Abstract
Biotin synthase (BioB) is an S-adenosylmethionine radical enzyme that catalyzes addition of sulfur to dethiobiotin to form the biotin thiophane ring. In vitro, E. coli BioB is active for only one turnover, during which the [2Fe-2S]2+ cluster is destroyed, one sulfide from the cluster is incorporated as the biotin thiophane sulfur, while Fe2+ ions and the remaining S2- ion are released from the protein. The present work examines the fate of the protein following the loss of the FeS clusters. We examine the quaternary structure and thermal stability of active and inactive states of BioB, and find that loss of either the [4Fe-4S]2+ or [2Fe-2S]2+ clusters results in destabilization but not global unfolding of BioB. Using susceptibility to limited proteolysis as a guide, we find that specific regions of the protein appear to be transiently unfolded following loss of these clusters. We also examine the in vivo degradation of biotin synthase during growth in low-iron minimal media and find that BioB is degraded by an apparent ATP-dependent proteolysis mechanism that sequentially cleaves small fragments starting at the C-terminus. BioB appears to be resistant to degradation and capable of multiple turnovers only under high-iron conditions that favor repair of the FeS clusters, a process most likely mediated by the Isc or Suf iron-sulfur cluster assembly systems.
Keywords: Biotin synthase, BioB, Iron-Sulfur Cluster, Protein Degradation
Biotin synthase (BioB) is an S-adenosylmethionine (AdoMet) radical enzyme that catalyzes oxidation of dethiobiotin and the addition of sulfur to form the biotin thiophane ring [1]. The enzyme is a homodimeric iron-sulfur protein in which each monomer contains both a [4Fe-4S]2+ cluster and a [2Fe-2S]2+ cluster [2]. The [4Fe-4S]2+/+ cluster is bound to a conserved CxxxCxxC motif that is common to a large superfamily of AdoMet-dependent radical enzymes, while the [2Fe-2S]2+ cluster, found only in BioB, is bound to 3 cysteines and an arginine that are located throughout the primary sequence [3-5]. The structure of E. coli BioB [6] shows that AdoMet is covalently coordinated to a unique iron in the catalytic [4Fe-4S]2+ cluster, and both are bound to an extended loop at the C-terminal end of an (αβ)8 barrel, with the [4Fe-4S]2+ cluster ∼5-7 Å from the exterior of the protein. In contrast, the [2Fe-2S]2+ cluster is bound within the core of the (αβ)8 barrel, in close proximity to dethiobiotin and ∼15-20 Å from the surface of the protein.
We have proposed a mechanism for the formation of biotin in which each FeS cluster plays a distinct catalytic role [4, 7]. The [4Fe-4S]2+ cluster accepts a low potential electron from flavodoxin and transfers this electron to the AdoMet sulfonium. The one-electron reduced sulfonium spontaneously cleaves to yield methionine and a 5′-deoxyadenosyl radical, which then abstracts a hydrogen atom from a methyl group at C9 position of dethiobiotin [8]. The substrate-centered carbon radical is then quenched by the bridging sulfide of the [2Fe-2S]2+ cluster [9], forming 9-mercaptodethiobiotin as a discrete tightly-bound intermediate [10]. Following dissociation of methionine and 5′-deoxyadenosine and binding of a second equivalent of AdoMet, a similar sequence of events leads to formation of a C-S bond at the C6 position of dethiobiotin, completing formation of the biotin thiophane ring. The overall reaction involves oxidation of dethiobiotin through H atom transfer to two transient 5′-deoxyadenosyl radicals and oxidation of sulfide through electron transfer to the Fe3+ ions within the [2Fe-2S]2+ cluster.
Several studies have supported a role for the [2Fe-2S]2+ cluster in providing the sulfur for biotin formation. BioB expressed in media containing 35S-cysteine [11], or apoBioB reconstituted with Fe3+ and 34S-sulfide or selenide [12-14], results in an enzyme with an isotope- or selenide-labeled [2Fe-2S]2+ cluster whose turnover produces biotin that incorporates the heavy-atom label in 60-80% yield. When turnover of BioB is monitored by UV/visible spectroscopy, the absorption band at ∼450 nm associated with the [2Fe-2S]2+ cluster disappears at a rate that is similar to the rate of biotin formation [9]. When 57Fe-labeled enzyme is analyzed by Mössbauer spectroscopy prior to and during turnover, the most notable changes are the conversion of a narrow quadropole doublet associated with the protein-bound [2Fe-2S]2+ cluster to overlapping broad quadropole doublets associated with Fe2+ ions coordinated to various buffer components [15, 16]. Finally, the most compelling evidence comes from the crystal structure of BioB, which shows that a bridging sulfide of the [2Fe-2S]2+ cluster is found in very close proximity to the dethiobiotin C9 methyl (∼4.7 Å) and there is no room for any other sulfur containing substrates to bind within the active site; this space is completely filled by the two substrates and the two FeS clusters [6].
Following a single turnover, both UV/visible and Mössbauer spectra reveal that the [2Fe-2S]2+ cluster is destroyed and dissociates from the enzyme [9, 15]. In addition, the [4Fe-4S]2+ cluster is rapidly oxidized in the absence of the [2Fe-2S]2+ cluster and bound substrates [17], most likely resulting in the formation of an inactive apoenzyme following turnover in aerobic bacteria. Mössbauer spectra of whole cell preparations of E. coli expressing recombinant BioB indicate that <35% of the enzyme contains a [4Fe-4S]2+ cluster, probably due to the air-sensitivity of this cluster [18, 19]. Since the biotin requirements of bacteria are very low, on the order of 100-1000 molecules per cell [20], it has been suggested that BioB may be a stoichiometric reactant that is degraded after a single turnover. However, Choi-Rhee and Cronan have recently used a combination of 35S-methionine protein labeling and Western blots to analyze the production of biotin from BioB under conditions that suppress new protein synthesis [21]. They find that each BioB polypeptide chain produces, on average, ∼20 biotin molecules in 4 hrs (kcat ≈ 0.08 min-1), which therefore requires that active BioB is regenerated after each turnover. Since turnover is accompanied by loss of the [2Fe-2S]2+ cluster, and possibly also the [4Fe-4S]2+ cluster, and recent studies have demonstrated that BioB is inactive in the absence of these clusters [3, 9, 16], reactivation of the enzyme likely involves cluster repair or reassembly. In vivo, this process is may be mediated by the iron-sulfur cluster assembly systems (isc and/or suf operons in E. coli). However, the binding site for the [2Fe-2S]2+ cluster is located deep within the (αβ)8 barrel [6], >15 Å from solvent, and therefore access to this site could limit the cluster assembly process. Opening the BioB enzyme active site to facilitate reassembly of the [2Fe-2S]2+ cluster would require either a significant conformational change or would require localized or global unfolding of the polypeptide chain, a process that might render the protein susceptible to degradation. Indeed Chio-Rhee and Cronan detected a significant decrease in the in vivo BioB concentration when the enzyme was forced to undergo multiple turnovers over 24 hrs in dethiobiotin-supplemented minimal media [21].
