Abstract
The dimeric Mycobacterium tuberculosis FabH (mtFabH) catalyses a Claisen-type condensation between an acyl-CoA and malonyl-Acyl Carrier Protein (ACP) to initiate the Type II fatty acid synthase cycle. To analyze the initial covalent acylation of mtFabH with acyl-CoA, we challenged it with mixture of C6–C20 -CoAs and the ESI-MS analysis showed reaction at both subunits and a strict specificity for C12-CoA. Crystallographic and ESI-MS studies of mtFabH with decyl-CoA disulfide inhibitor revealed decyl chain bound in acyl-binding channels of both subunits through disulfide linkage to the active site cysteine. These data provide the first unequivocal evidence that both subunits of mtFabH can react with substrates or inhibitor. The discrepancy between the observed C12-CoA substrate specificity in the initial acylation step and the higher catalytic efficiency of mtFabH for C18–C20 -CoA substrates in the overall mtFabH catalyzed reaction suggests a role for M. tuberculosis ACP as a specificity determinant in this reaction.
Keywords: mycolic acids, Mycobacterium tuberculosis, fatty acid biosynthesis, FabH, alkyl-CoA disulfide, mass spectrometry, crystal structure
1. Introduction
Mycolic acids (α-alkyl-β-hydroxy long chain fatty acids) are essential components of the lipid-rich cell-envelope of M. tuberculosis [1] and are biosynthesized through the sequential activities of both type I and type II fatty acid synthases (FASs) [2]. The type I FAS is a homodimer of a multifunctional polypeptide that carries all of the catalytic activities for de novo synthesis of coenzyme A-linked C16–C26 long-chain fatty acids. Further elongation to C56 meromycolic acids is carried out by a type II FAS, whose multiple catalytic activities are each associated with separate polypeptides that utilize acyl-carrier protein (ACP) as shuttle in the extension cycle. Mycolic acids are formed through condensation of these meromycolic acids with long chain fatty acids [3, 4].
M. tuberculosis FabH (mtFabH) is an important link between the type I and type II FAS and has garnered significant attention as a target for developing new treatments for tuberculosis [5–8]. This enzyme catalyzes a decarboxylative condensation of malonyl-ACP with the long chain (C16 – C20) acyl-CoA products of the type I FAS. The resulting 3-ketoacyl-ACP product is reduced to an acyl-ACP (extended by two carbons) by the action of three enzymes, and this acyl-ACP is elongated by KasA and KasB [9].
Enzymological studies have shown that mtFabH can utilize a wide range of C12–C20 acyl-CoA substrates [5–7], which distinguishes it from FabH in other type II FAS systems that typically utilize C2–C6 acyl-CoA substrates [6, 10–12]. Initial studies using malonyl-E. coli ACP (ecACP) as the malonyl-ACP substrate of mtFabH indicated a broad C8–C20 acyl-CoA substrate specificity [6]. The C10–C12 acyl-CoA substrates were processed most efficiently (6–8 nmol/min/mg), while the reaction rates using the putative, physiological longer chain (C18–C20) acyl-CoA substrates were significantly lower (<0.6 nmol/min/mg)[7]. This same pattern has recently been observed in assays using malonyl-CoA in place of malonyl-ACP [data in publication]. However, mtFabH has a higher turnover number toward longer chain C14–C20 acyl-CoA substrates (9–10.5 nmol/min/mg) in the overall reaction when cognate AcpM is used in the assay [7]. The basis for this differential efficiency in processing longer chain acyl-CoA substrates remains unclear.
