and Δhsd cells were infected with D364, and the kinetics of both genome penetration and DNA replication were monitored by assaying the time of methylation of GATC sites on the infecting genome. recD cells, defective in RecBCD nuclease, were used to minimize further degradation of products of EcoKI cleavage (23). Neither overproduction of Dam nor the absence of RecBCD affects the plating efficiency of D364 on strains containing EcoKI.
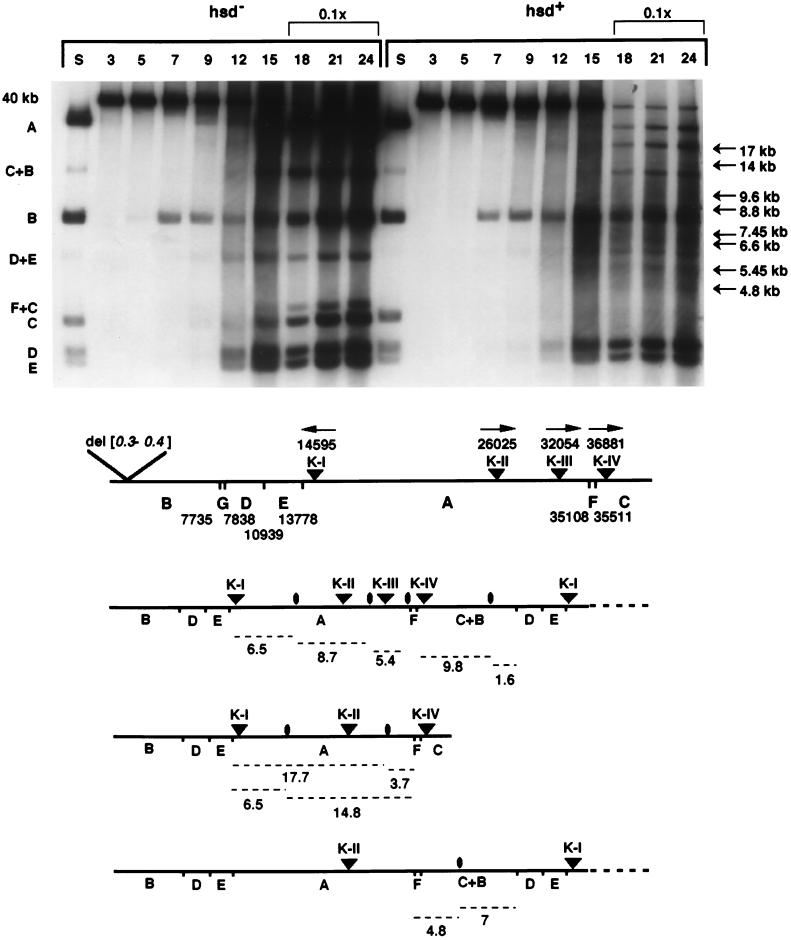
The first GATC sequence to enter the cell and be methylated by Dam in vivo, which makes it sensitive to cutting in vitro by DpnI, results in fragment B (Fig. 1). In both hsd+- and Δhsd-infected cells, this fragment can be visualized by 7 min of infection, indicating that at least the first 20% of phage DNA has penetrated the cells. In Δhsd-infected cells, the appearance of fragments D, E, A, and C by 9 min indicates that at least 90% of the chromosome has been internalized; however, in the infection of hsd+ cells, fragments A and C are very faint. Because EcoKI recognition sites are located within DNA represented by fragments A and C, the low intensity of these bands suggests that the majority of DNA fragments from this region of the T7 genome has been degraded by EcoKI. By 12 min, the increase in intensity of DNA bands, the appearance of the concatemer junction fragment C+B, and the heavy background signal all signify that phage DNA replication is occurring in both infections. From 15 to 24 min of infection in hsd+ cells, the intensity of fragments B, D, and E increases but to a lesser extent than in the infection of Δhsd cells. Fragments A, C, and the concatemer junction fragment C+B can also be visualized by 18 min in the restrictive infection, but again, their intensity is far less than in permissively infected cells. Nevertheless, the amplification of fragments corresponding to all regions of the T7 genome, including the concatemer junction, means that the DNA of a subpopulation of infecting phages is still biologically active in the late stages of the restrictive infection.
Figure 1.