Very little is known about how BioB responds to loss of the iron-sulfur clusters. There are several distinct possibilities, of which global unfolding is the most severe. It is also possible that smaller regions of the protein are destabilized, leading to unfolding of a specific subdomain. If this localized unfolding were to involve the dimer interface, one might observe dissociation into monomer subunits. A final possibility is that nothing happens; the structure and integrity of the protein could be maintained, with absence of the substrates providing sufficient access to the FeS cluster binding sites. Ideally, to address these possibilities in a manner most relevant to catalysis, one would like to compare the physical status of the form of the enzyme prior to turnover, containing both [2Fe-2S]2+ and [4Fe-4S]2+ clusters, to that found immediately after turnover, containing only a [4Fe-4S]2+ cluster. However, this latter form of the enzyme is exquisitely air sensitive, and we were not able to measure the properties of 4Fe-BioB without simultaneous oxidative destruction of the [4Fe-4S]2+ cluster. This air-sensitivity most likely results in a similar loss of this cluster in vivo in aerobic E. coli, and therefore a comparison of the holoenzyme to the apoenzyme may in fact be relevant. We have inferred the consequences of the loss of each FeS cluster by comparing the properties of three forms of BioB that are reasonably air stable: apoBioB, lacking both FeS clusters, 2Fe-BioB, containing only the [2Fe-2S]2+ cluster, and 2Fe4Fe-BioB, containing both the [2Fe-2S]2+ and [4Fe-4S]2+ clusters, as well as the two substrates dethiobiotin and AdoMet to inhibit air oxidation. We first used analytical ultracentrifugation and circular dichroism spectroscopy to examine the integrity of the quaternary and tertiary/secondary structures of the protein. Since the latter studies suggested a partial unfolding of apoBioB at 37 °C, we then used limited tryptic proteolysis to probe for regions of the protein that are more exposed in the apoprotein. These studies suggest a transient localized unfolding that may expose the FeS cluster binding sites to solvent for cluster reassembly. Lastly, we turned our attention to the in vivo fate of cluster-deficient BioB. Our data supports a model in which FeS cluster assembly or repair and targeted protein degradation compete to balance the various needs of the cell.
Experimental methods
Materials
Unless otherwise stated, all reagents were obtained from commercial sources and used without further purification. N-Terminal His6-BioB was purified as previously described [22]. For aerobically purified BioB, protein concentration was determined immediately after purification using ε452 = 8400 M-1 cm-1 per monomer [4] and all protein concentrations are given as the monomer concentration. For other samples, protein concentration was determined using the Bradford assay with BSA as a standard and divided by a correction factor of 1.1 [4]. Rabbit polyclonal antibodies to His6-BioB were obtained from Rockland Biochemical and specificity assessed against authentic BioB and crude E. coli B lysates. Several contaminating bands are observed in lanes loaded with lysates at greater than 100 μg total protein, corresponding to a detection limit of ∼0.1 ng per mg of total protein. Blots generated in this study use 20-fold lower protein levels to ensure that the observed bands correspond to authentic biotin synthase and biotin synthase-derived fragments. Western blots were detected with peroxidase-fused donkey anti-rabbit IgG (Amersham) and developed with tetramethylbenzidene (Promega). 2Fe4Fe-BioB was generated by incubation of 2Fe-BioB with 4 equivalents FeCl3, Na2S, and 5 mM DTT as previously described [4]. ApoBioB was generated by a modification of the procedure of Fontecave and coworkers [23]. Briefly, 2Fe-BioB (200 μM) was incubated with 10 mM EDTA, 50 mM Tris HCl, pH 8 under anaerobic conditions, sodium dithionite (1 mM) was added, and the visible spectrum monitored to verify loss of the [2Fe-2S]2+ cluster. The excess reagents and iron were removed by desalting in 50 mM Tris, pH 8 on a Sephadex G25 column (1 × 30 cm) under anaerobic conditions, and the protein concentrated in a 10 kDa centrifugal concentrator. Iron analysis indicates that <5% of the original Fe content remains in apoBioB generated in this manner
Analytical centrifugation
Native molecular weights were determined for apoBioB and 2Fe-BioB by equilibrium analytical centrifugation. Experiments were performed in a Beckman XLA analytical centrifuge equipped with an AN-60 Ti rotor. Cells were filled with each protein (100 μl at 5 μM) in 50 mM Tris HCl, 1 mM tris(2-carboxyethyl)-phosphine HCl (TCEP), pH 7.5. Samples were subjected to centrifugation at speeds of 10000, 15000, and 20000 rpm at 20 °C, and following equilibration for 2 hrs, five replicate radial scans at 280 nm were averaged for each data set. Data sets collected after 2 and 4 hrs were indistinguishable indicating that equilibrium was achieved and that the samples were stable over several hours. Absorbance data collected at 3 speeds were globally fit using the program Sedphat [24] to a mixture of non-equilibrating species. Initial estimated values of Abstotal = 0.3, dimer MW = 80000 and s = 5, tetramer MW = 160000 and s = 7, were provided by prior velocity centrifugation experiments (data not shown). Following multiple rounds of global fitting with all parameters allowed to vary, a consensus parameter set was derived. Attempts to fit to an equilibrium between monomer, dimer, and tetramer species, or just dimer and tetramer species, gave poor quality fits and were disregarded.
Circular dichroism spectroscopy
Circular dichroism spectra were recorded from 190 to 260 nm on a Jasco J-810 spectrapolarimeter equipped with a Jasco PFD-425S temperature controller. Protein samples (5 μM) in 10 mM MOPS, 25 mM NaF, pH 7.5 were placed in a rectangular quartz CD cell (1.0 mm pathlength) under nitrogen, the cell tightly sealed, and spectra were recorded at 25 °C. The secondary structure composition was estimated from each spectrum using DICHROWEB [25]. Samples were then subjected to a thermal melt from 4 to 98 °C in 2 °C increments with temperature slope of 20 °C/hour. A reverse temperature scan was also performed on each sample to monitor for possible refolding during sample cooling, but irreversible protein aggregation at high temperature prevented significant refolding. Thermal melts were fit to the Gibbs-Helmholtz equation as described by Fairman and coworkers [26].
Limited tryptic proteolysis
The transient unfolding of apoBioB, 2Fe-BioB, and 2Fe4Fe-BioB was assessed by monitoring susceptibility to proteolysis with trace levels of trypsin. Each protein (1 mg/ml) was incubated with trypsin (10 ng/ml) in 25 mM Tris HCl, pH 8.0, and at intervals 50-100 μl samples were quenched with Nα-tosyl-lysyl-chloroketone (10 μg/ml). Samples were analyzed by SDS polyacrylamide gel electrophoresis on 4-20% gradient gels (BioRad) and stained with Coomassie Brilliant Blue R-250 (BioRad). Wet gels sandwiched between glass plates were scanned at high resolution using a standard flatbed optical scanner. The relative optical density of major bands was determined using NIH Image 1.61 (available at http://rsb.info.nih.gov/nih-image/). Select samples were also analyzed by MALDI-TOF mass spectrometry (supplementary data).
Proteolysis by endogenous E. coli proteases was also assessed by incubating each protein (1 mg/ml) with a cell-free lysate (0.5 mg/ml) at 37 °C in 25 mM Tris HCl, pH 8.0 in the presence or absence of ATP (1 mM) and MgCl2 (5 mM). The wild-type strain E. coli B was grown in Luria broth to an OD600 = 4 and cells collected by centrifugation. Cells were resuspended in 25 mM Tris HCl, pH 8.0, lysed by sonication, and pelleted for 20 min at 18,000 ×g to produce the fresh cell-free lysate. No attempt was made to remove ATP or other possible substrates and cofactors. Following incubation with this lysate, samples of biotin synthase were quenched by addition of a protease inhibitor cocktail (Sigma) and frozen at -80 °C until further analysis. Samples were analyzed by SDS polyacrylamide gel electrophoresis on 4-20% gradient gels (BioRad) and stained with Coomassie Brilliant Blue R-250.