The crystal structures of mtFabH and an array of mutants have raised other questions relating to processing of longer chain C18–C20 acyl-CoA substrates [5, 7, 8]. mtFabH has a closely similar topology and active site architecture to that of E. coli FabH (ecFabH). However, whereas ecFabH has a small acyl binding pocket contiguous with the pantetheinate binding channel, the substrate binding pocket of mtFabH is L-shaped, consisting of a solvent accessible pantetheinate binding channel connected to a closed, elongated acyl binding channel. The active site catalytic triad (Cys112-His244-Asn274) lies at the junction of the two arms of the “L”. The closed distal end of the acyl binding channel imposes an upper length limit of C16 for acyl-CoA substrates in the static mtFabH structure. An mtFabH crystal structure of the Michaelis complex of dodecanoyl-CoA with an inactive Cys112Ala mutant [8] clearly shows substrate bound in the “L” shaped channel of each monomer, with the dodecanoyl group in the acyl binding channel. The length of this channel raises the as yet unanswered question of how longer C18–C20 acyl-CoA substrates bind and acylate mtFabH.
Previous observations made with both the ecFabH and mtFabH raise a third question of whether both monomers of the mtFabH homodimer are acylated. In the case of the ecFabH crystal structures of ecFabH [13, 14], including an unliganded tetragonal form of ecFabH in which several polypeptide segments that contribute to the structure of the substrate binding site and to the dimer interface are disordered, indicate structural identity of the two monomers. However, we have recently obtained a crystal structure of the inhibition complex of ecFabH with a methyl-CoA disulfide (MeSSCoA) in which there is a methyl disulfide linkage to the active site cysteine in only one monomer of the dimer [15]. Attempts to obtain a crystal structure in which both subunits have been modified have thus far been unsuccessful. Kinetic analysis of MeSSCoA inhibition suggests that both subunits of ecFabH are modified via a biphasic process in which one subunit is modified quickly and the second more slowly. A similar biphasic acylation of the ecFabH with acetyl-CoA substrate was also observed. These observations have led to the proposal that ecFabH exists in an open form that orders around either a substrate or inhibitor in one subunit, and in so doing retards binding of ligand to the second subunit [15]. Similar asymmetry of ligand binding and disorder→order transitions may occur in mtFabH and may severely hamper ligand binding and reaction at the second subunit. Indeed, it has been reported that acylation studies carried out using the mtFabH and radiolabeled decanoyl-CoA (in a molar ratio of 1:2.3) result in only 23% acylation of the protein [7]. Our complex of the mtFabH Cys122Ala mutant with lauroyl-CoA indicates structural identity of the two monomers (as seen for all mtFabH structures) and also that under prolonged incubation dodecanoyl CoA can bind in both subunits of the enzyme [5, 8].
In this work we have addressed these questions of mtFabH specificity and mechanism using ESI-MS and crystallography to monitor the reaction of the wild type mtFabH with both a decyl-CoA disulfide inhibitor and a wide range of C6–C20 acyl-CoA substrates. We have shown conclusively that substrate and inhibitor react with the active site cysteines in both subunits of the mtFabH dimer. We have also probed for the first time the substrate specificity of the acylation step catalyzed by mtFabH (previous analyses have only assessed this step indirectly by analysis of the overall reaction), and shown clear preferential formation of a modified enzyme using a C12 dodecanoyl-CoA. These findings imply a role for AcpM in determining the specificity of the mtFabH reaction and the profile of the product pool and provide new and essential data for the formulation of a model for mtFabH catalysis.
2. Materials and methods
Materials
long chain acyl-CoAs, coenzyme-A, imidazole, dithiothreitol (DTT) (Sigma); decylmethanethiolsulfonate (TRC Biomedical research Chemicals); Crystal screens I and II (Hampton Research).
2.1. Enzyme expression and purification
The wild type and C122A mtFabH proteins were overexpressed in E. coli with N-terminal polyhistidine tag and subsequently purified by metal chelation chromatography as described previously [5, 8].
2.2 Synthesis of decylSSCoA
DecylSSCoA was synthesized by reacting coenzyme A with decylmethanethiosulfonate as described previously [15].
