Abstract
Background
DNA double-strand breaks (DSBs) are induced by exogenous insults such as ionizing radiation and chemical exposure, and they can also arise as a consequence of stalled or collapsed DNA replication forks. Failure to repair DSBs can lead to genomic instability or cell death and cancer in higher eukaryotes. The Schizosaccharomyces pombe fbh1 gene encodes an F-box DNA helicase previously described to play a role in the Rhp51 (an orthologue of S. cerevisiae RAD51)-dependent recombinational repair of DSBs. Fbh1 fused to GFP localizes to discrete nuclear foci following DNA damage.
Results
To determine the functional roles of the highly conserved F-box and helicase domains, we have characterized fbh1 mutants carrying specific mutations in these domains. We show that the F-box mutation fbh1-fb disturbs the nuclear localization of Fbh1, conferring an fbh1 null-like phenotype. Moreover, nuclear foci do not form in fbh1-fb cells with DNA damage even if Fbh1-fb is targeted to the nucleus by fusion to a nuclear localization signal sequence. In contrast, the helicase mutation fbh1-hl causes the accumulation of Fbh1 foci irrespective of the presence of DNA damage and confers damage sensitivity greater than that conferred by the null allele. Additional mutation of the F-box alleviates the hypermorphic phenotype of the fbh1-hl mutant.
Conclusion
These results suggest that the F-box and DNA helicase domains play indispensable but distinct roles in Fbh1 function. Assembly of the SCFFbh1 complex is required for both the nuclear localization and DNA damage-induced focus formation of Fbh1 and is therefore prerequisite for the Fbh1 recombination function.
Background
Homologous recombination (HR) is a major error-free pathway of DSB repair found in all organisms thus far examined (for reviews, see [1-3]). Extensive studies of HR repair mechanisms in the budding yeast Saccharomyces cerevisiae have shown that HR requires members of the RAD52 epistasis group, including RAD50, MRE11, XRS2, RAD51, RAD54, RAD55, RAD57, and RAD59 [4-6]. More recent studies of HR mechanisms in the fission yeast Schizosaccharomyces pombe have revealed many similarities with HR in S. cerevisiae and have led to many insights into the mechanisms of HR-dependent DSB repair and the identification of novel genes with homologues in higher eukaryotes [7,8]. For example, Rhp51-interacting proteins such as the Swi5-Sfr1 mediator complex function in a separate pathway from Rhp55-Rhp57 to promote an Rhp51 strand exchange reaction [9,10]. Furthermore, the fbh1 gene encodes a protein consisting of a unique domain architecture, with N-terminal F-box and C-terminal DNA helicase domains [11,12], which is conserved in mammals but not in S. cerevisiae. The Fbh1 protein was originally identified as a 3' to 5' DNA helicase that is stimulated by RPA at low ATP concentrations [13]. The helicase domain of Fbh1 is structurally related to the Rep, UvrD, PcrA, and Srs2 family of helicases [14]. Previous studies have shown that S. cerevisiae Srs2 regulates RAD52-dependent HR by actively disrupting the Rad51 nucleoprotein filament [15,16]. Interestingly, in contrast to Fbh1, Srs2 is conserved in budding and fission yeasts but not in mammals. In S. pombe, the fbh1Δ mutation is lethal when combined with the srs2Δ mutation, and this synthetic lethality can be suppressed by a loss of HR functions [11,12]. Recently, Chiolo et al. reported that human FBH1 (hFBH1) suppresses specific recombination defects of S. cerevisiae srs2 mutants and that the F-box domain is essential for hFBH1 functions in this respect [17]. Thus, the Fbh1 and Srs2 helicases appear to have only partially analogous functions in controlling HR after the formation of Rhp51 nucleoprotein filaments.
F-box proteins were first characterized as components of SCF ubiquitin-ligase complexes containing Skp1, Cullin, and F-box proteins, in which they bind substrates for ubiquitin-mediated proteolysis [18-21]. The F-box motif consists of 40–50 amino acids and is required for binding to SKP1. Therefore, the F-box motif links F-box proteins to other components of the SCF complex. Indeed, hFBH1 was shown to form an SCF complex and to have ubiquitin ligase activity in vitro [14,22]. However, the physiological substrates of SCFFbh1 are still unknown.