In vivo proteolytic degradation
Proteolysis of biotin synthase was examined during growth and expression in E. coli strain BL21(DE3)pLysS carrying plasmid pJJ15-4A expressing the E. coli bioB gene with an N-terminal hexahistidine tag [22]. The procedure employed was a slight modification of that described by Choi-Rhee and Cronan [21]. For all steps, medium A is defined as M9 medium with glucose (0.2% w/v), vitamin-depleted casamino acids (1% w/v), ampicillin (50 μg/ml), and chloramphenicol (30 μg/ml). Overnight cultures (50 ml) in medium A were pelleted at 1500 ×g, washed once with medium A, resuspended in medium A with isopropylthiogalactoside (0.5 mM), and then incubated with gentle shaking for 3 h at 25 °C to allow expression of His6-BioB. The cells were pelleted and resuspended in medium A (50 ml) with tetracycline (60 μg/ml) to inhibit further protein synthesis, and the cells were incubated for 30 min at 37 °C. Biotin or dethiobiotin (5 μM) were then added and the cells further incubated with gentle shaking for 8 h at 37 °C. At intervals, 10 ml of the culture was removed, pelleted by centrifugation at 1500 ×g, and the cell pellet frozen at -80 °C. The pellet was later resuspended in 50 mM Tris, pH 8 containing a protease inhibitor cocktail (Sigma), lysed by brief sonication, and the soluble fraction examined by SDS polyacrylamide gel electrophoresis. Polyacrylamide gels were visualized using either Coomassie Brilliant Blue R-250 stain or by transferring the protein bands to nitrocellulose and developing with rabbit anti-BioB polyclonal antibodies, peroxidase-fused donkey anti-rabbit IgG, and tetramethylbenzidene stain. Immediately after developing with TMB, wet blots were scanned at high resolution using a standard flatbed optical scanner. The relative optical densities of major bands were determined using NIH Image.
Mass spectrometry
ApoBioB and 2Fe-BioB samples were treated with 0.001% trypsin for 30 and 60 minutes. These samples are identical to those shown in Figure 3, lane 6 and 7. MALDI-TOF mass spectra were recorded on a Ciphergen PBSIIC mass spectrometer with the assistance of Dr. Michael Rosenblatt at the Protein Core Facility of the Children’s Hospital of Philadelphia. Spectra were collected by averaging 100 scans using a laser energy of 200 and the sensitivity set at 10. Digests were diluted 1:1, 1:5, 1:10, and 1:50 in either α-cyano-4-hydroxycinnamic acid (for peptide fragments from 1-9 kD) or sinnapinic acid (for fragments from 10-100 kD). The concentration of both matrices was 3.5 mg/mL (in 50:50:0.1 acetonitrile/water/TFA). In separate controls, samples were desalted using Millipore Ziptips using the manufacturers protocols, but this did not affect the results.
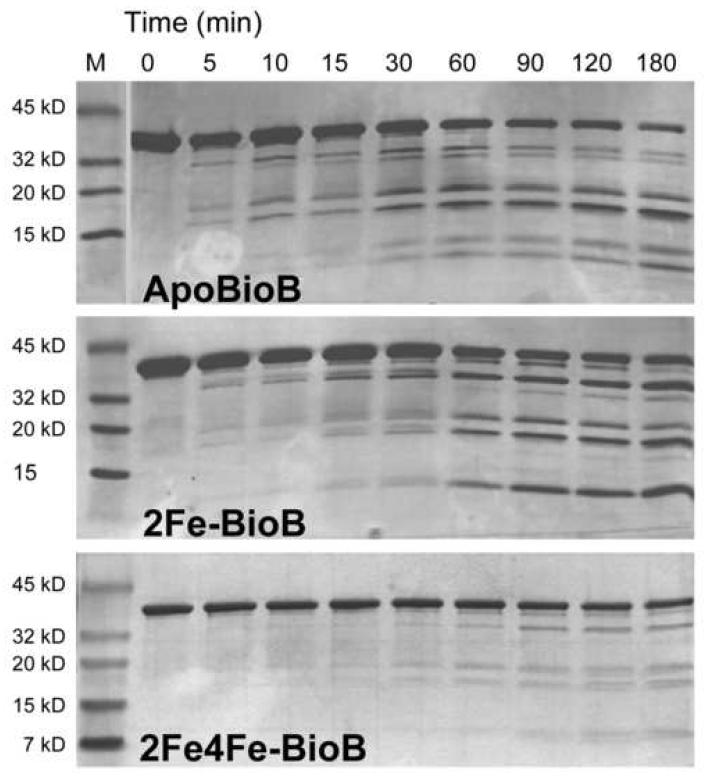
Figure 3.
Limited proteolysis of ApoBioB, 2Fe-BioB, and 2Fe4Fe-BioB. Each protein (1 mg/ml) was incubated with trypsin (0.001% for apoBioB and 2Fe-BioB, 0.01% for 2Fe4Fe-BioB) in 25 mM Tris HCl, pH 8 at 37 °C. At intervals, samples were removed, quenched with TLCK (10 μg/ml), and analyzed by SDS polyacrylamide electrophoresis on 4-20% gradient gels visualized with Coomassie R-250 stain. Samples removed at 30 and 60 min were also subjected to mass spectrometry (supplementary data) to facilitate identification of the major fragments produced.
Results
The quaternary structure of apoBioB and 2Fe-BioB
The quaternary structure of aerobically-purified BioB has been previously examined using native gel electrophoresis by Flint and coworkers. Although the gene sequence predicts a monomer molecular weight of 38.7 kDa, they report a native molecular weight of 104 kDa (n = 2.7) and conclude that the native protein is most likely a dimer. The crystal structure of BioB shows two possible monomer-monomer interface regions, consistent with either a dimer or tetramer, and the major interface is reported as the most likely dimer interface by Berkovitch, et al. [6]. This dimer interface involves two N-terminal helices and helices 7 and 8 from the core (αβ)8 barrel [6]. If loss of the [2Fe-2S]2+ cluster were to significantly alter the barrel fold, then one consequence might be disruption of the dimer interface.
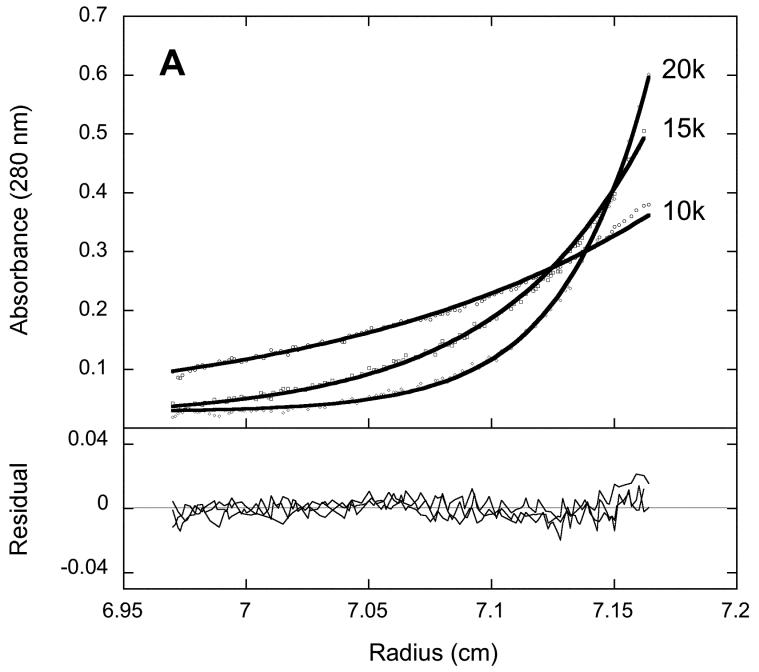
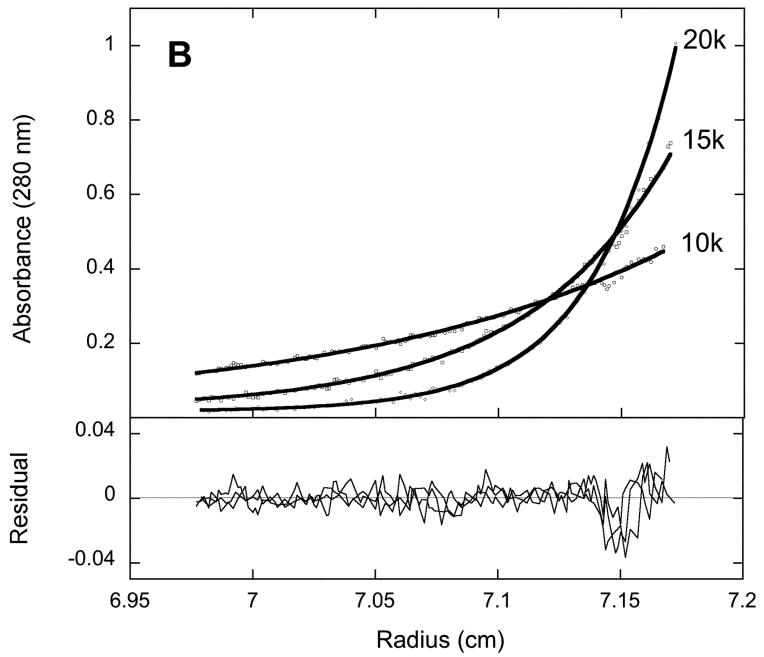
To examine the quaternary structure of 2Fe-BioB and apoBioB, we used equilibrium analytical centrifugation monitored by radial UV scans at 280 nm (Figure 1). Each protein was subjected to centrifugation at 10000, 15000, and 20000 rpm for 2-4 hrs, achieving an equilibrium radial concentration distribution that depends on the native molecular weight and centrifugation speed. Using the program Sedphat [24], the data at three speeds were globally fit to a non-equilibrating mixture of monomer, dimer, and tetramer. The centrifugation data for 2Fe-BioB are fit best to a mixture of 86 % dimer (MW = 92.8 kD and S = 4.97) and 14 % tetramer (MW = 188.2 kD and S = 6.86). We have previously detected the presence of a minor tetramer form in overexpressed recombinant BioB using gel filtration chromatography (J. Jarrett, unpublished data); the tetramer is not converted to dimer by reductants (TCEP was included in the buffer) or mild detergent and may possibly be due to UV-induced covalent crosslinks. Tetrameric BioB is less active than dimeric enzyme, and we have assumed that the more abundant and active dimer reflects the native quaternary structure. The centrifugation data for apoBioB are fit best to a nonequilibrating mixture of 88 % dimer (MW = 94.4 kD and S = 4.97) and 12 % tetramer (MW = 186.5 kD and S = 6.86). There is no evidence for the presence of BioB monomer in either 2Fe-BioB or apoBioB, nor is there any evidence for equilibration between dimer and tetramer forms. Since monomeric BioB is not detected in apoBioB samples, we can conclude that loss of the [2Fe-2S]2+ cluster from BioB does not result in global unfolding of the polypeptide chain at 20 °C and does not result in significant destabilization of the dimer interface.