2.3. ESI-MS analysis
A solution of wild type or mutant mtFabH in sodium phosphate buffer (50 mM, pH 7.0, 15 % glycerol) was incubated at room temperature for 40 min with either an equimolar mixture of C6, C8, C10, C14, C16, C18, and C20 acyl-CoAs (ratio of mtFabH monomer to each substrate was 1:5) or decylSSCoA (ratio of mtFabH monomer: inhibitor of 1:5). Excess ligand in both incubations was then removed using Pierce spin desalting columns and the enzyme concentrated in a Microcon concentrator (50 kD cutoff) and stored at – 20°C until the ESI-MS analysis was performed. The thawed samples were diluted to a concentration of approximately 1 pm/µl with 50% methanol/0.1% formic acid and analyzed by ESI-MS using a Quadrupole Time-of-flight mass spectrometer under standard procedures and the peaks were deconvoluted by the Bayesian protein reconstruct tool (in the Bioanalyst QS 1.1 software package).
2.4. Crystallization conditions
Prior to crystallization trials, the protein solution was concentrated to approximately 15mg/ml by ultrafiltration using a Microcon concentrator (Amicon) with a 30kDa cutoff. The enzyme (15mg/ml in 100mM Na phosphate buffer, pH 7.0) and inhibitor (13mM in methanol) solutions were mixed in a 1:3 molar ratio and incubated on ice for 2 hours. Crystallization of the mtFabH- decylSSCoA was achieved using the hanging-drop vapour-diffusion technique. Initial crystallization conditions were screened using Crystal Screen and Crystal Screen II from Hampton Research at 290K. Crystals grew within two-three weeks from PEG-4K and PEG-10K and further variation of conditions yielded two optimal crystal forms. Both were obtained by mixing 2µl of protein-inhibitor complex solution with an equal volume of reservoir solution and equilibrated over the precipitant well at 295K. The first crystal form was obtained in 25% (w/v) PEG-8K, 0.1 M ammonium sulfate and 100mM Na cacodylate, pH 6.5 and grew to maximum dimensions of ~ 0.04 × 0.02 × 0.06 mm (triclinic crystal form) within three weeks. These crystals have space group P1, with unit cell parameters a = 49.06, b = 55.76, c = 63.16 A, α = 92.8, β = 98.8, γ = 111.80. The second crystal form was crystallized in the presence of 18% PEG-10K (w/v) and 100mM Na Hepes, pH 7.0 and grew to 0.05 × 0.03 × 0.3mm within three weeks. These crystals have space group C2221 with unit cell parameters a = 67.65, b = 89.36, c = 232.01 A.
Data sets for both crystal forms were collected at 100°K on an R-Axis IV++ image-plate detector using CuKα X-rays (λ = 1.54 A) from a Rigaku MicroMax™ - 0007 X-ray source equipped with MSC Varimax confocal optics operating at 40kV and 20mA. The crystals were transferred with a cryoloop into corresponding mother liquor solutions containing additional 15–20% glycerol and 0.1mM inhibitor for ~10s and subsequently flash-cooled in a cold nitrogen-gas stream from an X-stream cryosystem (MSC). Both crystal forms diffracted to 2.5Å resolution, but the data-collection statistics were better for the second crystal form, which was used for structure determination and refinement. Coordinates of the final refined structure were deposited in the RCSB with accession number 2QX1.
