Abstract
Fungi are the principal degraders of biomass in most terrestrial ecosystems. In contrast to surface environments, deep-sea environmental gene libraries have suggested that fungi are rare and non-diverse in high-pressure marine environments. Here, we report the diversity of fungi from 11 deep-sea samples from around the world representing depths from 1500 to 4000 m (146–388 atm) and two shallower water column samples (250 and 500 m). We sequenced 239 clones from 10 fungal-specific 18S rRNA gene libraries constructed from these samples, from which we detected only 18 fungal 18S-types in deep-sea samples. Our phylogenetic analyses show that a total of only 32 fungal 18S-types have so far been recovered from deep-sea habitats, and our results suggest that fungi, in general, are relatively rare in the deep-sea habitats we sampled. The fungal diversity detected suggests that deep-sea environments host an evolutionarily diverse array of fungi dominated by groups of distantly related yeasts, although four putative filamentous fungal 18S-types were detected. The majority of our new sequences branch close to known fungi found in surface environments. This pattern contradicts the proposal that deep-sea and hydrothermal vent habitats represent ancient ecosystems, and demonstrates a history of frequent dispersal between terrestrial and deep-sea habitats.
Keywords: life under huge barometric pressures, osmotrophy, environmental gene library, microbial diversity, SSU rDNA phylogeny
1. Introduction
Environmental gene library methods are constantly expanding our understanding of microbial diversity and evolution, enabling scientists to sample previously unreachable environments and unculturable microbes. However, this research has so far failed to reach sampling saturation for either eukaryotes or prokaryotes, and therefore the extent and distribution of microbial life on Earth remains unknown (López-García et al. 2001; Moon-van der Staay et al. 2001; Edgcomb et al. 2002; López-García et al. 2003; Sogin et al. 2006; Stoeck et al. 2006). Fungal microbes encompass a large proportion of total microbial diversity (Lawley et al. 2004; Richards & Bass 2005) and biomass in terrestrial environments, and include key biological components in ecologically important symbioses, chemical cycles and food webs (Gadd et al. 2007). Despite the approximately 100 000 fungal species currently described, some estimates suggest that over 1.5 million fungal ‘species’ may exist (Hawksworth 2001). However, comparatively very few fungal lineages have been detected in deep oceanic environments (e.g. López-García et al. 2001, 2003, 2007; Edgcomb et al. 2002). Group-specific environmental PCR followed by cloning and gene library construction provides a means of comparing subsets of microbial diversity between environments, detecting low-abundance microbes and improving sampling efficiency (Vandenkoornhuyse et al. 2002; Bass & Cavalier-Smith 2004). Such methods enable us to describe fungal diversity even in habitats where it is apparently very limited.
Here, we use a primer set designed to detect the full taxonomic diversity of fungal small subunit ribosomal RNA genes (18S rDNA; Vandenkoornhuyse et al. 2002). We constructed 10 environmental gene libraries from marine environments including 11 deep-sea environmental DNA samples from sites in excess of 1500 m below the sea surface, and two from 250 and 500 m deep (electronic supplementary material, table 1). Phylogenetic analyses of the resulting SSU rDNA dataset reveal that the diversity of fungi in the deep sea is dominated by several groups of evolutionarily unrelated yeasts, with few 18S-types grouping with fungi known to be exclusively filamentous.
2. Material and methods
(a) Environmental sampling and DNA extraction
Environmental DNA was sampled as follows.
Deep sea. (i) Water column from near the wreck of Bismarck (3000 and 4000 m deep; 48°10′ N, 16°12′ W). Two DNA extractions each of 2 l of filtered water (pore size 0.2 μm). (ii) Water column from near the wreck of Titanic (3000 and 3700 m deep; 41°43′ N, 49°56′ W). Two DNA extractions each of 2 l of filtered water (pore size 0.2 μm). (iii) Mid-Atlantic Ridge: Rainbow hydrothermal sediment (metal-rich; 2264 m deep; 36°6′ N, 33°11′ W). DNA extracted from 200 mg sediment, as described by López-García et al. (2003). (iv) Mid-Atlantic Ridge: sterile microcolonizer deployed adjacent to a fluid emission at the Tour Eiffel chimney (Lucky Strike site: 37°17′ N, 32°16′ W, 1695 m deep; López-García et al. 2003). DNA was extracted from 100 mg of chemically inert plastic mesh, as described by López-García et al. (2001). (v) Water column from Drake Passage at 2000 and 3000 m deep (Antarctic polar front limit, 59°19′48″ S, 55°45′11″ W; sea floor at 3671 m). Sequentially filtered through 5 μl and then 0.2 μm filters, as described by López-García et al. (2001). (vi) Anoxic white bacterial mat cores (0–10 cm sediment depth) from Gulf of California (27°35′ N 111°28 W, 1575 m deep). DNA extracted from three separate samples, as described by Edgcomb et al. (2002).