In this study, we characterize the in vivo function of Fbh1, focusing on the role of its F-box and helicase domains. Our results demonstrate that the F-box domain of Fbh1 is required for its recruitment to the nucleus and to DNA damage sites, whereas the helicase domain is involved in DNA processing after the Rhp51-dependent step of HR. Thus, both domains have indispensable but distinct roles in Fbh1 functions, and assembly of the SCFFbh1 complex is a prerequisite for its DNA recombination activities.
Results and Discussion
Construction of fbh1 mutants with substitutions in the F-box or helicase motif
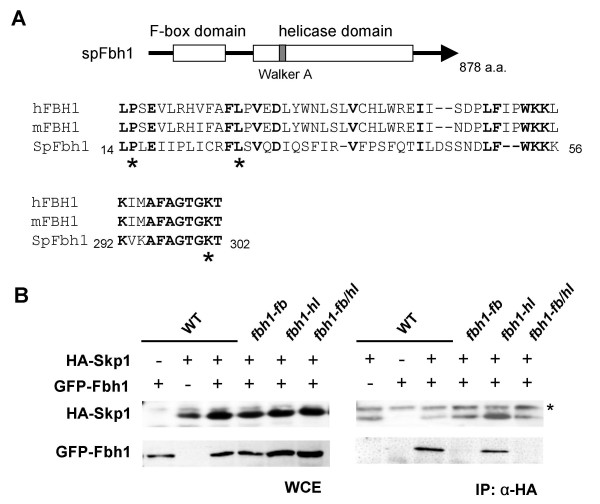
Fbh1 has a highly conserved N-terminal F-box motif and seven C-terminal helicase motifs. To gain insights into the roles of the F-box and helicase motifs in DNA repair, we constructed two fbh1 mutants, fbh1-fb and fbh1-hl, in which alanine replaces the Pro15 and Leu26 residues within the F-box motif and the Lys301 residue within the Walker A motif of the helicase domain, respectively (Fig. 1A). The highly conserved Pro15 and Leu26 residues in the F-box motif are essential for binding to Skp1 [21,23-25], and Lys301 is a conserved catalytic residue in the Walker A motif essential for ATPase activity [26]. To determine whether the fbh1-fb mutant is defective in binding to Skp1, we performed a co-immunoprecipitation assay using HA-tagged Skp1 and GFP-tagged versions of Fbh1. GFP-fused fbh1, fbh1-fb, fbh1-hl, or fbh1-fb/hl alleles were integrated into the genome at the ars1 locus in an fbh1Δ strain and expressed under the control of the nmt1 promoter. GFP-Fbh1 complemented the repair deficiency of the fbh1 deletion strain (data not shown) [11], indicating that GFP-Fbh1 and Fbh1 function similarly in vivo. We found that wild type Fbh1 and Fbh1-hl, but not Fbh1-fb, co-immunoprecipitated with HA-tagged Skp1 (Fig. 1B), indicating that the fbh1-fb mutation prevents association with SCF components.
Figure 1.
Construction of fbh1 mutants with substitutions in the F-box or helicase motif. (A) Schematic structure of the Fbh1 protein with the F-box and helicase domains. Sequence alignments of F-box (upper panel) and Walker A (bottom panel) motifs of S. pombe (SpFbh1), mouse (mFbh1), and human Fbh1 (hFbh1). Identical amino acids are shown in bold. Asterisks show the positions of the P15A/L26A and K301A mutations. (B) Co-immunoprecipitation of Skp1 with Fbh1. Cells expressing HA-Skp1 and GFP-Fbh1 were cultured as described in the Methods, and cellular extracts were immunoprecipitated with anti-HA antibodies. Samples were resolved by SDS-PAGE followed by western blotting with anti-GFP or anti-HA antibodies. *, non-specific bands.