3. Results and discussion
3.1. Acylation of mtFabH by a mixture of acyl-CoA substrates
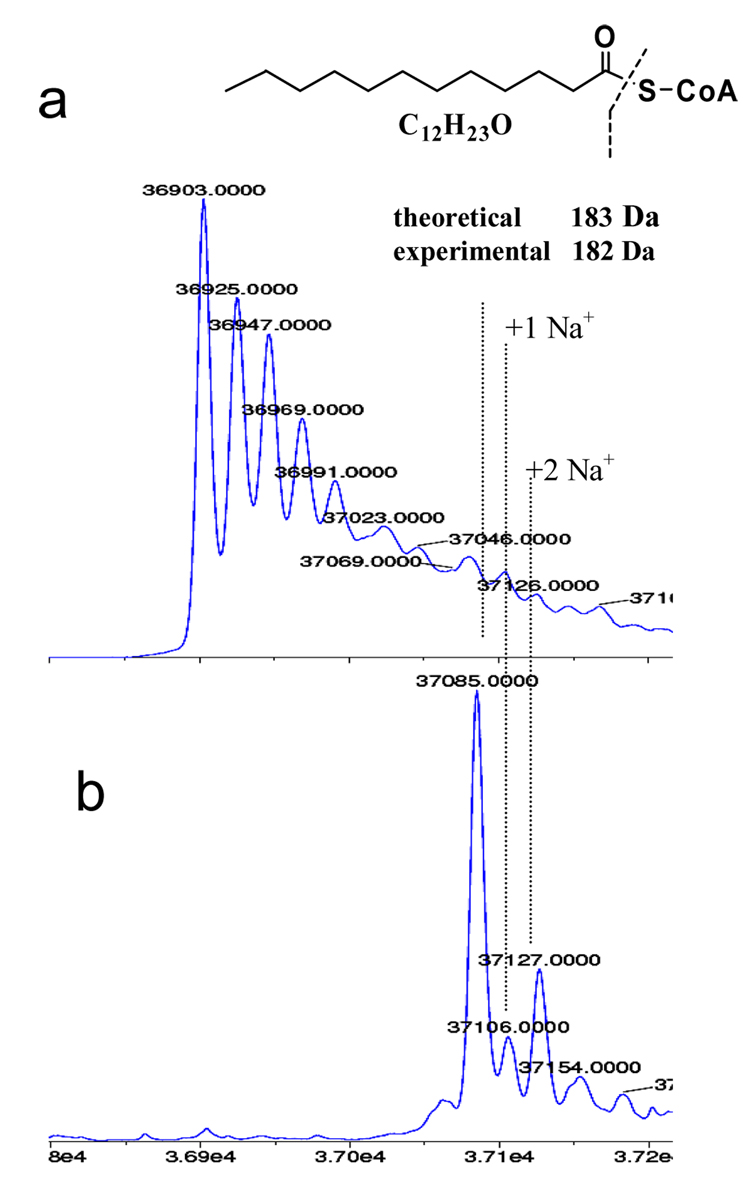
mtFabH was treated with a mixture of acyl-CoA substrates ([C6–C20]-CoA) in the absence of ACP and the resultant acylated mtFabH was analyzed using ESI-MS. The ratio of each acyl-CoA to the mtFabH monomer is ~ 5:1. The experimental ESI mass value for native mtFabH monomer is 36903 Da (compared to a theoretical value of 36904.7 Da) (Figure 1a). Treatment of mtFabH with this mixture of acyl-CoAs for 40 minutes at room temperature resulted in total modification of the enzyme as evident from the complete absence of native mtFabH at 36903 Da (Figure 1b). These data provide the first evidence that both monomers of the mtFabH dimer are acylated during reaction. The appearance of a major peak at 37085.0 Da corresponds to the addition of one dodecanoyl group (from dodecanoyl-CoA) to the mtFabH enzyme (experimental mass shift of +182 Da compared to theoretical mass value of +183 Da). Surprisingly, under our assay conditions, we did not observe mtFabH acylation with any other acyl-CoAs.
Figure 1.
Reconstructed ESI-TOF mass spectra of the incubation products of WT mtFabH with a mixture of acyl-CoAs (C6–C20).
a) Mass spectra of untreated WT mtFabH (control)
b) Mass spectra of WT mtFabH treated with the acyl-CoA mixture.