Figure 1.
Equilibrium analytical ultracentrifugation of (A) 2Fe-BioB and (B) apoBioB. Each protein was diluted to 5 μM in 50 mM Tris HCl, 1 mM TCEP, pH 7.5, at 20°C and subjected to centrifugation at 10000, 15000, and 20000 rpm for 2-4 hrs. Five radial scans at 280 nm were averaged for each data set. Data are globally fit using Sedphat [24] to a mixture of nonequilibrating BioB dimer and tetramer.
Loss of the FeS clusters decreases the thermal stability of BioB
Although ApoBioB remains folded and retains the dimeric quaternary structure of the active enzyme, changes in local conformation or stability following loss of the FeS clusters could be important to allow reassembly of these clusters. In principle, we would like to compare the folding and stability of active enzyme, 2Fe4Fe-BioB, with enzyme lacking only the [2Fe-2S]2+ cluster, 4Fe-BioB. Unfortunately, this form of the enzyme is very air sensitive, and we cannot record even a single CD spectrum under N2 atmosphere without simultaneous oxidation and loss of the [4Fe-4S]2+ cluster. In light of this problem, we have chosen to infer the changes in the protein fold for each cluster by comparing three relatively air-stable states of BioB. We have determined the consequences of the loss of the [4Fe-4S]2+ cluster by comparing the properties of anaerobically reconstituted 2Fe4Fe-BioB with those of aerobically purified 2Fe-BioB. Similarly, we have determined the consequences of the loss of the [2Fe-2S]2+ cluster by comparing the properties of 2Fe-BioB with those of apoBioB, prepared by chemical reduction in the presence of an iron chelator.
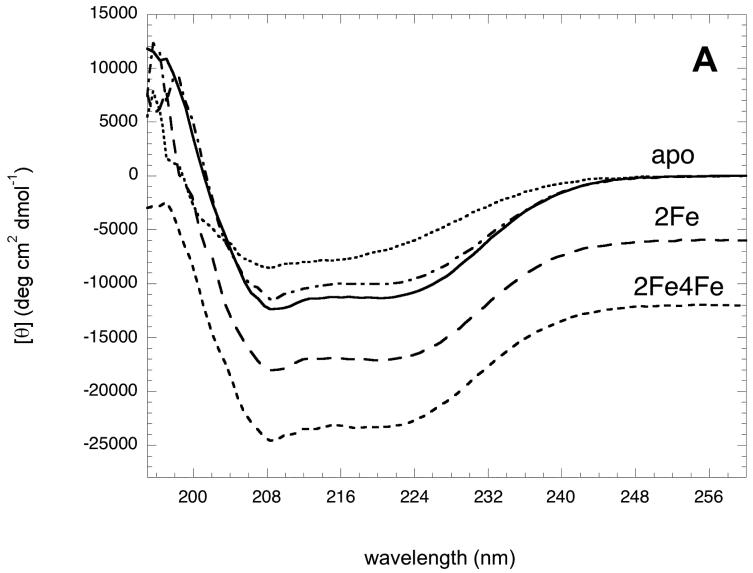
To examine possible changes in the gross secondary structure of BioB, we collected accurately calibrated circular dichroism (CD) spectra of apoBioB, 2Fe-BioB, and 2Fe4Fe-BioB at 25 °C (Figure 2). Since the [4Fe-4S]2+ cluster is bound within an extended loop between β strand 1 and α helix 1 found close to the surface of the protein [6], it is perhaps not surprising that loss of this cluster has no effect on the overall secondary structure content of BioB. Both spectra are superimposable and are well-fit to a mixture of 38 % α helix, 14 % β sheet, and 48 % turns and random coil [25], values that are within ±3 % of those predicted on a per residue basis from the crystal structure [6]. However, apoBioB does show a modest decrease in structure as compared to 2Fe-BioB, detectable as a decrease in intensity of the CD spectrum at 25 °C over the range 205-230 nm, corresponding to a 3 - 4 % decrease in α helix and β sheet composition, and a corresponding increase in random coil. When the temperature of this sample is lowered to 4°C, this lost intensity is regained, suggesting that a small region of the protein is reversibly unfolded at moderate temperatures but is stabilized at low temperature.
Figure 2.
Circular dichroism spectra of apoBioB, 2Fe-BioB, and 2Fe4Fe-BioB. (A) Circular dichroism spectra of 2Fe4Fe-BioB ( ), 2Fe-BioB (
), 2Fe-BioB ( ), and apoBioB (
), and apoBioB ( ) in 10 mM MOPS, 25 mM NaF, pH 7.5 at 25 °C. The spectrum of apoBioB is also shown at 4 °C (
) in 10 mM MOPS, 25 mM NaF, pH 7.5 at 25 °C. The spectrum of apoBioB is also shown at 4 °C ( ) and 95 °C (
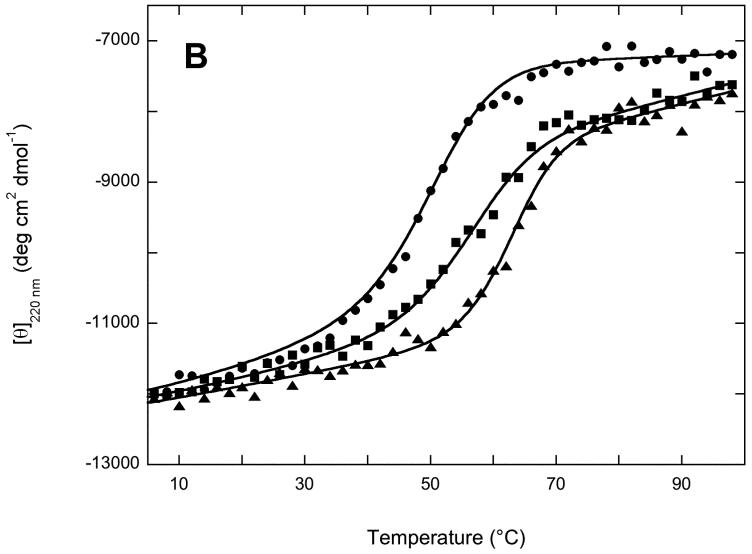
) and 95 °C ( ) (B) Thermal unfolding of 2Fe4Fe-BioB ( ▲ ), 2Fe-BioB ( ■ ), and apoBioB ( ● ) as monitored by circular dichroism at 220 nm. Each protein was incubated at 4 °C for 6 min, a spectral reading was taken, and the temperature was raised by 2 °C, repeating up to 98 °C. Attempts to measure a comparable curve for refolding during cooling proved unsuccessful due to aggregation of the unfolded protein. Data are fit to the Gibbs-Helmholtz equation as described in Materials and Methods.