Shallow marine site. (i) Two water column samples from Drake Passage at 250 and 500 m deep (Antarctic polar front limit, 59°19′48″ S, 55°45′11″ W; sea floor at 3671 m), as described by López-García et al. (2001).
(b) Environmental PCR and clone library analyses
Molecular screening. Fungal-specific primers AU2 and AU4 were used to amplify partial SSU rDNA sequences as described by Vandenkoornhuyse et al. (2002). Multiple PCR products for each DNA sample (electronic supplementary material, table 1) at two dilutions were pooled prior to separation on a 1% agarose gel from which they were excised and cleaned using the GFX gel purification kit (GE Healthcare). The resulting fragments were cloned using the TOPO TA cloning kit (Invitrogen) and sequenced using dye terminators and separated through capillaries on an automated 3730xl DNA analyzer (Applied Biosystems). Primers AU2 and AU4 were used for sequencing, ensuring that the variable V4 SSU rDNA region (Wuyts et al. 2000) used for defining unique sequences (see below) was double-strand sequenced. Sequences were aligned manually using SE-AL (http://evolve.zoo.ox.ac.uk/software.html?id=seal) and preliminary BioNJ trees were constructed. These were used to refine the environmental sequence alignment and determine 18S-type boundaries. The standard requirement for two genotypes to be considered distinct from one another was at least three differences within the V4 SSU rDNA region (Wuyts et al. 2000). This was reduced to a single difference where the same sequence character was shared by sequences deriving from more than one library. Bass et al. (2007) describe this strategy and justification for grouping sequences into genotypes in more detail. Thus, 239 sequences were reduced to 19 unique fungal SSU rDNA sequences and 7 non-fungal 18S rDNA sequence types. All 22 opisthokont (Baldauf 1999) sequences reported in figures 1–4 have been submitted to GenBank (accession nos. EU154971–EU154992 and EU158830–EU158831).
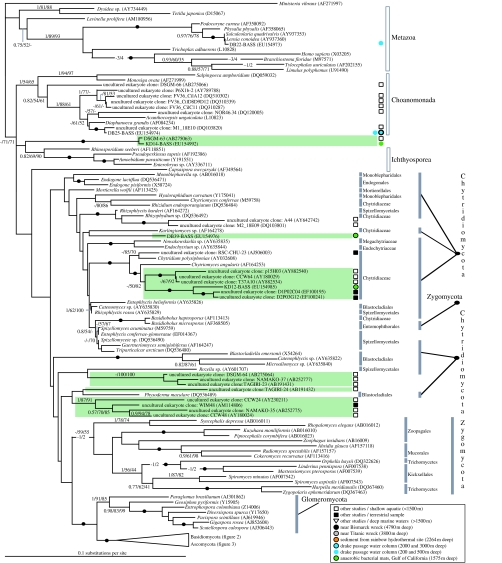
Figure 1.
Phylogeny demonstrating the branching position of deep-sea SSU types with a wide diversity of opisthokont sequences and sequences from general eukaryotic SSU eDNA surveys. A subsection of the fungal SSU rDNA phylogeny is shown, including other opisthokont groups such as chytrids, Zygomycetes and Glomeromycetes. Tree topology shown was calculated using PHYML (Guindon et al. 2005) from an alignment of 220 sequences and 1098 characters. Nodes in the phylogeny supported by in excess of 95% bootstrap support and 0.95 posterior probabilities in all the three analysis methods are represented by a black circle on the relevant node. Topology support values are given in the following order: Bayesian posterior probabilities calculated using MrBayes/100 PHYML (Guindon et al. 2005) bootstraps and 1000 LogDet distance bootstraps, and are shown when in excess of 0.75 and 50%, respectively. Non-fungal opisthokonts (Adl et al. 2005) are marked with a grey box bar. Higher fungal taxonomic groups labelled, according to Adl et al. (2005) or as listed in GenBank taxonomy databases, are marked with a shaded grey bar. Highly novel sequences potentially representing highly divergent lineages or unidentified taxonomic groups are placed within green boxes. All sequences are listed with GenBank accession numbers and environmental gene library sequences are marked according to crude environmental type listed in the key.
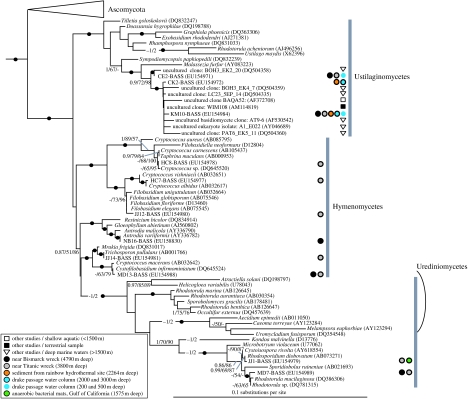
Figure 2.
Subsection of the fungal SSU rDNA focusing on the basidiomycete section of the phylogeny. See figure legend 1 for details on diagrammatic conventions. A wide diversity of basidiomycete fungi detected in deep-sea environments are shown. The diversity detected tends to branch very close to basidiomycete fungi with an yeast growth form with notable exceptions, including a close deep-sea relative of the fruiting bodied brown-rot fungi Antrodia.