The apparent selective formation of the dodecanoyl product is puzzling given the relatively similar rates of reaction for the overall mtFabH catalyzed reaction with a range of acyl-CoA substrates [7]. However, it is consistent with our repeated observation of adventitiously bound dodecanoic acid in the substrate binding channel of crystal structures of recombinant mtFabH expressed in E. coli. Also, in trying to prepare Michaelis complexes of mtFabH Cys112Ala mutant with either tetradecanoyl-CoA or hexadecanoyl-CoA for co-crystallization, we have invariably obtained a complex with dodecanoyl-CoA in the binding site (unpublished data). We cannot exclude the possibility that the tetradecanoyl- or hexadecanoyl- groups are present in the determined crystal structures but are disordered at their terminal pairs of methylene groups. However, this is made less likely by the determination from HPLC analyses of these longer chain acyl-CoA substrate preparations, that a dodecanoyl-CoA impurity (≤2%) is present, which we infer binds preferentially to the mtFabH during crystallization. Together these observations suggest that the mtFabH Michaelis complex and subsequent acyl-enzyme intermediate thermodymamically favor the dodecanoyl-CoA substrate.
3.2. thio-Alkylation of mtFabH by decylSSCoA
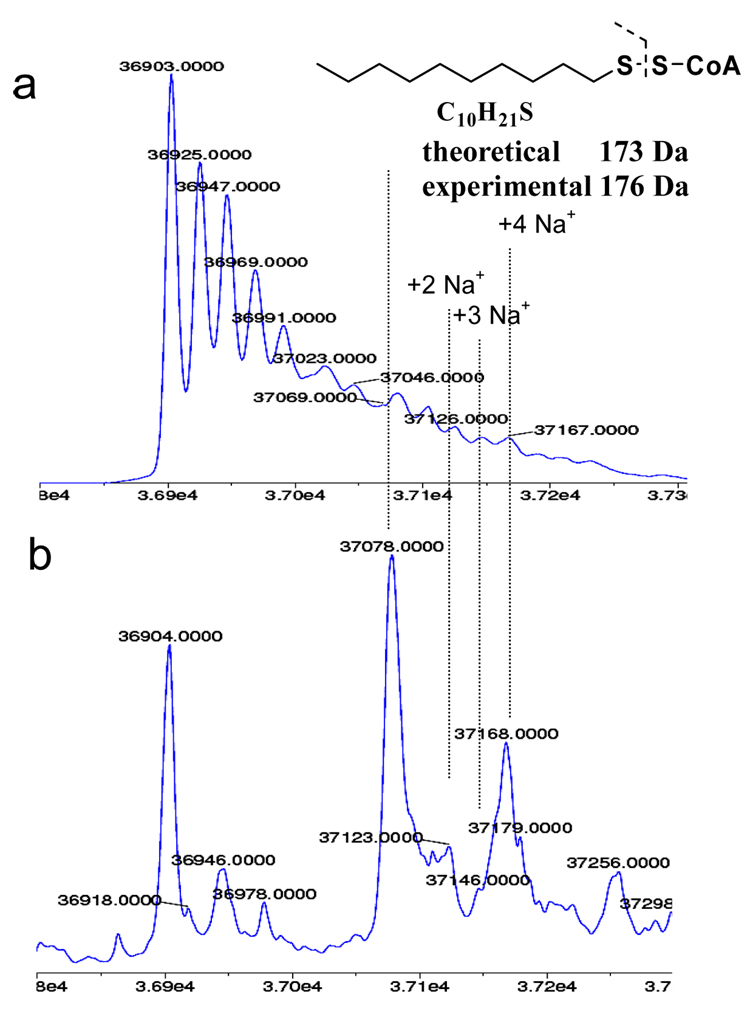
As noted above, MeSSCoA inhibits ecFabH through a disulfide exchange mediated transfer of the MeS- group to the sidechain sulfur of the active site Cys112 of the enzyme [15]. We also prepared decylSSCoA, which in preliminary experiments did not inhibit ecFabH but was effective against mtFabH [15]. To determine whether this class of inhibitors acts by the same mechanism on mtFabH as on ecFabH, the inhibition of mtFabH by decylSSCoA was analyzed using ESI-MS under similar conditions to those described above for the acylation reaction (Figure 2).
Figure 2.