The role of the F-box domain
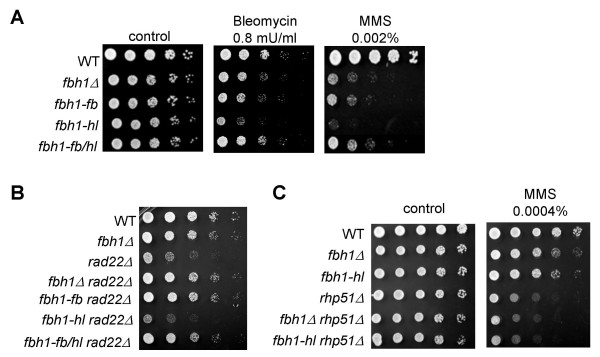
The fbh1Δ mutation confers hypersensitivity to DNA damaging agents and suppresses the slow growth of a rad22Δ strain, which is defective in an orthologue of S. cerevisiae RAD52 [11,12]. To examine the effect of the fbh1 mutations on these phenotypes, they were introduced into the S. pombe genome at the endogenous fbh1 locus. The fbh1-fb mutation conferred methyl methanesulfonate (MMS) and bleomycin sensitivities similar to those of the fbh1Δ mutant, and it suppressed the poor growth phenotype of the rad22Δ strain to a similar extent as the fbh1Δ mutation (Fig. 2A and 2B). Thus, the fbh1-fb mutant is defective in binding to Skp1, and the fbh1-fb mutation behaves like the fbh1Δ mutation with respect to DNA damage sensitivity and the suppression of the poor growth of rad22Δ cells. These results are consistent with a recent study showing that hFBH1 suppresses the hypersensitivity of S. cerevisiae srs2Δ cells to DNA damaging agents and that the F-box domain of hFBH1 is essential for this effect [17]. In another study, Osman et al. showed that the F-box domain plays a minor role in Fbh1 function because an F-box mutant (L14A/P15A) created in a previous study has no or little sensitivity to DNA damaging agents [12]. This discrepancy may arise from the use of different F-box mutants. Our F-box mutant (P15A/L26A) is completely defective in binding to Skp1, while in the previous study, the fbh1 L14A/P15A mutant had not been characterized in this regard. We suppose that the fbh1 L14A/P15A mutation may not fully inactivate the F-box, as previously discussed [12].
Figure 2.
F-box and helicase mutants are deficient in DNA damage repair. (A) Wild type and fbh1Δ, fbh1-fb, fbh1-hl, and fbh1-fb/hl mutant cells were grown in liquid YES. Cells were diluted and spotted onto YES plates with the indicated DNA damaging agents as described in the Methods. The plates were incubated for 3 days at 30°C. (B) Cells were grown and spotted onto YES plates. The plates were incubated for 4 days at 30°C. (C) Cells were grown and spotted onto YES plates containing the indicated concentrations of MMS. The plates were incubated for 3 days at 30°C.
The role of the DNA helicase domain
We next examined the effect of the fbh1-hl mutation on Fbh1 function. The fbh1-hl mutant shows greater sensitivity to DNA damaging agents than does the fbh1Δ mutant (Fig. 2A). In addition, ectopic expression of the fbh1-hl allele renders wild type cells sensitive to MMS, suggesting that the fbh1-hl alteration is a dominant mutation (see Additional file 1). Previous studies suggest that Fbh1 promotes HR repair by controlling the action of Rhp51 [11,12], which contributes to the suppression of inappropriate recombination events. Therefore, one possibility to explain the fbh1-hl-dependent toxic phenotype is that toxic recombination intermediates caused by faulty Rhp51-dependent HR accumulate to a greater extent in the fbh1-hl mutant than in the fbh1Δ mutant. To test this possibility, we examined the MMS sensitivity of the fbh1-hl strain in an rhp51Δ background. As expected, fbh1-hl cells were as sensitive to MMS as fbh1Δ cells with the rhp51Δ background (Fig. 2C). These results indicate that Fbh1-hl is not only defective in Fbh1 function but also that it interferes with Rhp51-dependent HR. In addition, the fbh1-hl mutation does not suppress the poor growth of rad22Δ cells, but rather, it exacerbates their decreased growth rate (Fig. 2B). However, the fbh1-fb/hl mutant, which has alterations within both the F-box and Walker A motifs, is as sensitive to DNA damaging agents as the fbh1-fb or fbh1Δ strains (Fig. 2A), and mutations affecting both domains suppress the slow growth phenotype of the rad22Δ strain (Fig. 2B), suggesting that F-box activity is likely to be a prerequisite for helicase activity. Thus, the F-box and DNA helicase domains play indispensable but distinct roles in Fbh1 function.