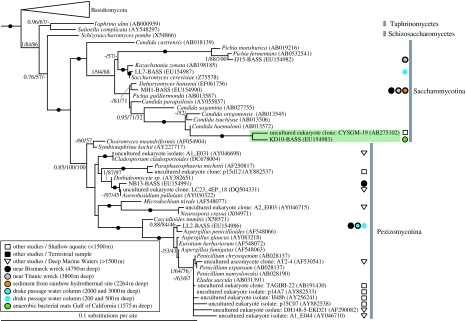
Figure 3.
Subsection of the fungal SSU rDNA focusing on the ascomycete section of the phylogeny. See figure legend 1 for details on diagrammatic conventions. Similar to the basidiomycete analyses (figure 2), a wide diversity of ascomycete fungi detected in deep-sea environments are shown, with the deep-sea ascomycete fungi detected generally grouping with microbes with an yeast growth form. However, again, we see exceptions to this, including the detection of a deep-sea Cladosporium and Aspergillus SSU type.
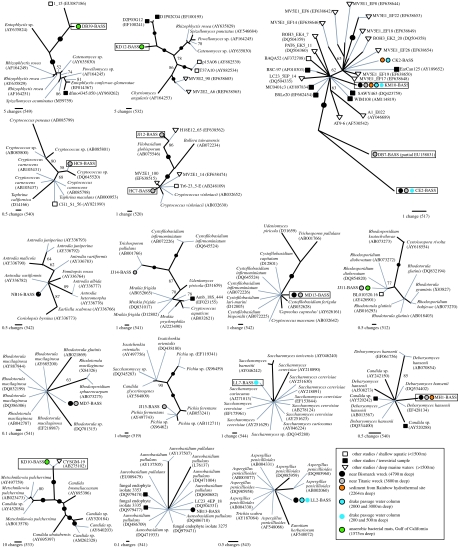
Figure 4.
Phylogenies of fungal sequences generated by this study in the context of their closest BLASTn matches. Boxed sequences are unique 18S-types (as defined by the fragment analysed: V4–V5 of 18S rDNA) detected in this study from deep-sea and marine environments. Phylogenies were calculated from highly similar sequences, therefore parsimony methods were used with gaps treated as a fifth character state. One thousand bootstrap replicates were calculated using the same parsimony methods and branches with 95% support are represented by a black circle on the relevant node. All trees are shown unrooted. Grey lines are only for labelling purposes. The scale bar for each phylogeny denotes the number of changes across the given branch length. The number of positions used to calculate each phylogeny is given in brackets below the scale bar.
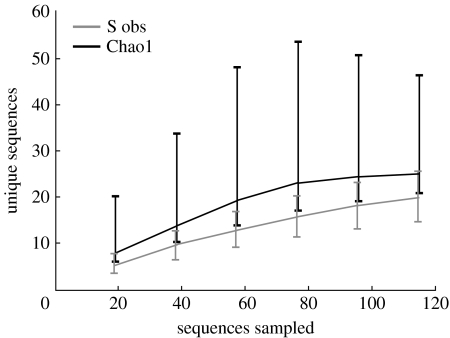
Species accumulation curves and Chao1 total diversity estimates for all deep-sea libraries were calculated in EstimateS (Colwell 2006) and plotted on a graph (figure 5).
Figure 5.
Sampling saturation analyses of our marine fungal molecular survey demonstrate that the sampling of our deep-sea libraries was close to saturation. Species accumulation curve and cumulative Chao1 total diversity estimate for all deep-sea libraries are plotted.
(c) Bioinformatics and phylogenetic analyses
All successful sequences were searched against NCBI–GenBank using the BLASTn (Altschul et al. 1990) database search method to identify crude taxonomic groupings. Our 22 new sequences were aligned with a comprehensive sampling of opisthokont sequences and sequences with close BLASTn hits for our marine sequences. Additional putative fungal SSU rDNA sequences from GenBank, sampled from general eukaryotic primer environmental gene libraries of deep-sea environments (López-García et al. 2001, 2003, 2007; Edgcomb et al. 2002), were also included (as available at the end of April 2007). These sequences were also subjected to the same process of Blast resampling (see above) and chimaera analyses (see below).
All SSU rDNA sequences were manually aligned using SeaView (Galtier et al. 1996). The alignment was closely inspected in the 5′ and 3′ regions in order to identify sequences with inconsistent patterns of similarity across the sequence alignment—potentially the product of PCR chimaera events (Berney et al. 2004). Alignment positions that were hypervariable or high in insertions/deletions, and so could not be aligned with confidence, were removed from all sequences in the alignment (masked) and preliminary phylogenetic analyses were conducted using distance settings (BioNJ) in PAUP (Swofford 2002) with 1000 bootstrap replicates. Initially, all highly similar sequences not closely clustering with any of the environmental gene library sequences were reduced to representative taxa. The PAUP analyses were repeated for four extra phylogenies using shorter sections of the full alignment: (i) the first 30% (positions 1–330), (ii) the first 40% (positions 1–440), (iii) the last 30% (768–1098), and (iv) the last 40% (658–1098). These four phylogenies were compared with each other and with the phylogenetic tree produced from the fully sampled sequence alignment in order to identify alternative branching orders and contrasting bootstrap supports that could be indicative of PCR chimaera events. These analyses demonstrated no candidate chimaera sequences.