Reconstructed ESI-TOF mass spectra of the incubation mixture of WT mtFabH and decylSSCoA.
a) Mass spectrum of untreated WT mtFabH (control)
b) Mass spectrum of WT mtFabH treated with the decylSSCoA disulfide inhibitor.
Treatment of mtFabH with decylSSCoA resulted in formation of a modified mtFabH species (Figure 2b) that correlates well with the addition of a C10S- fragment (a theoretical mass increase of 173 Da to the mtFabH monomer fragment). The absence of this peak in C112A mtFabH mutant confirmed that this modification occurs at the active site cysteine in WT mtFabH (data not shown). Only a fractional modification of the mtFabH with decylSSCoA was observed, in contrast to the full modification observed using dodecanoyl-CoA. This observation might indicate a slower reaction rate of the inhibitor relative to the substrate. This latter is the case for acetyl-CoA and methylSSCoA inhibitors with ecFabH. [15].
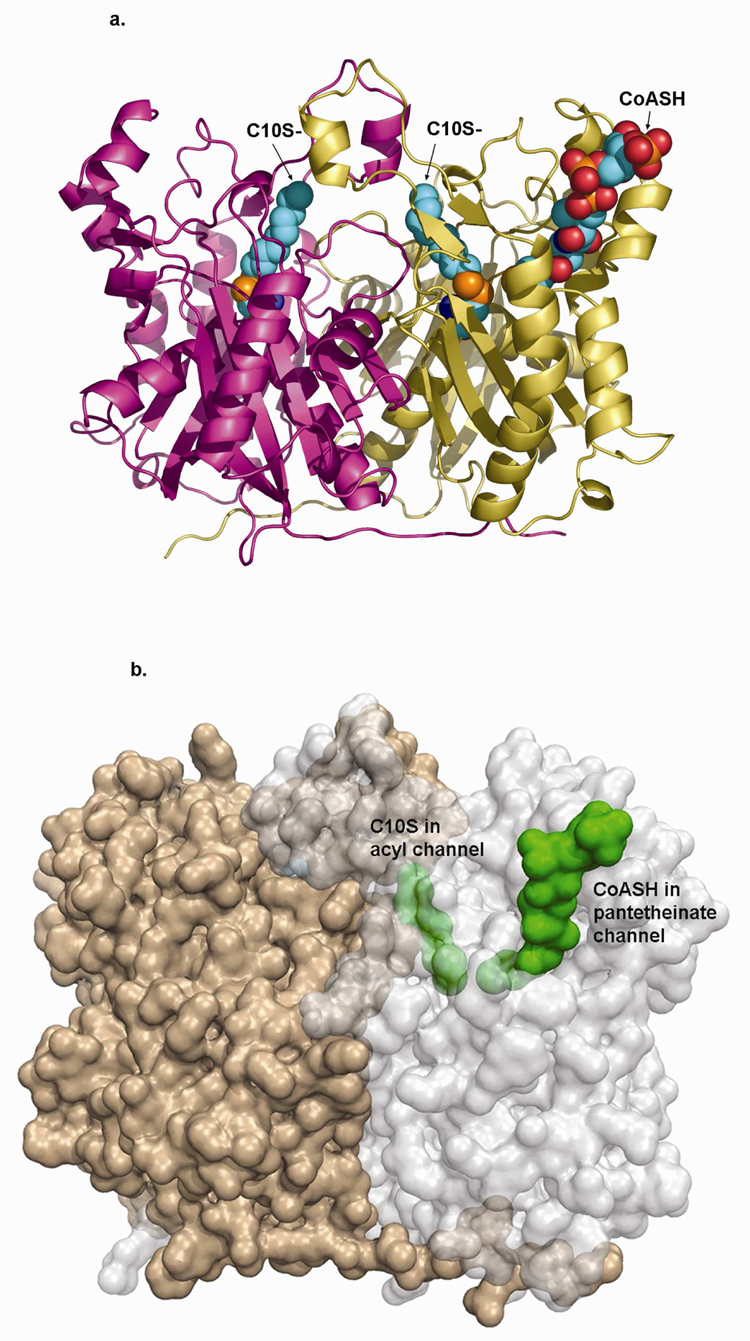
A co-crystal structure of mtFabH and decylSSCoA showed both of the active site cysteines of the mtFabH dimer to be modified through disulfide linkage to the decylthio-group (Figure 3). The decyl group has full occupancy in one subunit and slightly less than full occupancy in the second subunit. As we and others have frequently observed, CoASH product is only weakly retained in the pantetheinate channel of FabH – ligand complex structures. No interpretable density for CoASH was observed in one subunit of the decylSSCoA complex, while density in the other subunit was largely complete except for breaks around the pyrophosphate group. CoASH is probably preferentially retained in this latter channel as a consequence of an interaction of the pyrophosphate group of the nucleotide with Arg214 of a symmetry-related molecule. The terminal thiol of the pantetheinate group of this CoASH is 6.1Å from the Cys112 decyl-disulfide and the terminal adenine nucleotide group stacks on Trp32 and Arg151 at the mouth of the binding pocket (Figure 3b), which represents the only obvious access path to the buried acyl-binding channel.
Figure 3.
a) Backbone structure of mtFabH homodimer (subunits in gold and magenta ribbon) in complex with C10S- (space filling) covalently linked to the sidechain sulfur of Cys112 (also space filling) in both subunits. CoASH is shown bound in only the A-subunit (gold), also in space filling model.