) (B) Thermal unfolding of 2Fe4Fe-BioB ( ▲ ), 2Fe-BioB ( ■ ), and apoBioB ( ● ) as monitored by circular dichroism at 220 nm. Each protein was incubated at 4 °C for 6 min, a spectral reading was taken, and the temperature was raised by 2 °C, repeating up to 98 °C. Attempts to measure a comparable curve for refolding during cooling proved unsuccessful due to aggregation of the unfolded protein. Data are fit to the Gibbs-Helmholtz equation as described in Materials and Methods.
The decrease in intensity of the CD spectrum for apoBioB suggests a decrease in the stability of at least a small region of the protein, and perhaps of the entire folded structure. We subjected each protein to increasing temperatures from 4 to 98 °C and monitored the molar ellipticity at 220 nm, a wavelength that detects changes in both α helix and β sheet content. In each case, we did not detect significant refolding when the temperature was slowly lowered back to 4 °C, and instead observed turbidity that suggested irreversible aggregation of the unfolded proteins (data not shown). Although strictly speaking, the Gibbs-Helmholtz relationship is valid only for reversible two-state unfolding/folding, we used this equation [26] to estimate an approximate Tm for each melt: apoBioB, Tm = 49 °C; 2Fe-BioB, Tm = 56 °C; 2Fe4Fe-BioB, Tm = 63 °C. The loss of each FeS cluster results in a corresponding moderate decrease in the thermal stability. Loss of the [4Fe-4S]2+ cluster and the bound substrates results in a 7 °C decrease in stability, not surprising when one considers the number of hydrogen bonds and hydrophobic contacts between [4Fe-4S]2+ cluster, the substrates, and the protein core. However, 2Fe-BioB still remains >98% folded at 37 °C. Loss of the [2Fe-2S]2+ cluster results in a further 7 °C decrease in thermal stability, which is perhaps more than one might expect given that only 4 protein residues contact this cluster. More importantly, at 37 °C, apoBioB contains only ∼80% of the secondary structure found in 2Fe-BioB at the same temperature. This suggests that either a small region of apoBioB is reversibly unfolded at moderate temperature, or that the entire apoprotein is undergoing reversible global unfolding.
Limited proteolysis suggests apoBioB is unfolded near specific arginine residues
Although circular dichroism spectra had indicated that a small region of apoBioB may be unfolded at 37 °C, the direct observation of changes in the local structure by solution methods such as NMR was likely to be very difficult due to the large size of dimeric BioB. Susceptibility to limited proteolysis has been previously used to detect large conformational changes in folded proteins, and depends on the fact that trypsin has a strong preference for locally unfolded substrates in which both amino acid side-chains and amide bonds are accessible to the protease active site. BioB contains 36 Arg and Lys residues that are spread throughout the primary sequence, and most are found within discrete secondary structure elements. If we observe an increase in the cleavage at any specific residue in apoBioB or 2Fe-BioB as compared to 2Fe4Fe-BioB, we can conclude that the solvent exposure of that Arg or Lys residue increases upon loss of one or more FeS clusters. To facilitate identification of the fragments by mass spectrometry, the N-terminal His6-tag was removed by treatment with TEV protease, followed by repurification of the protein by passing through a Ni-NTA agarose column to remove the His6-tag and any uncleaved protein.
Each protein was incubated with trypsin (0.001 - 0.01 % (w/w)) at 37 °C (Figure 3). In the presence of the full complement of FeS clusters and substrates, 2Fe4Fe-BioB is surprisingly stable to proteolysis. Although the concentration of trypsin was increased to 0.01 %, we observed <20 % cleavage of the full length protein after 3 hrs. In fact, due to the prolonged incubation time, we suspect that the cleavage we did observe may correspond to protein in which the [4Fe-4S]2+ cluster is slowly oxidized and damaged. The trace fragments are identical to those observed for 2Fe-BioB.
BioB containing only the [2Fe-2S]2+ cluster is more susceptible to trypsin at four sites. The identity of proteolysis fragments was established by obtaining MALDI mass spectra of the 30 min sample for apoBioB and the 60 min sample for 2Fe-BioB and comparing these to predicted fragments based on 1 or 2 simultaneous cleavage events (supplementary data). In the presence of 0.001 % trypsin, 2Fe-BioB (38.5 kD) is initially partially cleaved at Arg65 and Arg327, resulting in formation of 36 and 31 kD fragments (residues 2-327 and 66-346). Arg327 is found in an unstructured extension at the C-terminus and would be expected to be highly susceptible to proteolysis, and rapid cleavage at this site is observed in all BioB forms. Arg65 is found in the loop that binds the [4Fe-4S]2+ cluster, and rapid cleavage indicates that this catalytically important loop becomes unstructured and exposed in the absence of this cluster. Two additional competing cleavage events occur at a slower rate. Cleavage at Arg245 results in the formation of fragments at 27 and 11 kD (residues 2-245 and 246-346), and cleavage at Arg168 results in formation of two fragments at ∼20 kD (residues 2-168 and 169-346). Arg168 and Arg245 are found in helices 4 and 6 and are flanked by conserved residues that bind the substrates and the [2Fe-2S]2+ cluster. The four fragments at 27, 20, and 11 kD appear to be relatively stable to further proteolysis, suggesting that they retain a stable well-folded structure, perhaps in a native conformation bridged by the [2Fe-2S]2+ cluster.
Overall, apoBioB is cleaved 3-fold more rapidly than 2Fe-BioB, yielding some similar fragments but with an altered time-course and significantly decreased stability. In apoBioB, the first cleavage detected within 5 min results in formation of fragments at ∼20 kD, again corresponding to cleavage at Arg168. Cleavage at Arg245 is also detected, resulting in fragments at 27 and 11 kD, but these fragments are further cleaved to fragments at 25 kD and 9 kD (possibly residues 2-227 and 260-346). Finally, the 27 and 25 kD fragments are much less stable and are mostly degraded to small peptides after 3 hrs. Rapid cleavage at Arg168 and Arg245 suggests that these regions of the protein become unstructured and exposed to solvent in the absence of the FeS clusters.
In vivo degradation of BioB occurs under iron-limited conditions
Loss of the FeS clusters results in decreased thermal stability and apparent localized unfolding of BioB, particularly in the regions around Arg168 and Arg245. These structural changes could play an important role in facilitating cluster reassembly or repair by allowing access to the cluster binding site. However, if cluster reassembly does not readily occur, then the same destabilization that facilitates cluster reassembly could also render the protein susceptible to endogenous proteolysis. Of course, E. coli cytosol does not contain trypsin, and we would not expect that in vivo degradation would occur through the same fragments as observed in figure 3. We have taken two different approaches to understanding the in vivo degradation of BioB. First, we examined the in vitro degradation of purified 2Fe-BioB by cell-free extracts of E. coli B. This in vitro experiment allowed us to follow the sequential production of major fragments from BioB, and provided an experimental basis for interpreting the subsequent in vivo studies. Second, we used polyclonal antibodies to BioB to probe for the presence of major degradation products of BioB in cells grown in a defined minimal medium containing either high or low iron concentrations. In these in vivo studies, tetracycline was added to inhibit fresh BioB expression and we were able to detect fragments similar to those observed in cell-free extracts.