Autoradiogram depicting the time course of methylation of D364 DNA during an infection, at a multiplicity of 0.4, of the Δhsd recD strain IJ1190 and the hsd+ recD strain IJ1409, both containing the dam plasmid pTP166. Time (in minutes) after infection is shown above the lanes. Lanes marked S contain DNA digested with Sau3AI; DNA in other lanes was digested with DpnI. Sau3AI or DpnI fragments are indicated by letters at the left of the panel, and their location on the phage genome is shown on the map below; nucleotide numbers reflect the 578-bp deletion in D364 DNA. The DpnI fragments D+E and F+C arise when the GATC sequences that separate the D and E or F and C fragments are not methylated completely in vivo (18). Arrows at the right of the panel point to bands unique to the infection of IJ1409; fragment sizes were estimated by using a commercial DNA ladder (not shown) and by D364 DNA DpnI fragments. EcoKI recognition sites are shown as triangles, arrows reflect the orientation of the asymmetric EcoKI recognition sequence 5′-AACN6GTGC, with respect to the sense, or l strand of T7 DNA. To visualize clearly the methylation profile of DNA before and during replication on the same autoradiogram, only 1/10 of the total DNA isolated at time points 18, 21, and 24 min was loaded on the agarose gel. Schematic diagrams of D364 DNA depict GATC sites, EcoKI sites, and predicted sites of EcoKI cleavage (ovals). Predicted DNA fragments (dashed lines) and their approximate sizes (in kilobases) generated by the combination of cleavage at the midpoint between two EcoKI recognition sites and by DpnI digestion are shown at the bottom.
It was proposed that EcoKI cleaves DNA when two translocating EcoKI complexes meet (7). In agreement with this model, the presence of intense fragments of the B, D, and E fragments during the late stages of DNA entry through DNA replication in the restrictive infection of hsd+ cells indicates that cleavage of D364 DNA does not occur appreciably at random sites to the left of the first restriction site K-I. More surprising is that several faint but discrete bands appear by 18 min in the restrictive infection (Fig. 1). These bands are not products of DpnI alone, and their sizes are inconsistent with a partial digest. They are also not seen in the permissive infection, suggesting that they might be discrete cleavage products of EcoKI. The 9.6-, 8.8-, 6.6-, and 5.45-kb fragments are consistent with the sizes predicted if cleavage occurred at the midpoint between two recognition sites (Fig. 1 and Table 2). Other bands can be rationalized by assuming that two EcoKI complexes, bound at nonneighboring sites on a subpopulation of D364 DNA molecules, simultaneously translocated DNA through themselves; the intervening site either could simply not be recognized within the requisite time frame or could have been modified by the EcoKI methyltransferase. The latter possibility is quite likely; relative to HsdM and HsdS, the HsdR subunit is thought to be limiting in wild-type cells (8, 24). In addition, because phage DNA replication is occurring, there is likely an excess of substrate for the enzyme.
Table 2.
Fragments predicted from combined DpnI and EcoKI cutting on concatemeric D364 DNA, assuming the midpoint cleavage model for EcoKI
| EcoK sites | Midpoint* | Affected DpnI fragment | Predicted fragment size, kb | Closest size fragment observed, kb |
|---|---|---|---|---|
| K-I → K-II | 20,310 | A | 6.5 | 6.6 |
| K-IV → K-I | 6,058 | C + B | 9.8, 1.6 | 9.6 |
| K-II → K-III | 29,040 | A | 8.7 | 8.8 |
| K-III → K-IV | 34,468 | A | 5.4 | 5.45 |
| K-II → K-IV | 20,310 | A | 14.8, 6.5 | 14, 6.6 |
| (not at K-III) | ||||
| K-II → K-IV | 31,453 | A | 17.7, 3.7† | 17 |
| (not at K-I, K-III) | ||||
| K-II → K-I | 711 | F + C + B‡ | 7.0, 4.8 | 7.45, 4.8 |
| (not at K-III, K-IV) |
Nucleotide position in D364 DNA.
Not resolved from the DpnI C fragment.
Assumes the GATC sequence between the F and C fragments is not methylated fully in vivo.