The final masked alignment was analysed using Modelgenerator (Keane et al. 2004) to identify the most appropriate model for phylogenetic analyses (GTR+Γ (four discrete categories, α=0.57)+proportion of invariant sites (0.49)). Then, the phylogeny was calculated using two methods: (i) PHYML (Guindon et al. 2005) using the model parameters estimated using Modelgenerator and 100 bootstrap replicates, and (ii) MrBayes3 (Ronquist & Huelsenbeck 2003) analysis was conducted for 1 000 000 generation samples using model of site rate variation as mentioned above but allowing the MCMCMC to search alternative model parameter values. The tree search included two MCMCMC searches with four chains (heat parameters set to default) with a sampling frequency of 100 generations. The likelihood values of the two MCMCMC searches were compared to check whether they had converged and reached a plateau. A ‘burn-in’ of 3000 samples (300 000 generations) was excluded and the remaining plateaus sampled for the final tree. To further investigate topology support, we used LogDet-NJ distance methods with 0.366 proportion of sites assumed to be invariable (calculated from an ML tree score of a preliminary BioNJ tree in PAUP). This approach enabled us to observe support for the ML tree topology using a method that accounts for potential base-compositional heterogeneity biases (Lockhart et al. 1994).
To investigate further the evolutionary relationship between our marine and deep-sea fungi and other fungal sequences available in GenBank, we generated 16 separate alignments and phylogenetic trees. For each of our fungal sequences, we Blast searched approximately 500 positions, including the V4 and V5 18S rDNA regions (Wuyts et al. 2000), and constructed trees comprising our sequences and a minimum of the top 10 Blast matches. The only positions omitted from these analyses were the cases of single nucleotide ‘insertions’, which occurred in less variable regions of the alignment and were judged to be clearly sequence-read errors; the vast majority of these were instances of an additional nucleotide of the same kind in only one sequence (e.g. three Ts where all other sequences had two). The V4 region is generally the fastest evolving region of eukaryotic SSU rDNA genes (Wuyts et al. 2000) and is the most variable region of the reads generated by our primers. Therefore, we included this region and the same downstream stretch for each of our sequences to ensure that our new trees derive from alignments closely similar in length and that are long enough to give good phylogenetic signal, but as short as possible to minimize the effects of randomly distributed sequencing errors. Such sequencing errors could give inflated values for sequence variance, increasing diversity estimates and values of difference. Since all 16 alignments each encompassed highly similar sequences, we used parsimony methods to construct a phylogeny with gaps treated as a fifth character state. Parsimony analyses were conducted using 20 heuristic searches with the stepwise and TBR settings in PAUP. Bootstrapping was conducted with the same method but with 1000 pseudoreplicates.
3. Results and discussion
(a) Fungal diversity detected
From the 10 environmental gene libraries, we successfully sequenced 239 clones resulting in 26 eukaryotic 18S-types (figures 1–4). Of the successful sequences, 115 were fungal, the rest were SSU sequences from other eukaryotes (see below and non-rDNA amplicons. Of the 26 eukaryotic 18S-types, 19 were fungal (electronic supplementary material, table 2). Figure 5 shows that the sampling of fungi in our deep-sea libraries is close to saturation—the cumulative Chao1 diversity estimate curve is levelling off at a predicted 25 18S-types after 115 fungal sequences were sampled. Although sampling more eDNAs (particularly from other deep-sea habitat types) would be likely to reveal more taxa, the 13 eDNAs (electronic supplementary material, table 1) we screened harbour relatively few fungal genotypes. The number of 18S-types recovered from each sampling site ranged from 3 to 11 (electronic supplementary material, table 2). Interestingly, the sites with the highest diversity were also the deepest (Titanic and Bismarck wreck sites: 3000–4000 m).
No single clone library was dominated by a single 18S-type while eight of our deep-sea 18S-types were detected in one or more of our deep-sea libraries figures 2–4, electronic supplementary material, table 2; Furthermore, in five cases (CE2, CK2, KM10, NB13 and DB7; figure 4), the deep-sea fungal 18S-types that were detected grouped close to environmental clones sampled from existing general eukaryotic deep-sea environmental gene libraries (López-García et al. 2001, 2003, 2007; Edgcomb et al. 2002; figures 2–4). Given the breadth of diversity sampled in each clone library, the absence of some terrestrially common fungal taxonomic groups (see below) and the detection of several 18S-types in several different independently generated deep-sea environmental gene libraries, we do not suspect that our results have been compromised by contaminating sequences from non-target environments (see also §3b). Of the 19 fungal 18S-types detected, only three (CE2, KM10 and LL2) were detected in both shallow and deep sites (figures 2–4; electronic supplementary material, table 2); a further one was detected in shallower water column, but not below 1500 m (LL7).