b) Semi-transparent surface figure of the mtFabH homodimer rotated slightly about the x-axis relative to A. Subunits are gold and light gray, C10S- and CoASH are green space filling and shown bound in only one subunit. The C10S- group in the acyl channel is completely buried (partly obscured by semi-transparent protein surface envelope); the distal end of the CoASH pantetheinate is buried, while the adenosine pyrophosphate end is exposed at the mouth of the pantetheinate binding channel.
4. Conclusion
We provide the first evidence that the active site cysteine in both subunits of the wild type mtFabH dimer is able to react with either a dodecanoyl-CoA substrate or a decylSSCoA inhibitor. The data clearly demonstrate that binding and reaction at one subunit does not preclude reaction at the other, but we cannot determine whether binding and reaction occur at the same rate in both subunits. In the case of ecFabH, reaction at the second subunit is slower than that at the first [15].
In the static crystal structure of wild type mtFabH modified by formation of a disulfide product with the decylSSCoA inhibitor, the CoASH binding channel provides an access to the buried acyl-binding channel (Figure 3b). Binding and reaction of one of the mtFabH subunits with substrate or inhibitors via a reptational threading through this pantetheinate channel and past the active site residues would be energetically unfavorable. The data however show that this occurs readily to both subunits suggesting that there is likely an alternative more favorable pathway for binding of substrate or inhibitor ligand to mtFabH. These observations support the existence of a hitherto unobserved ‘open’ form of mtFabH, like that proposed for ecFabH [14, 15], which provides a direct, low energy path for substrate or inhibitor entry to the acyl binding channel.
The overall mtFabH catalyzed reaction using malonyl-AcpM has been shown to use C14–C20 acyl-CoAs at least as efficiently as dodecanoyl-CoA [7]. In this current work we have shown that in the acylation reaction performed with a range of acyl-CoAs (carried out in the absence of malonyl AcpM) the mtFabH selectively forms a product with the dodecanoyl CoA. It has been previously proposed that AcpM modulates the substrate specificity of the mtFabH [7]. Our current data supports this hypothesis and we propose that AcpM assists in formation of an open mtFabH structure, which facilitates both binding of longer chain acyl-CoA substrates and also release of the long chain 3-ketoacyl-ACP products.
In conclusion, this and other recent analyses of FabH enzymes suggest that the model for catalysis provided by current crystal structures is incomplete, and must be expanded to include significant protein conformational changes which are associated with the acyl–CoA and specific malonyl-ACP substrates.