If EcoKI fails to recognize the K-III site, a 14.8-kb band would be formed between the DpnI site at position 35,108 and cleavage at the midpoint of K-I and K-II; similarly, if EcoKI fails to recognize K-I and K-III, a 17.7-kb band would be formed if D364 DNA were cut at the midpoint of K-II and K-IV (Fig. 1 and Table 2). When the GATC sequence at position 35,511 is not methylated fully in vivo by Dam, the DpnI F+C fragment becomes apparent (infection of Δhsd cells; Fig. 1); assuming that incomplete methylation also occurs during infection of hsd+ cells, we can also account for the 4.8-kb band and, with less confidence, the 7.45-kb band. Fragments of comparable size should result if EcoKI cuts D364 DNA midway between K-II on one genome and K-I on its neighbor in a replicating molecule, close to the concatemer junction. All discrete bands seen only in the infection of hsd+ cells can be accounted for remarkably well by the hypothesis that EcoKI cleavage in vivo occurs at the midpoint of two recognition sites (7). Other bands expected by this model may also be present but are obscured by normal DpnI fragments or were not resolved in this experiment.
The intensities of those bands that have no counterparts in the equivalent infection of a Δhsd strain are much less than the intensities of the DpnI bands D and E, which are not predicted to be affected by EcoKI. However, the fact that these additonal bands can be observed at all necessitates that at least two requirements are met. Fragments generated by EcoKI cutting in vivo must not be degraded further by a nonprocessive nuclease, which would convert any primary restriction products into DNAs with a broad size distribution. Although the infected cells used lack RecBCD, other host nucleases are still present, and the T7 gene 6 product is a potent exonuclease. Indeed, a high background is present in the 12–24 min lanes of the infection of hsd+ cells shown in Fig. 1, indicating that such secondary degradation is occurring.
A much more rigorous requirement for the appearance of discrete EcoKI-induced fragments is that two EcoKI molecules must bind to their recognition sites and initiate DNA translocation simultaneously and at the same rate. Asynchronous initiation of translocation by EcoKI, different rates of translocation, or interference of translocation by other enzymes that also use T7 DNA as a template would generate fragments with variable sizes, precluding the appearance of discrete bands. The presence of such bands in the D364 infection of hsd+ cells means that the above requirements can be met in vivo, albeit only rarely. Nevertheless, translocation of EcoKI from distinct and well separated sites on the D364 genome clearly can be a coordinated event.
DNA Ejection from a Bacteriophage λ Particle Results in Immediate Cleavage of 0.3+ Phage DNA by EcoKI.
The above data suggest that cleavage of D364 DNA occurs only after two or more recognition sites are accessible to EcoKI. The time when restriction is first observed is consistent with the idea that, in an infection by wild-type T7, transcription-coupled DNA entry allows gp0.3 to be synthesized before two recognition sites become accessible to EcoKI. To test this model directly, the effect of rapidly introducing T7 DNA into an hsd+ host was determined.
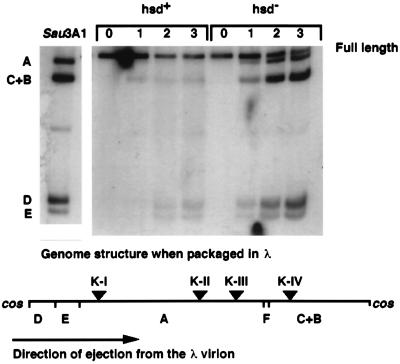
T71.7∷λcosΔ10-NB1 can be propagated as a T7 mutant on cells containing a gene 10-complementing plasmid, and the resulting stock is then conveniently termed T7[T7]. The phage can also be packaged into λ particles in vivo, in which case it is referred to as T7[λ] (18). T7[λ] productively infects hsd− λS cells containing gene 10 but plates at an efficiency less than 0.01 on hsd+ strains, even though the phage is 0.3+ (not shown). The low plating efficiency of T7[λ] on hsd+ hosts is caused by degradation of the phage genome after one-step DNA ejection from the λ virion. Ejection of DNA from a λ particle is fast, and the fragments C+B, D, E, and A appear by 1–2 min after infection of the Δhsd strain IJ1133 (Fig. 2). However, in the infection of the hsd+ strain IJ891, cleavage of T7[λ] DNA by EcoKI results in immediate loss of DNA. Although in a permissive infection, T7[λ] synthesizes gp0.3 within 1–2 min of infection (18), T7 DNA ejection from a λ virion must allow EcoKI immediate access to all four recognition sites on the genome before the inhibitor gp0.3 can be made.