Phylogenies including marine fungal sequences from previous studies (López-García et al. 2001, 2003, 2007; Dawson & Pace 2002; Edgcomb et al. 2002; Stoeck & Epstein 2003; Stoeck et al. 2003, 2006) revealed seven clusters of highly novel sequences (boxed in green; figures 1 and 3) grouping separately from known opisthokont groups or forming a long branch within identified fungal clades. Note that our phylogenetic analyses failed to recover strong bootstrap support among the lower branches of the fungi, consistent with other SSU rDNA analyses of fungal phylogeny, where multiple concatenated genes are required to improve resolution (James et al. 2006). In the absence of phylogenetic resolution among the lower branches of the fungi, we could not robustly classify KD14, DB39 or several other environmental 18S-types as fungi (boxed in green; figure 1).
We are not aware of any other published study using environmental 18S rDNA libraries to investigate deep-sea fungi, other than those using primers to detect all eukaryotes (López-García et al. 2001, 2003, 2007; Edgcomb et al. 2002), the data from which have been included in our trees (figures 1–4). Two main fungal groups were detected by those studies: Ustilaginomycetes and Pezizomycotina. Our new sequences are concordant with these findings. We discovered three new ustilaginomycete sequences (CE2, DB7 and KM10; figures 2,4) related but not identical to those from other deep-sea libraries, each of which we recovered from more than one library. Similarly, we found two pezizomycote sequences (NB13 and LL2), one of which was recovered from three libraries. Previous deep-sea studies (above 1500 m) did not recover any Saccharomycotina, Hymenomycetes or Urediniomycetes, whereas we detected four, six and two representatives of these groups, respectively. Thus, our use of group-specific primers has revealed fungi not previously found in deep-sea environments in several cases from independently collected and purified deep-sea samples from globally dispersed sites. In total, 11 of the 18S-types we detected have so far only been found in deep-sea habitats.
(b) Divergence between terrestrial surface and deep-sea fungi
All of our new 18S-types were robustly identified as fungal grouping, with known ascomycetes and basidiomycetes with moderate to strong bootstrap support and, significantly, with only short genetic distances separating them from known fungi sampled from surface environments. The trees in figure 4 show that some of our sequences have clearly different 18S sequences from their closest matches in GenBank, whereas others are identical (in terms of 18S rDNA sequence) to strains from surface or non-marine environments. In the former case, it is possible that the organisms represented by these sequences are specifically adapted to deep-sea habitats, but this can only be confirmed with more detailed genomic and/or culture-based analyses (the suggestion of deep-sea specificity may be an artefact caused by relatively low sampling and representation of such sequences in the database). The fact that some of our 18S-types are identical to those in GenBank does not necessarily imply that the organisms represented belong to the same strain or species, as faster-evolving genes than 18S rRNA (e.g. ITS rDNA) are required as markers for species boundaries in many eukaryotes and to distinguish between lineages with different ecological traits (Coleman 2000, 2007; Amato et al. 2007; Bass et al. 2007). Furthermore, it has been shown that Saccharomyces cerevisiae can alter its membrane composition to tolerate high hydrostatic pressure under short-term experimental conditions (Simonato et al. 2006), which implies that, for some fungi, rapid colonization of deep-sea habitats by surface-dwelling strains might be possible. While it is possible that sequences from abundant and ubiquitous taxa in our study such as Saccharomyces (LL7) and Aspergillus (LL2) are contaminants from surface environments, this is unsupported by any evidence, and arguably unlikely in the case of Aspergillus as sequence LL2 was recovered from three separate libraries constructed from DNA extracted by different, globally disparate laboratories. Interestingly, LL7 was only found in the 250–500 m water column library, not true deep-sea samples, and therefore is perhaps likely to be more closely related to surface strains.
It is therefore clear that some, if not all, fungi in deep-sea and surface environments have not been isolated from each other for long periods of evolutionary history, and that there is relatively frequent interchange between the two habitats. This pattern of microbial dispersal is inconsistent with the proposal that the deep-sea and hydrothermal vents represent ancient ecosystems harbouring ancient microbial lineages stemming from the tree of life before the Earth's atmospheric oxygen content increased (Horikoshi 1998; Reysenbach et al. 2000; López-García et al. 2003).