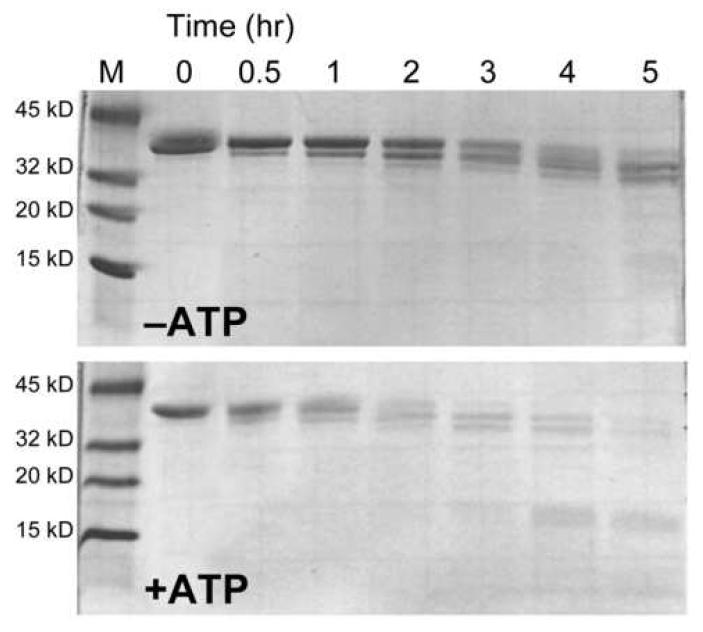
To characterize the fragments generated during cytosolic degradation of BioB, we incubated 2Fe-BioB (1 mg/ml) with a cell free extract (0.5 mg/ml) prepared from E. coli B grown in Luria broth (Figure 4). In these studies, 2Fe-BioB was used because apoBioB was degraded too rapidly to detect intermediate fragments. Proteolysis samples removed at intervals over 5 hrs show the sequential loss of ∼4 kD fragments (∼35 residues each), until the protein reaches ∼30 kD, at which point the entire protein is rapidly degraded. This sequential cleavage pattern is a hallmark of ATP-dependent targeted protein degradation; E. coli cytosol contains several ATP-dependent proteases, including ClpXP, ClpAP, Lon, HslV, and HflB [27]. To test whether one of these proteases might be involved in BioB degradation, ATP (1 mM) and MgCl2 (5 mM) were added to the proteolysis mixture (Figure 4). In this case, a similar stepwise fragmentation is observed, but at a rate that is ∼4-fold faster than in the absence of ATP, with little full length 2Fe-BioB remaining after 1 hr.
Figure 4.
Proteolytic degradation of 2Fe-BioB in cell-free extracts of E. coli B. 2Fe-BioB (1 mg/ml) was incubated with a cell-free extract prepared from E. coli B (0.5 mg/ml total protein) in 25 mM Tris HCl, pH 8 at 37 °C. Samples were removed at intervals, quenched with a protease inhibitor cocktail, and analyzed by SDS polyacrylamide gel electrophoresis on 4-20% gradient gels. In the lower panel, ATP (1 mM) and MgCl2 (5 mM) were added to the buffer prior to the addition of proteins.
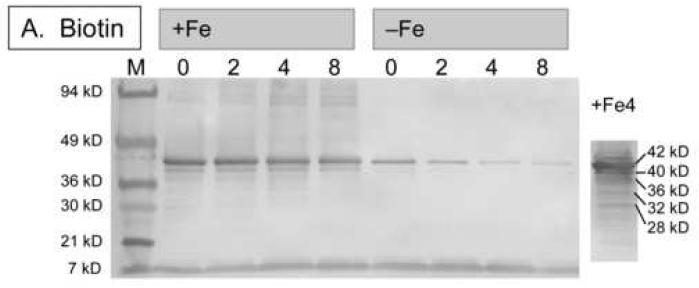
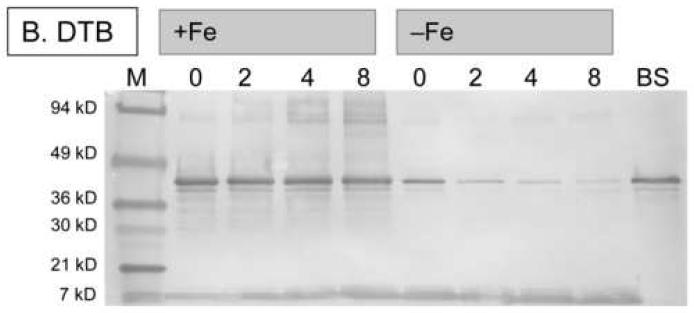
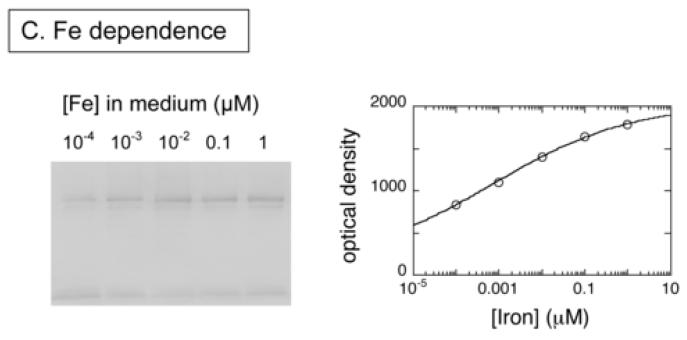
To observe true in vivo degradation of BioB, we induced expression of His6-BioB in E. coli BL21(DE3)pLysS cells grown in M9 medium containing glucose, vitamin-free casamino acids, ampicillin, chloramphenicol, and a trace metal supplement that contained 10-4 - 1 μM Fe(NH3)2SO4 (medium A). The medium was prepared using analytical grade trace-metal analyzed salts and Milli-Q purified water to prevent additional iron contamination, and culture flasks that were washed with nitric acid and rinsed with Milli-Q water prior to use. After 3 h, the cells were pelleted and resuspended in fresh medium A containing tetracycline, which inhibits further protein synthesis. Dethiobiotin or biotin (5 μM) were then added to initiate or inhibit enzyme turnover and cells were collected at various time intervals for Western blot analysis. Full length His6-BioB and protein fragments were detected by developing nitrocellulose blots with rabbit anti-BioB polyclonal antibodies (Figures 5A & 5B). In those samples grown in medium containing high iron levels (100 nM Fe2+), we observe little degradation of His6-BioB after 8 h, suggesting that BioB was maintained with a full complement of FeS clusters and was resistant to degradation. The minor degradation that was observed appears to be occurring via an ATP-dependent proteolysis mechanism originating at the C-terminus. When the sample grown for 4 hours was extracted with Ni-NTA agarose, eluted with imidazole, and then overloaded on the gel (Figure 5A, lane 10), a series of BioB fragments were detected whose mass decreases at ∼4 kD intervals, similar to the cleavage pattern observed in E. coli B extracts. In contrast, those samples grown in medium with low iron levels (0.1 nM Fe2+) show >90% degradation after 8 h, suggesting that a failure to maintain the FeS cluster content of BioB rendered the protein susceptible to complete proteolytic degradation. To confirm this protective effect of iron, we repeated this study using increasing amounts of Fe2+ in the medium (Figure 5C) and analyzed the amount of full length His6-BioB remaining after incubation of the cells for 4 h with dethiobiotin. Again, we found that BioB is significantly degraded in low iron medium and is protected in high iron medium.
Figure 5.