Figure 2.
Autoradiogram depicting the time course of methylation of T7 DNA ejected from a λ virion during the infection of the hsd+ strain IJ891(pTP166) and of the Δhsd strain IJ1133(pTP166). Time (in minutes) after infection is shown above the lanes. T71.7∷λcosΔ10-NB1[λ] was added to cells at a multiplicity of 0.4. The lane marked Sau3AI illustrates the pattern of cleavage when each GATC site in the phage genome is cut (GATC sites are present in the λ cos DNA; any circular form of intracellular T7 DNA caused by annealing and ligation of the single-stranded ends of T7[λ] DNA cannot be detected in this experiment). DNA in other lanes was cleaved by DpnI; fragments are indicated by letters to the left of the panel, and their location on the phage genome is indicated on the map below; Dam methylation sites (vertical bars) and EcoKI recognition sites (triangles) are also shown. The 103-bp G fragment is not indicated.
EcoKI Can Catalyze Translocation of the Entire T7 Genome into the Cell.
The Studier model (7) predicts that restriction of a linear DNA by EcoKI requires two recognition sites; however, a single site might suffice for DNA translocation. We reasoned that, if this site were positioned within the leading 850 bp of T7 DNA, EcoKI might translocate the remainder of the genome into the cell. We isolated a derivative of the 0.3 mutant D364 that is insensitive to EcoKI (D364sK0; Table 1). The mutant likely has lost all EcoKI recognition sites, because it grows normally in cells containing the hsdR plasmid pSB2, which enhances restriction of λ (8); restriction of T7 may be even more enhanced, because hsdR is under T7 promoter control in pSB2. Even a single EcoKI site on an infecting T7 genome could lead to restriction by EcoKI in vivo, because replicating concatemers contain multiple recognition sites. A single EcoKI site was then introduced into D364sK0 by swapping its left end with that of sRK836, which contains an EcoKI recognition site at position 836 (17). This site is therefore part of the DNA ejected into the cell by a transcription-independent mechanism (18, 19). The resulting phage, sRK836sK0, is 0.3+ and is thus insensitive to both EcoKI and EcoBI during normal growth (Table 1).
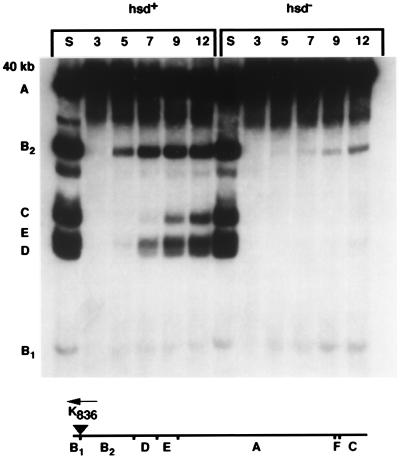
Infecting sRK836sK0 into rifampin-treated cells that lack EcoKI (Δhsd) results in very little DNA entering the cell. The 836-bp fragment B1 is visible by 5 min, but only a small fraction of infecting genomes penetrate far enough in vivo for methylation at position 8,312, which allows the B2 fragment to become apparent (Fig. 3). In contrast, infection of rifampin-treated hsd+ cells leads to complete genome internalization within 9 min. Fragments B1, B2, and traces of fragment D and E can be visualized by 5 min; by 7 min, fragments D and E are very intense, and by 9 min after infection, fragments A and C are both clearly visible. Based on the times of appearance of the B2, E, and C fragments, the approximate rate of translocation by EcoKI at 30°C in vivo is 100 bp/s, a value that agrees well with the estimated 200 bp/s at 37°C in vitro (7). No significant degradation of sRK836sK0 was observed in this experiment, in contrast to a control infection (in rifampin) by sRK836, whose genome contains five EcoKI sites (data not shown). All experiments described in this report were conducted at a low multiplicity of infection, allowing us to conclude that EcoKI requires only a single site on a linear molecule to catalyze T7 DNA translocation in vivo but requires more than one site to effect efficient cleavage.
Figure 3.