(c) Relative abundance of fungi in the deep sea
Our experimental approach set out to sample fungal diversity from the deep oceans. However, owing to the broad selectivity of the primer set used, we also detected six clearly non-fungal 18S-types. These sequences comprised four Cercozoa (Bass & Cavalier-Smith 2004; not included in further analyses) and two opisthokont sequences (Metazoa and a choanoflagellate; figure 1). The other sequence, KD14, groups close to the base of the fungi, but its taxonomic affinity is unclear. However, the pattern of broad fungal/opisthokont genotype detection is encouraging, suggesting that our methods are capable of detecting a broad evolutionary diversity of opisthokonts, including the majority of fungal SSU types. In support of this conclusion, we detected a diverse array of fungi including ascomycetes, basidiomycetes and chytrids from our marine samples (figures 1–3). However, we detected no putative Glomeromycetes or Zygomycetes from any of our marine samples. We do not believe that our methods prevented detection of these higher taxonomic groups because the same primer set has readily detected a wide diversity of these taxa from terrestrial environments (Vandenkoornhuyse et al. 2002). This suggests that fungal diversity in the deep seas tends to be dominated by ascomycetes and basidiomycetes, while other fungal taxonomic groups may be rare or absent. For some of these groups, this is unsurprising. For example, known Glomeromycetes form symbiotic relationships with aerobic phototrophic plants and algae (James et al. 2006); these are organisms, niches and lifestyles generally absent from deep-sea environments. However, the absence of Zygomycetes suggests that deep-sea high-pressure environments are not easily colonized by these filamentous fungi.
Our results also suggest that fungal diversity in deep-sea habitats is relatively low. In the previous study using the AU2/AU4 primer pair employed here (Vandenkoornhuyse et al. 2002), 200 clones produced 49 fungal sequences, with no other eukaryotes detected, whereas in our study 239 clones resulted in only 18 fungal sequences from the deep sea. Moreover, the demonstration that our methodology was not entirely fungal-specific is likely to provide information about the relatively high abundance of fungal cells in the sites sampled: the fact that 29% of the 18S-types we detected were non-fungal suggests that fungi are either rare in deep-sea habitats, or that other organisms are relatively much more abundant than fungi in deep-sea than terrestrial habitats, and therefore appear in our cloned sequences because the relative abundance of non-fungal DNA promotes primer binding errors by our ‘fungal-specific’ primers. By comparing the ease with which fungi were detected in our libraries, we make some tentative suggestions about the distribution of fungal diversity in deep-sea environments (electronic supplementary material, table 1). Libraries with high proportions of non-rDNA sequences might also be indicative of low levels of fungi (we carried out cloning quality control experiments to verify the efficiency of our cloning procedure and methods). Assuming that these inferences are correct, our results are consistent with the fact that the seas of the Drake Passage, particularly at depth, are low in biomass (López-García et al. 2001). The colonization module (López-García et al. 2003) is an even more extreme case—no fungal sequences were recovered from this sample at all. It is possible that a deployment time of 15 days is too short a time for fungal colonization under the prevailing conditions and/or that potentially colonizing fungal cells were sparsely distributed at this site. The shallow bacterial mat libraries also had a high proportion of non-rDNA sequences, perhaps due to the extreme dominance of non-fungal DNA in these samples. However, the deepest sites sampled (Titanic and Bismarck wreck areas) were both relatively rich in fungi and produced the highest ratios of fungal: non-fungal sequences in this study (electronic supplementary material, tables 1 and 2).
(d) Dominant fungal form in the deep seas
Species with yeast and filamentous growth forms are distributed as an overlapping mosaic throughout ascomycete and basidiomycete phylogenies (James et al. 2006). Of the 32 deep-sea (below 1500 m) fungal lineages identified from our study and previous deep-sea general eukaryotic surveys (López-García et al. 2001, 2003, 2007; Edgcomb et al. 2002), 23 18S-types branched very close to known fungi with yeast lifestyles (electronic supplementary material, table 3; figures 1, 2 and 4). The presence of multiple representatives from these distantly related yeast clades suggests that the yeast growth form is the dominant and most successful fungal form in the deep seas, and that the requirement for osmotrophy is not a barrier to life under huge hydrostatic pressures. Interestingly, eukaryote-wide SSU libraries have shown that a basidiomycete yeast, Cryptococcus curvatus, is the dominant microbial eukaryote 640 m deep at a cold methane seep site near Ishigaki Island, Japan (Takishita et al. 2006). Remarkably, however, we identified four fungal branches (NB16, LL2 (this study), A1_E031 and A2_E003 (Edgcomb et al. 2002)) likely to represent exclusively filamentous fungi presumably capable of forming hyphal structures at up to 388 atm (4000 m).
Many of the fungal types detected branched close to known pathogens, suggesting that they may be parasites or opportunistic pathogens of deep-sea animals. Such a scenario is consistent with the hypothesis that deep-sea and hydrothermal vents host a unique array of animal parasites (Moreira & López-García 2003).
Our results demonstrate the presence of an evolutionarily wide diversity of fungi in deep-sea environments (figures 1–4). The use of fungal-specific environmental gene library methods has identified that the dominant fungi in deep-sea environments are ascomycetes and basidiomycetes closely related to those capable of an yeast lifestyle and often branching close to opportunistic animal pathogens, suggesting that elements of the biology of these fungi predispose them to success in deep-sea ecosystems. The exact biological nature of these deep-sea fungi will provide interesting tools for understanding adaptive radiation of fungi into different environment types, survival of microbes in deep-sea high-pressure environments and use of fungi in high-pressure industrial processes (Simonato et al. 2006).