Acknowledgments
This work was supported by a grant form the NIH (AI52230)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Molle V, et al. The condensing activities of the Mycobacterium tuberculosis type II fatty acid synthase are differentially regulated by phosphorylation. J Biol Chem. 2006;281(40):30094–30103. doi: 10.1074/jbc.M601691200. [DOI] [PubMed] [Google Scholar]
- 2.Asselineau C, et al. The biosynthesis of mycolic acids by Mycobacteria: current and alternative hypotheses. Prog Lipid Res. 2002;41(6):501–523. doi: 10.1016/s0163-7827(02)00008-5. [DOI] [PubMed] [Google Scholar]
- 3.Portevin D, et al. A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proc Natl Acad Sci U S A. 2004;101(1):314–319. doi: 10.1073/pnas.0305439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takayama K, Wang C, Besra GS. Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin Microbiol Rev. 2005;18(1):81–101. doi: 10.1128/CMR.18.1.81-101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scarsdale JN, et al. Crystal structure of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III. J Biol Chem. 2001;276(23):20516–20522. doi: 10.1074/jbc.M010762200. [DOI] [PubMed] [Google Scholar]
- 6.Choi KH, et al. Identification and substrate specificity of beta -ketoacyl (acyl carrier protein) synthase III (mtFabH) from Mycobacterium tuberculosis. J Biol Chem. 2000;275(36):28201–28207. doi: 10.1074/jbc.M003241200. [DOI] [PubMed] [Google Scholar]
- 7.Brown AK, et al. Probing the mechanism of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III mtFabH: factors influencing catalysis and substrate specificity. J Biol Chem. 2005;280(37):32539–32547. doi: 10.1074/jbc.M413216200. [DOI] [PubMed] [Google Scholar]
- 8.Musayev F, et al. Crystal structure of a substrate complex of Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III (FabH) with lauroyl-coenzyme A. J Mol Biol. 2005;346(5):1313–1321. doi: 10.1016/j.jmb.2004.12.044. [DOI] [PubMed] [Google Scholar]
- 9.Sridharan S, et al. X-ray crystal structure of Mycobacterium tuberculosis beta-ketoacyl acyl carrier protein synthase II (mtKasB) J Mol Biol. 2007;366(2):469–480. doi: 10.1016/j.jmb.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He X, Reynolds KA. Purification, characterization, and identification of novel inhibitors of the beta-ketoacyl-acyl carrier protein synthase III (FabH) from Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46(5):1310–1318. doi: 10.1128/AAC.46.5.1310-1318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsay JT, et al. Isolation and characterization of the beta-ketoacyl-acyl carrier protein synthase III gene (fabH) from Escherichia coli K-12. J Biol Chem. 1992;267(10):6807–6814. [PubMed] [Google Scholar]
- 12.Han L, Lobo S, Reynolds KA. Characterization of beta-ketoacyl-acyl carrier protein synthase III from Streptomyces glaucescens and its role in initiation of fatty acid biosynthesis. J Bacteriol. 1998;180(17):4481–4486. doi: 10.1128/jb.180.17.4481-4486.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu X, et al. Crystal structure of beta-ketoacyl-acyl carrier protein synthase III. A key condensing enzyme in bacterial fatty acid biosynthesis. J Biol Chem. 1999;274(51):36465–36471. doi: 10.1074/jbc.274.51.36465. [DOI] [PubMed] [Google Scholar]
- 14.Qiu X, et al. Refined structures of beta-ketoacyl-acyl carrier protein synthase III. J Mol Biol. 2001;307(1):341–356. doi: 10.1006/jmbi.2000.4457. [DOI] [PubMed] [Google Scholar]
- 15.Alhamadsheh MM, et al. Alkyl-CoA disulfides as inhibitors and mechanistic probes for FabH enzymes. Chem Biol. 2007;14(5):513–524. doi: 10.1016/j.chembiol.2007.03.013. [DOI] [PubMed] [Google Scholar]