Autoradiogram depicting the time course of methylation of sRK836sK0 DNA during the infection of the hsd+ strain IJ891(pTP166) and of the isogenic Δhsd strain IJ1133(pTP166). Cells were pretreated with rifampin and infected at a multiplicity of 0.4. Lanes marked S correspond to sRK836sK0 DNA digested with Sau3AI and illustrate the cleavage pattern when every GATC site in the genome is cut. Time (in minutes) after infection is shown above the lanes. DpnI fragments are indicated by letters to the left, and their location on the phage genome is shown on the map below the panel; the single EcoKI recognition site (triangle) is also indicated. The 103-bp G fragment is not indicated.
Discussion
Several characteristics of T7 biology play supporting roles in reducing restriction by EcoKI. Transcription-dependent DNA translocation from the phage head into the cell ensures that gene 0.3 is necessarily transcribed. The product of gene 0.3 is the primary factor that allows the phage to escape restriction by binding directly to the enzyme and inhibiting its activity. Gene 0.3 is transcribed from the strong E. coli promoters A1, A2, and A3 and is the first T7 gene to be expressed; the 0.3 mRNA is also efficiently translated (20), and large amounts of gp0.3 are synthesized rapidly. Because rapid ejection from a λ particle allows restriction of 0.3+ phage, transcription-coupled DNA internalization is also crucial in avoiding restriction both by delaying access of EcoKI to its recognition sites and by allowing gp0.3 to accumulate.
Selective recognition of the T7 genome by E. coli and T7 RNA polymerases at early times seems logical. The E. coli enzyme normally transcribes the early region of the genome, simultaneously bringing it into the cell. In the absence of transcription, the infection is not established. gp0.3 can be detected within 2 min after infection, a time earlier than type I or type III restriction nucleases (17) or Dam (18, 19) can access the leading end of the infecting genome. Interestingly, the only enzymes known to be capable of immediately recognizing the leading 850 bp of the genome are the enzymes that catalyze entry of the DNA into the cell. Even after E. coli RNA polymerase is inactivated, superinfecting T7 phage genomes can be brought into the cell, because T7 RNA polymerase can transcribe from the øOL promoter, also located on the 850 bp directly ejected into the cell. Evolution of the process by which T7 DNA enters an infected cell likely has selected not only for specific recognition of the leading end of the DNA by the two RNA polymerases but also for the exclusion of other enzymes.
It is significant that the first 850 bp of the T7 genome enters the cell at a faster rate than that catalyzed by E. coli RNA polymerase (18, 19). In the latter study, phage mutants were described whose entire genomes were translocated into the cell at this faster rate in the absence of transcription. It is only logical then to argue that T7 has evolved its peculiar mode of genome internalization specifically to avoid restriction by type I enzymes and other potentially inhibitory cellular proteins. Only the T7 family of phages has been shown to internalize their genomes slowly. Other phages that avoid type I restriction do so by modifying their DNA or, in the case of phage P1, by coejecting antirestriction proteins together with the genome into the cell (1, 9, 10, 25). Like the entry of the T7 genome, conjugal DNA transfer is a slow process and some broad-host-range plasmids express antirestriction functions early during DNA transfer (11). Unlike T7, single-stranded DNA is transferred by conjugation, although it is rapidly made double-stranded. Both the IncN plasmid pKM101 and the IncI1 plasmid ColIb-P9 code for acidic proteins that may bind directly to the restriction enzyme, like T7 gp0.3. Conjugal transposons are also likely to express antirestriction functions early, because they transfer between genera (26).
Transcription-dependent internalization of the T7 genome also helps protect from type I restriction enzymes because of their mode of action. Because T7 DNA is linear, the Studier model (7) requires that two recognition sites become accessible to EcoKI for restriction to occur. During infection by T7, the first of these enters the cell after 7 min; at this time, the genome is being translocated into the cell at approximately 250 bp/s (18). At this rate, the second EcoKI site enters the cell 45 s later, consistent with in vivo observations that cleavage of an infecting 0.3− T7 chromosome by EcoKI begins 7.5–10 min after infection (ref. 17 and this work). Because it takes only 9 or 10 min for the entire T7 genome to be internalized (18) and because DNA replication is initiated by about 12 min (Fig. 1), there is only a narrow window of opportunity for EcoKI to initiate degradation of the infecting DNA before it is amplified by replication. Even when lacking the inhibitor gene 0.3, a significant fraction of infecting T7 genomes still make a burst of progeny phages in cells containing EcoKI (Table 1), reinforcing the suggestion that T7 DNA replication outpaces the restriction system (12).