Acknowledgments
We thank N. J. Talbot for critical appraisal of the manuscript and Steven Hallam and David Moreira for providing marine environmental DNA. D.B. and A.H. are supported by NERC. T.A.R. is supported by a Leverhulme Early Career Fellowship. We also thank Nancy Burns, Helen Carter, Jasper Johnson, Matthew Hodges, Karen Mecz, Edward Mitchard, Cerys Evans and Christina Kimeze for their assistance in sequencing clones.
Supplementary Material
Supplementary tables 1–3
References
- Adl S.M, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. doi:10.1111/j.1550-7408.2005.00053.x [DOI] [PubMed] [Google Scholar]
- Altschul S.F, Gish W, Miller W, Myers E.W, Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amato A, Kooistra W.H, Ghiron J.H, Mann D.G, Proschold T, Montresor M. Reproductive isolation among sympatric cryptic species in marine diatoms. Protist. 2007;158:193–207. doi: 10.1016/j.protis.2006.10.001. doi:10.1016/j.protis.2006.10.001 [DOI] [PubMed] [Google Scholar]
- Baldauf S.L. A search for the origins of animals and fungi: comparing and combining molecular data. Am. Nat. 1999;154:178–188. doi: 10.1086/303292. doi:10.1086/303292 [DOI] [PubMed] [Google Scholar]
- Bass D, Cavalier-Smith T. Phylum-specific environmental DNA analysis reveals remarkably high global biodiversity of Cercozoa (Protozoa) Int. J. Syst. Evol. Microbiol. 2004;54:2393–2404. doi: 10.1099/ijs.0.63229-0. doi:10.1099/ijs.0.63229-0 [DOI] [PubMed] [Google Scholar]
- Bass D, Richards T.A, Matthai L, Marsh V, Cavalier-Smith T. DNA evidence for global dispersal and probable endemicity of protozoa. BMC Evol. Biol. 2007;7:162. doi: 10.1186/1471-2148-7-162. doi:10.1186/1471-2148-7-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berney C, Fahrni J, Pawlowski J. How many novel eukaryotic ’kingdoms’? Pitfalls and limitations of environmental DNA surveys. BMC Biol. 2004;2:13. doi: 10.1186/1741-7007-2-13. doi:10.1186/1741-7007-2-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman A.W. The significance of a coincidence between evolutionary landmarks found in mating affinity and a DNA sequence. Protist. 2000;151:1–9. doi: 10.1078/1434-4610-00002. doi:10.1078/1434-4610-00002 [DOI] [PubMed] [Google Scholar]
- Coleman A.W. Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Res. 2007;35:3322–3329. doi: 10.1093/nar/gkm233. doi:10.1093/nar/gkm233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell, R. K. 2006 EstimateS: statistical estimation of species richness and shared species from samples, version 8. Persistent URL: purl.oclc.org.estimates
- Dawson S.C, Pace N.R. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl Acad. Sci. USA. 2002;99:8324–8329. doi: 10.1073/pnas.062169599. doi:10.1073/pnas.062169599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgcomb V.P, Kysela D.T, Teske A, de Vera Gomez A, Sogin M.L. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc. Natl Acad. Sci. USA. 2002;99:7658–7662. doi: 10.1073/pnas.062186399. doi:10.1073/pnas.062186399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadd G, Watkinson S.C, Dyer P.S. Cambridge University Press; Cambridge, UK: 2007. Fungi in the environment. [Google Scholar]
- Galtier N, Gouy M, Gautier C. Seaview and Phylo_Win: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. doi:10.1093/nar/gki352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth D.L. The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol. Res. 2001;105:1422–1432. [Google Scholar]
- Horikoshi K. Barophiles: deep-sea microorganisms adapted to an extreme environment. Curr. Opin. Microbiol. 1998;14:291–295. doi: 10.1016/s1369-5274(98)80032-5. doi:10.1016/S1369-5274(98)80032-5 [DOI] [PubMed] [Google Scholar]
- James T.Y, et al. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. doi:10.1038/nature05110 [DOI] [PubMed] [Google Scholar]
- Keane T.M, Creevey C.J, Naughton T.J, Pentony M.M, Naughton T.J, Mcinerney J.O. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 2004;6:29. doi: 10.1186/1471-2148-6-29. doi:10.1186/1471-2148-6-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley B, Ripley S, Bridge P, Convey P. Molecular analysis of geographic patterns of eukaryotic diversity in Antarctic soils. Appl. Environ. Microbiol. 2004;70:5963–5972. doi: 10.1128/AEM.70.10.5963-5972.2004. doi:10.1128/AEM.70.10.5963-5972.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart P.J, Steel M.A, Hendy M.D, Penny D. Recovering evolutionary trees under a more realistic model of sequence evolution. Mol. Biol. Evol. 1994;11:605–612. doi: 10.1093/oxfordjournals.molbev.a040136. [DOI] [PubMed] [Google Scholar]