Focus formation of fbh1 mutants in response to DNA damage
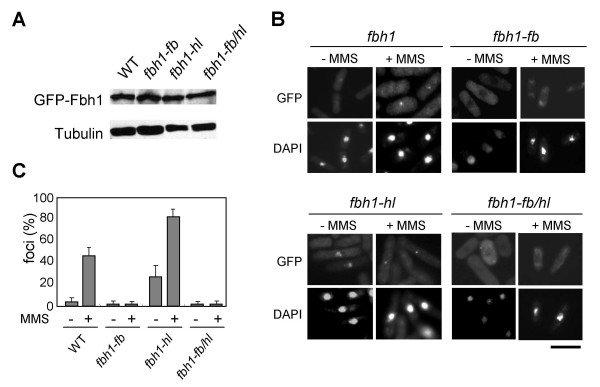
Since GFP-Fbh1 is predominantly detected in the nucleus and forms foci in response to DNA damage [11], we assessed DNA damage-induced focus formation in the fbh1 mutants. Exponentially growing cells were incubated in EMM2 medium containing 0.1% MMS in the absence of thiamine for 2 h, and GFP-Fbh1 was localized by fluorescence microscopy. The levels of expression of the GFP-fused wild type Fbh1 and of the three mutant Fbh1 proteins were comparable (Fig. 3A). Five percent of untreated cells expressing wild type GFP-Fbh1 contained foci, and 47% of these cells contained foci following MMS treatment (Fig. 3B and 3C). Interestingly, no foci were visible in cells expressing GFP-Fbh1-fb, even after exposure to MMS (Fig. 3B and 3C), indicating that the F-box domain is required for Fbh1 focus formation. In contrast, 27% of untreated cells expressing GFP-Fbh1-hl had foci, and this percentage was dramatically larger than that of cells expressing wild type GFP-Fbh1 (Fig. 3B and 3C). Following MMS treatment, the proportion of cells with foci further increased to 78% (Figure 3B and 3C). One possible explanation for the increased focus formation of fbh1-hl cells is that Fbh1-hl can localize to DNA damage sites but not complete DNA processing because it lacks helicase activity, leading to its accumulation at these sites and interference with the HR pathway. Moreover, the additional presence of the F-box mutation almost abolished focus formation in Fbh1-hl cells irrespective of the presence of DNA damage (Fig. 3B and 3C). Taken together, these results suggest that the F-box domain is required for the recruitment of Fbh1 to DNA damage sites and that the helicase domain is required to mediate the HR process.
Figure 3.
DNA damage-induced focus formation of Fbh1 mutant proteins. (A) Protein extracts were prepared from the indicated strains expressing GFP-wild type or mutant fbh1. Samples were analyzed by SDS-PAGE followed by western blotting with anti-GFP or anti-α-tubulin antibodies. (B) Cells expressing GFP-wild type or mutant fbh1 were incubated in EMM2 without or with MMS (0.1%) for 2 h at 30°C and observed by fluorescence microscopy. The upper and lower panels show GFP and DAPI images, respectively. The scale bar indicates 10 μm. (C) Quantitative analysis of GFP-Fbh1 foci. Cells with Fbh1 foci were counted and divided by the total number of cells. More than 150 individual cells were scored for each strain. The result represents the average of three independent measurements.
Fbh1 is required for the DNA damage-induced formation of Skp1 nuclear foci
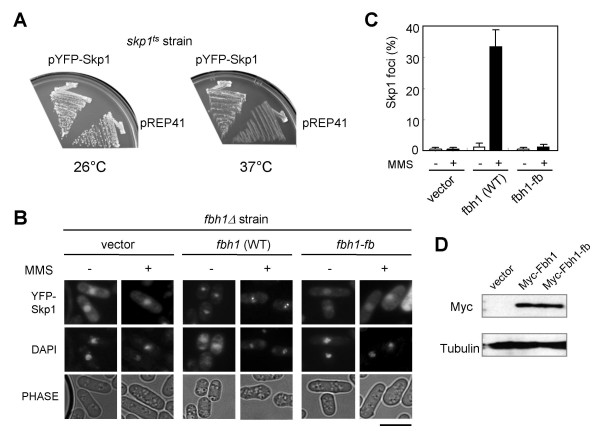
Since Fbh1 is assembled into the SCF complex, we next examined the subcellular localization of Skp1 by fusing YFP to its N terminus. The resulting fusion protein was expressed from a plasmid under the control of the nmt1 promoter. YFP-Skp1 functions normally in vivo, since it fully complements the temperature sensitivity of an skp1ts mutant (Fig. 4A). When the YFP-Skp1 was expressed in fbh1Δ cells expressing wild-type fbh1 or fbh1-fb, it was detected in both the nucleus and cytoplasm, with a higher level of the protein in the nucleus. YFP-Skp1 foci were not detected in fbh1Δ cells expressing wild type fbh1, but following 1 h exposure to MMS, most of the cells had nuclear foci (Fig. 4B and 4C). Remarkably, fbh1Δ cells expressing the fbh1-fb mutant did not have any YFP-Skp1 foci, even in the presence of MMS (Fig. 4B and 4C). Wild-type fbh1 and fbh1-fb were expressed at a similar level (Fig. 4D). Thus, these results indicate that Skp1 focus formation is dependent on the F-box domain of Fbh1.