In the experiment shown in Fig. 1, the paucity of fragment A, the persistence of fragments B, D, and E, and the appearance of discrete cleavage fragments during infection into a restricting host when gene 0.3 is not expressed are all consistent with the model that cleavage of DNA occurs when two bidirectionally translocating enzymes come into contact. However, fragment C, which contains the rightmost EcoKI site K-IV, is also largely degraded. According to the model (7), cleavage should occur midway between K-III and K-IV, which lies within the DpnI A fragment, leading to the expectation that the C fragment remains intact. The discrepancy likely can also be explained by the process of T7 genome entry, in which recognition sites become accessible to EcoKI in a temporal fashion. T7 RNA polymerase brings the site K-III into the cell, and K-IV enters about 20 s later, during which time EcoKI could bind to K-III and spool about 2 kb through itself. The cutting site predicted by the bidirectional translocation model is thus skewed from the midpoint of K-III and K-IV and is estimated to be very close to the DpnI site that defines fragment C. Alternatively, when the enzyme bound to K-III spools all the DNA corresponding to fragment C through itself and reaches the genome end, it may simply pause, allowing a second translocating enzyme molecule, bound to K-IV, to come into contact.
Cleavage between recognition sequences on λ DNA by EcoKI was first inferred purely from genetic data, long before DNA sequence information was obtained (27). Numerous in vitro studies of EcoKI eventually led to the bidirectional translocation model (7), which is now shown to fit in vivo data. The sizes of the discrete T7 DNA fragments resulting from restriction in vivo are most easily explained by EcoKI translocating DNA bidirectionally from a recognition site and catalyzing cleavage when two enzyme molecules collide. By using the type IC enzyme EcoRI24I on small plasmid DNAs in vitro, it was concluded that cutting also occurred at low efficiency within 250 bp of the recognition site (28). No comparable activity has ever been observed with the type IA enzyme EcoKI in vitro, including when T7 DNA was the substrate (7), and the sizes of bands expected if EcoKI did cut T7 DNA near its recognition sites in vivo cannot easily be fitted to the data shown in Fig. 1. Rare cleavage near a type I enzyme recognition site in vitro may simply reflect a proportion of enzyme molecules bound to their recognition site being incompetent for translocation, thereby requiring a second enzyme molecule to translocate the whole distance between two sites.
The appearance of discrete fragments requires that translocation of DNA through two distinct enzyme molecules that are bound to their recognition sites be synchronized. Synchronization is easy to establish in vitro; saturating levels of enzyme are added to substrate while translocation is prevented by omitting the cofactor ATP. Obviously, this procedure is not possible in vivo. How, then, can synchrony be achieved? Although EcoKI in solution is ≈473 kDa, consistent with the structure HsdR2:HsdM2:HsdS1 (29), it was proposed recently that two EcoKI molecules rapidly dimerize after they have bound to separate recognition sites (30). A dimeric form of EcoKI would seem to be the simplest way to achieve synchrony of translocation and cleavage in vivo. However, although dimerization may occur in vivo, it is clear that dimerization across two recognition sites is not necessary for DNA translocation. Only a single recognition site is necessary for EcoKI to translocate 39 kb of DNA through itself. It should be noted that, in addition to spooling DNA through itself, the ATPase activity associated with EcoKI also provides sufficient energy for the mechanical work necessary to translocate DNA through a cell envelope channel from one compartment into another, i.e., between the phage head and the cell cytoplasm.
Acknowledgments
We thank F. W. Studier and S. Rosenberg for providing strains. This work was supported by Public Health Service Grant GM32095.