- López-García P, Rodriguez-Valera F, Pedros-Alio C, Moreira D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature. 2001;409:603–607. doi: 10.1038/35054537. doi:10.1038/35054537 [DOI] [PubMed] [Google Scholar]
- López-García P, Philippe H, Gail F, Moreira D. Autochthonous eukaryotic diversity in hydrothermal sediment and experimental microcolonizers at the Mid-Atlantic Ridge. Proc. Natl Acad. Sci. USA. 2003;100:697–702. doi: 10.1073/pnas.0235779100. doi:10.1073/pnas.0235779100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-García P, Vereshchaka A, Moreira D. Eukaryotic diversity associated with carbonates and fluid–seawater interface in Lost City hydrothermal field. Environ. Microbiol. 2007;9:546–554. doi: 10.1111/j.1462-2920.2006.01158.x. doi:10.1111/j.1462-2920.2006.01158.x [DOI] [PubMed] [Google Scholar]
- Moon-van der Staay S.Y, De Wachter R, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature. 2001;409:607–610. doi: 10.1038/35054541. doi:10.1038/35054541 [DOI] [PubMed] [Google Scholar]
- Moreira D, López-García P. Are hydrothermal vents oases for parasitic protists? Trends Parasitol. 2003;19:556–558. doi: 10.1016/j.pt.2003.09.013. doi:10.1016/j.pt.2003.09.013 [DOI] [PubMed] [Google Scholar]
- Reysenbach A.L, Banta A.B, Boone D.R, Cary S.C, Luther G.W. Microbial essentials at hydrothermal vents. Nature. 2000;404:835. doi: 10.1038/35009029. doi:10.1038/35009029 [DOI] [PubMed] [Google Scholar]
- Richards T.A, Bass D. Molecular screening of free-living microbial eukaryotes: diversity and distribution using a meta-analysis. Curr. Opin. Microbiol. 2005;8:240–252. doi: 10.1016/j.mib.2005.04.010. doi:10.1016/j.mib.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. doi:10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Simonato F, Campanaro S, Lauro F.M, Vezzi A, D'Angelo M, Vitulo N, Valle G, Bartlett D.H. Piezophilic adaptation: a genomic point of view. J. Biotechnol. 2006;126:11–25. doi: 10.1016/j.jbiotec.2006.03.038. doi:10.1016/j.jbiotec.2006.03.038 [DOI] [PubMed] [Google Scholar]
- Sogin M, Morrison H.G, Huber J.A, Welch D.M, Huse S.M, Neal P.R, Arrieta J.M, Herndl G.J. Microbial diversity in the deep sea and underexplored “rare biosphere”. Proc. Natl Acad. Sci. USA. 2006;103:12 115–12 120. doi: 10.1073/pnas.0605127103. doi:10.1073/pnas.0605127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck T, Epstein S. Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Appl. Environ. Microbiol. 2003;69:2657–2663. doi: 10.1128/AEM.69.5.2657-2663.2003. doi:10.1128/AEM.69.5.2657-2663.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck T, Taylor G.T, Epstein S.S. Novel eukaryotes from the permanently anoxic Cariaco Basin (Caribbean Sea) Appl. Environ. Microbiol. 2003;69:5656–5663. doi: 10.1128/AEM.69.9.5656-5663.2003. doi:10.1128/AEM.69.9.5656-5663.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck T, Hayward B, Taylor G.T, Varela R, Epstein S.S. A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples. Protist. 2006;157:31–43. doi: 10.1016/j.protis.2005.10.004. doi:10.1016/j.protis.2005.10.004 [DOI] [PubMed] [Google Scholar]
- Swofford, D. L. 2002 PAUP* Phylogenetic analysis using parsimony (*and other methods), version 4. Sunderland, MA: Sinauer Associates.
- Takishita K, Tsuchiya M, Reimer J.D, Maruyama T. Molecular evidence demonstrating the basidiomycetous fungus Cryptococcus curvatus is the dominant microbial eukaryotic in sediment at the Kuroshima Knoll methane seep. Extremophiles. 2006;10:165–169. doi: 10.1007/s00792-005-0495-7. doi:10.1007/s00792-005-0495-7 [DOI] [PubMed] [Google Scholar]
- Vandenkoornhuyse P, Baldauf S.L, Leyval C, Straczek J, Young J.P.W. Extensive fungal diversity in plant roots. Science. 2002;295:2051. doi: 10.1126/science.295.5562.2051. doi:10.1126/science.295.5562.2051 [DOI] [PubMed] [Google Scholar]
- Wuyts J, De Rijk P, Van de Peer Y, Pison G, Rousseeuw P, De Wachter R. Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eukaryotic small subunit ribosomal RNA. Nucleic Acids Res. 2000;28:4698–4708. doi: 10.1093/nar/28.23.4698. doi:10.1093/nar/28.23.4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables 1–3