Western blots showing in vivo degradation of His6-BioB. (A) and (B) His6-BioB was expressed from a pET21 plasmid in E. coli strain BL21(DE3)pLysS in M9 minimal media supplemented with 0.2 % glucose and 1 % casamino acids, as well as 100 nM Fe(NH4)2(SO4)2 (+Fe lanes) or 0.1 nM Fe(NH4)2(SO4)2 (-Fe lanes). Following expression of BioB for 3 hrs, cells were pelleted and resuspended in fresh media containing tetracycline (60 μg/ml) and BioB turnover was inhibited or initiated by addition of biotin (5 μM, panel A) or dethiobiotin (5 μM, panel B). Cells were withdrawn at intervals and extracts analyzed by SDS polyacrylamide electrophoresis on 4-20% gradient gels detected by Western blot analysis with rabbit anti-BioB polyclonal antibodies. Each lane contains 5 μg of total protein except the far right lanes: panel A, right lane contains the sample from lane 4 loaded at 100 μg total protein; panel B, right lane contains 50 ng of purified His6-BioB. (C) His6-BioB was expressed in M9/glucose/casamino acids as described above, except that the added Fe(NH4)2(SO4)2 was varied from 104 - 1 μM. Protein production was halted with tetracycline (60 μg/ml), and turnover of BioB was initiated by addition of dethiobiotin (5 μM). Cells were incubated for 4 hrs, pelleted, and analyzed by Western blot as described above. Blots were optically scanned and the relative optical density of full-length His6-BioB is shown for varying levels of iron in the growth medium.
Surprisingly, there are only minor differences in degradation patterns between BioB that has been forced to undergo turnover for 8 hrs by addition of excess dethiobiotin to the growth medium, and BioB in which turnover has been inhibited by addition of biotin to the growth medium (Figures 5A & 5B). In iron-rich medium, we observed <20% degradation relative to the initial BioB levels, suggesting that rapid FeS cluster reassembly may have prevented targeted protein degradation. In low iron medium, we observed ∼70% degradation in the presence of biotin, and ∼95% degradation in the presence of dethiobiotin. Thus under low iron conditions, BioB appears to be inherently unstable, perhaps due to a substoichiometric cluster content, and turnover increases this effect somewhat due to transient loss of the [2Fe-2S]2+ cluster during catalysis. These results are qualitatively consistent with the findings of Choi-Rhee and Cronan, who report that BioB is degraded following overnight incubation of cells with excess dethiobiotin [21]. In addition, however, it is clear from the data presented in Figure 5 that the iron level in the growth medium is critical in observing this effect. When sufficient iron is available, E. coli is capable of maintaining the FeS complement of BioB, even during continuous turnover, and the enzyme is rescued from targeted degradation.
Discussion
Biotin synthase catalyzes a novel reaction in which an iron-sulfur cluster functions as both catalyst and substrate. A [2Fe-2S]2+ cluster is bound in close proximity to dethiobiotin within the core of the (αβ)8 barrel fold. Both isotope labeling and spectroscopic data suggest that a sulfide from this cluster becomes incorporated into dethiobiotin [9, 13, 14], generating the thiophane ring of biotin. However, the cluster is more than just a binding site for sulfide. The sulfide must be oxidized during the formation of new C-S bonds, and UV/visible and Mössbauer spectra suggest that the Fe3+ ions from the [2Fe-2S]2+ cluster are reduced to Fe2+ and released into solution [9, 15]. Thus we propose that the [2Fe-2S]2+ cluster is both the sulfur-donating substrate and the sulfur-oxidizing cofactor, but each round of catalysis generates an inactive enzyme in which this cluster has been destroyed.
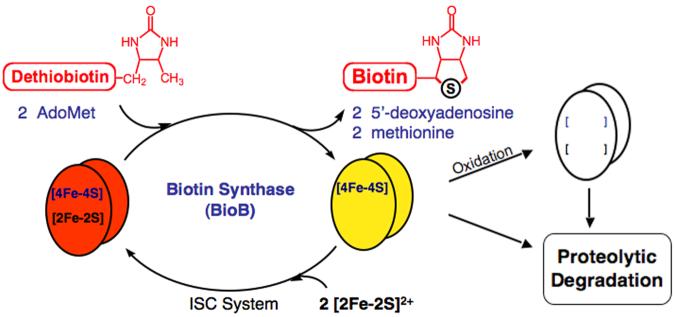
The resulting protein initially contains a [4Fe-4S]2+ cluster, and could have one of three possible fates in vivo (Figure 6). First, the protein could obtain a new [2Fe-2S]2+ cluster and return to catalysis. Although there is no data indicating how this occurs in E. coli, genetic experiments in S. cerevisiae suggest that the Isc iron-sulfur cluster assembly system, and IscU in particular, is essential for maintaining the FeS clusters in biotin synthase [28, 29]. When E. coli are incubated with excess dethiobiotin, measurements of BioB turnover and biotin production clearly indicate that each polypeptide is involved in multiple rounds of catalysis [21], and therefore E. coli must be capable of restoring the FeS cluster complement of BioB following each turnover when sufficient iron is available. A second possible fate immediately following turnover, particularly in aerobic organisms, is rapid oxidation and loss of the [4Fe-4S]2+ cluster resulting in formation of apoBioB. Since 4Fe-BioB does not bind substrates, exposure to even low concentrations of oxygen results in complete loss of this cluster within 1 min [17]. In the absence of rescue by either the Isc or Suf cluster assembly systems, the ultimate fate of both 4Fe-BioB and apoBioB is most likely complete proteolytic degradation.
Figure 6.
The fate of biotin synthase following catalysis. The active enzyme contains both [2Fe-2S]2+ and [4Fe-4S]2+/+ clusters; turnover with dethiobiotin and 2 equiv. of AdoMet results in the loss of the [2Fe-2S]2+ cluster. In the absence of substrates, the [4Fe-4S]2+/+ cluster is highly susceptible to oxidative degradation, leading to the formation of the less stable apoBioB, which is prone to ATP-dependent proteolytic degradation. In vivo, 4Fe-BioB could be rescued and returned to catalysis through reassembly of the [2Fe-2S]2+ cluster, a process that may be mediated by the Isc or Suf iron-sulfur cluster assembly systems.
Our laboratory is interested in understanding how E. coli sustains multiple rounds of biotin formation. In vitro, biotin synthase is capable of only one turnover, and we are unable to reconstitute an active [2Fe-2S]2+ cluster via standard chemical methods. In addition, we find that apoBioB is unstable during these reconstitution attempts, undergoing both slow proteolysis and precipitation. To better understand how to control these undesired effects, and to understand the fate of apoBioB following catalysis in vivo, we have examined the stability and the proteolytic fate of various forms of BioB, both in vitro and in vivo. We find that apoBioB is maintained as a stable dimeric protein, but that it has ∼20% less secondary structure at 37 °C, and is irreversibly thermally denatured with a Tm ≈ 49 °C. The partial loss of structure suggests that specific regions of the protein may be disordered. Susceptibility to tryptic proteolysis indicates that this disorder is localized in the vicinity of Arg168 and Arg245. Arg168 is found within helix 4 and Arg245 is found within helix 6 of the (αβ)8 barrel [6]. Both residues are flanked by nearby conserved residues that contact the [2Fe-2S]2+ cluster and form portions of the dethiobiotin and AdoMet binding sites, and therefore loss of the FeS clusters might be expected to alter the conformation of this region of the protein.
Increased disorder in these regions of the protein following loss of the FeS clusters also renders the protein more susceptible to in vivo proteolytic degradation. We monitored in vivo degradation during growth in iron-limited media using Western blot analysis, and find that BioB undergoes a stepwise fragmentation that suggests an ATP-dependent degradation at the C-terminus of the protein. With the recombinant enzyme, this degradation does not occur more rapidly in enzyme forced to undergo catalytic turnover by the addition of excess dethiobiotin to the media, provided that there is sufficient iron in the growth medium (compare left side of Figures 5A and 5B). However, in vivo degradation is enhanced in very low iron media and is decreased in high iron media, indicating that FeS cluster assembly (or repair) is the predominant means of preserving the catalytically active enzyme. In addition, there could be stress-response proteins or other factors that slow proteolytic degradation and favor cofactor reassembly under typical in vivo conditions, whereas experiments with recombinant enzyme might overwhelm these capabilities.