Abbreviation
- kb
kilobase
References
- 1.Bickle T A, Krüger D H. Microbiol Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson G G, Murray N E. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 3.Hadi S M, Bickle T A, Yuan R. J Biol Chem. 1975;250:4159–4164. [PubMed] [Google Scholar]
- 4.Kan N C, Lautenberger J A, Edgell M H, Hutchison C A., III J Mol Biol. 1979;130:191–209. doi: 10.1016/0022-2836(79)90426-1. [DOI] [PubMed] [Google Scholar]
- 5.Bickle T A, Brack C, Yuan R. Proc Natl Acad Sci USA. 1978;75:3099–3103. doi: 10.1073/pnas.75.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan R, Hamilton D L, Burkhardt J. Cell. 1980;20:237–244. doi: 10.1016/0092-8674(80)90251-2. [DOI] [PubMed] [Google Scholar]
- 7.Studier F W, Bandyopadhyay P K. Proc Natl Acad Sci USA. 1988;85:4677–4681. doi: 10.1073/pnas.85.13.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webb J L, King G, Ternent D, Titheradge A J B, Murray N E. EMBO J. 1996;15:2003–2009. [PMC free article] [PubMed] [Google Scholar]
- 9.Krüger D H, Bickle T A. Microbiol Rev. 1983;47:345–360. doi: 10.1128/mr.47.3.345-360.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warren R A J. Annu Rev Microbiol. 1980;34:137–158. doi: 10.1146/annurev.mi.34.100180.001033. [DOI] [PubMed] [Google Scholar]
- 11.Belogurov A A, Delver E P, Rodzevich O V. J Bacteriol. 1992;174:5079–5085. doi: 10.1128/jb.174.15.5079-5085.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Studier F W. J Mol Biol. 1975;94:283–295. doi: 10.1016/0022-2836(75)90083-2. [DOI] [PubMed] [Google Scholar]
- 13.Studier F W, Movva N R. J Virol. 1976;19:136–145. doi: 10.1128/jvi.19.1.136-145.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mark K K, Studier F W. J Biol Chem. 1981;256:2573–2578. [PubMed] [Google Scholar]
- 15.Bandyopadhyay P K, Studier F W, Hamilton D L, Yuan R. J Mol Biol. 1985;182:567–578. doi: 10.1016/0022-2836(85)90242-6. [DOI] [PubMed] [Google Scholar]
- 16.Zavriev S K, Shemyakin M F. Nucleic Acids Res. 1982;10:1635–1652. doi: 10.1093/nar/10.5.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moffatt B A, Studier F W. J Bacteriol. 1988;170:2095–2105. doi: 10.1128/jb.170.5.2095-2105.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García L R, Molineux I J. J Bacteriol. 1995;177:4066–4076. doi: 10.1128/jb.177.14.4066-4076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García L R, Molineux I J. J Bacteriol. 1996;178:6921–6929. doi: 10.1128/jb.178.23.6921-6929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunn J J, Studier F W. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 21.Studier F W. Science. 1972;176:367–376. doi: 10.1126/science.176.4033.367. [DOI] [PubMed] [Google Scholar]
- 22.Marinus M G, Poteete A, Arraj J A. Gene. 1984;28:123–125. doi: 10.1016/0378-1119(84)90095-7. [DOI] [PubMed] [Google Scholar]
- 23.Simmon V F, Lederberg S. J Bacteriol. 1972;112:161–169. doi: 10.1128/jb.112.1.161-169.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiserova M, Janscak P, Benada O, Hubacek J, Zinkevich V E, Glover S W, Firman K. Nucleic Acids Res. 1993;21:373–379. doi: 10.1093/nar/21.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iida S, Hiestand-Nauer R, Sandmeier H, Lehnherr H, Arber W. Virology. 1998;251:49–58. doi: 10.1006/viro.1998.9405. [DOI] [PubMed] [Google Scholar]
- 26.Scott J R. J Bacteriol. 1992;174:6005–6010. doi: 10.1128/jb.174.19.6005-6010.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brammar W J, Murray N E, Winston S. J Mol Biol. 1974;90:633–647. doi: 10.1016/0022-2836(74)90529-4. [DOI] [PubMed] [Google Scholar]
- 28.Szczelkun M D, Janscak J, Firman K, Halford S E. J Mol Biol. 1997;271:112–123. doi: 10.1006/jmbi.1997.1172. [DOI] [PubMed] [Google Scholar]
- 29.Dryden D T F, Cooper L P, Thorpe P H, Byron O. Biochemistry. 1997;36:1065–1076. doi: 10.1021/bi9619435. [DOI] [PubMed] [Google Scholar]
- 30.Ellis D J, Dryden D T F, Berge T, Edwardson J M, Henderson R M. Nat Struct Biol. 1999;6:15–18. doi: 10.1038/4882. [DOI] [PubMed] [Google Scholar]