In vivo, apoBioB cluster reassembly or repair is most likely mediated by the Isc or Suf cluster assembly systems [30]. Although genetic experiments in yeast suggest that the Isc system is essential for maintaining FeS clusters in biotin synthase [28], we have not been able to replicate these experiments in E. coli due to apparent rescue of function between the Isc and Suf systems and also due to the lethality of Isc/Suf double deletions. We have recently demonstrated a multi-component complex between apoBioB and proteins found within the Isc operon (M. Reyda and J. Jarrett, unpublished data), suggesting that the Isc system may play a role in initially generating the FeS clusters in BioB, and possibly in repairing these clusters following turnover. However, attempts to reconstruct the Isc-mediated cluster assembly process in vitro using apoBioB as a target protein have proven unsuccessful. The [2Fe-2S]2+ cluster binding site is deeply buried within the BioB (αβ)8 barrel, and reassembly of this cluster must require that this site becomes more exposed. Further studies are clearly needed to understand how the Isc system (or other proteins) identify deeply buried cluster binding sites in diverse enzymes, induce appropriate conformational changes, and deposit the correct FeS cluster form in each enzyme.
Supplementary Material
Acknowledgements
We thank Dr. James Lear (University of Pennsylvania) assistance in modeling the equilibrium analytical centrifugation data.
Footnotes
This research has been supported by the NIH (R01 GM059175 to J.T.J.) and a fellowship from the David and Lucille Packard Foundation (J.T.J.).
- AdoMet
- S-adenosyl-L-methionine
- BioB
- E. coli biotin synthase
- CD
- circular dichroism
- DTB
- dethiobiotin
- DTT
- dithiothreitol
- EDTA
- ethylenediamine-tetraacetic acid
- ISC
- E. coli iron-sulfur cluster assembly proteins
- MOPS
- 4-morpholinepropanesulfonic acid
- TCEP
- tris(2-carboxyethyl)phosphine hydrochloride
- TLCK
- Nα-tosyl-lysyl-chloroketone
- Tris
- tris(hydroxymethyl)aminomethane
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information Available
MALDI mass spectra of tryptic fragments derived from limited proteolysis of apoBioB and 2Fe-BioB.
References
- [1].Marquet A, Tse Sum Bui B, Florentin D. Vitam. Horm. 2001;61:51–101. doi: 10.1016/s0083-6729(01)61002-1. [DOI] [PubMed] [Google Scholar]
- [2].Sanyal I, Cohen G, Flint DH. Biochemistry. 1994;33:3625–3631. doi: 10.1021/bi00178a020. [DOI] [PubMed] [Google Scholar]
- [3].Cosper MM, Jameson GN, Hernandez HL, Krebs C, Huynh BH, Johnson MK. Biochemistry. 2004;43:2007–2021. doi: 10.1021/bi0356653. [DOI] [PubMed] [Google Scholar]
- [4].Ugulava NB, Gibney BR, Jarrett JT. Biochemistry. 2001;40:8343–8351. doi: 10.1021/bi0104625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ugulava NB, Surerus KK, Jarrett JT. J. Am. Chem. Soc. 2002;124:9050–9051. doi: 10.1021/ja027004j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Science. 2004;303:76–79. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jarrett JT. Arch Biochem Biophys. 2005;433:312–321. doi: 10.1016/j.abb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- [8].Guianvarc’h D, Florentin D, Tse Sum Bui B, Nunzi F, Marquet A. Biochem. Biophys. Res. Commun. 1997;236:402–406. doi: 10.1006/bbrc.1997.6952. [DOI] [PubMed] [Google Scholar]
- [9].Ugulava NB, Sacanell CJ, Jarrett JT. Biochemistry. 2001;40:8352–8358. doi: 10.1021/bi010463x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tse Sum Bui B, Lotierzo M, Escalettes F, Florentin D, Marquet A. Biochemistry. 2004;43:16432–16441. doi: 10.1021/bi048040t. [DOI] [PubMed] [Google Scholar]
- [11].Gibson KJ, Pelletier DA, Turner IM., Sr. Biochem. Biophys. Res. Commun. 1999;254:632–635. doi: 10.1006/bbrc.1998.9991. [DOI] [PubMed] [Google Scholar]
- [12].Tse Sum Bui B, Escalletes F, Chottard G, Florentin D, Marquet A. Eur. J. Biochem. 2000;267:2688–2694. doi: 10.1046/j.1432-1327.2000.01284.x. [DOI] [PubMed] [Google Scholar]
- [13].Tse Sum Bui B, Florentin D, Fournier F, Ploux O, Mejean A, Marquet A. FEBS Lett. 1998;440:226–230. doi: 10.1016/s0014-5793(98)01464-1. [DOI] [PubMed] [Google Scholar]
- [14].Tse Sum Bui B, Mattioli TA, Florentin D, Bolbach G, Marquet A. Biochemistry. 2006;45:3824–3834. doi: 10.1021/bi052388m. [DOI] [PubMed] [Google Scholar]
- [15].Jameson GN, Cosper MM, Hernandez HL, Johnson MK, Huynh BH. Biochemistry. 2004;43:2022–2031. doi: 10.1021/bi035666v. [DOI] [PubMed] [Google Scholar]
- [16].Tse Sum Bui B, Benda R, Schunemann V, Florentin D, Trautwein AX, Marquet A. Biochemistry. 2003;42:8791–8798. doi: 10.1021/bi034426c. [DOI] [PubMed] [Google Scholar]
- [17].Ugulava NB, Frederick KK, Jarrett JT. Biochemistry. 2003;42:2708–2719. doi: 10.1021/bi0261084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Benda R, Tse Sum Bui B, Schunemann V, Florentin D, Marquet A, Trautwein AX. Biochemistry. 2002;41:15000–15006. doi: 10.1021/bi026590q. [DOI] [PubMed] [Google Scholar]
- [19].Cosper MM, Jameson GNL, Eidsness MK, Huynh BH, Johnson MK. FEBS Lett. 2002;529:332–336. doi: 10.1016/s0014-5793(02)03390-2. [DOI] [PubMed] [Google Scholar]
- [20].Cronan JE., Jr. J Biol Chem. 2001;276:37355–37364. doi: 10.1074/jbc.M106353200. [DOI] [PubMed] [Google Scholar]
- [21].Choi-Rhee E, Cronan JE. Chem Biol. 2005;12:461–468. doi: 10.1016/j.chembiol.2005.02.006. [DOI] [PubMed] [Google Scholar]
- [22].Ugulava NB, Gibney BR, Jarrett JT. Biochemistry. 2000;39:5206–5214. doi: 10.1021/bi9926227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ollagnier-de Choudens S, Mulliez E, Hewiston KS, Fontecave M. Biochemistry. 2002;41:9145–9152. doi: 10.1021/bi0122011. [DOI] [PubMed] [Google Scholar]
- [24].Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P. Anal. Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- [25].Lobley A, Whitmore L, Wallace BA. Bioinformatics. 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- [26].Boice JA, Dieckmann GR, DeGrado WF, Fairman R. Biochemistry. 1996;35:14480–14485. doi: 10.1021/bi961831d. [DOI] [PubMed] [Google Scholar]
- [27].Miller CG. In: Escherichia coli and Salmonella typhimurium. Neidhardt FC, editor. ASM Press; Washington, DC: 1996. pp. 938–954. [Google Scholar]
- [28].Muhlenhoff U, Richhardt N, Gerber J, Lill R. J Biol Chem. 2002;277:29810–29816. doi: 10.1074/jbc.M204675200. [DOI] [PubMed] [Google Scholar]
- [29].Muhlenhoff U, Gerl MJ, Flauger B, Pirner HM, Balser S, Richhardt N, Lill R, Stolz J. Eukaryot Cell. 2007;6:495–504. doi: 10.1128/EC.00191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Frazzon J, Dean DR. Cuurent Opinion in Chemical Biology. 2003;7:166–173. doi: 10.1016/s1367-5931(03)00021-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.