Figure 4.
Skp1 forms DNA-damage-induced foci in an Fbh1-dependent manner. (A) YFP-Skp1 complements the skp1ts mutants. The skp1ts mutants expressing YFP-wild type skp1 or none in the absence of thiamine were streaked onto EMM2 plates and incubated at 26°C (left) or 37°C (right) for 3 days. (B) DNA damage-induced YFP-Skp1 foci. The fbh1Δ strains harbouring the vector pREP41, pREP42 Myc-fbh1 or pREP42 Myc-fbh1-fb were transformed with pREP42 YFP-skp1. Cells were treated with or without MMS (0.1%) for 2 h at 30°C and observed by fluorescence microscopy. The top, middle, and bottom panels show YFP, DAPI, and phase contrast images, respectively. The scale bar indicates 10 μm. (C) Quantitative analysis of YFP-Skp1 foci. Cells with Skp1 foci were counted and divided by the total number of cells. At least 100 cells were counted per strain. The result represents the average of three independent measurements. (D) Expression of Myc-tagged Fbh1. Cells were treated as in (B) and protein extracts were analysed by SDS-PAGE followed by Western blotting with anti-Myc or anti-α-tubulin antibodies.
The F-box domain is responsible for the nuclear localization of Fbh1
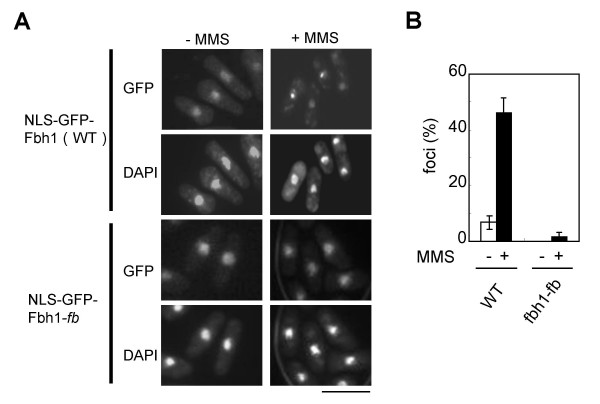
In the course of our studies, we noticed that the fbh1-fb mutation affected the subcellular localization of Fbh1. As shown in Fig. 3B, Fbh1-fb and Fbh1-fb/hl showed predominantly cytoplasmic localization and little GFP signal was seen in the nucleus, in striking contrast with the subcellular localization of wild type Fbh1. These results indicate that the fbh1-fb mutation disturbs the nuclear localization of Fbh1. Although this interpretation could explain why the fbh1-fb mutation confers an fbh1 null phenotype and suppresses the toxic phenotypes of the fbh1-hl mutation, it is still unknown whether SCFFbh1 complex formation is required for DNA damage-induced nuclear focus formation. To test this possibility, a NLS sequence from the simian virus 40 large-T antigen (PKKKRKV) [27] was fused to the GFP-Fbh1 and GFP-Fbh1-fb constructs at their N termini, and focus formation in response to DNA damage was examined. As with GFP-Fbh1 cells, these constructs were integrated into the genome at the ars1 locus in the fbh1Δ strain. Control experiments showed that NLS-GFP-Fbh1 but not NLS-GFP-Fbh1-fb complemented the MMS sensitivity of the fbh1Δ strain (data not shown). As expected, NLS-GFP-Fbh1 formed discrete foci in cells exposed to MMS (Fig. 5). NLS-GFP-Fbh1-fb was also detected in the nucleus like wild type Fbh1, but it still failed to form foci in response to DNA damage (Fig. 5). These results suggest that the F-box domain is required for the nuclear localization and DNA damage-induced focus formation of Fbh1. It should be noted that the GFP-F-box domain mutant could not enter the nucleus (see Additional file 2), suggesting that in addition to the F-box domain, another domain of Fbh1 might also be important for its nuclear targeting.
Figure 5.
The F-box motif is required for DNA damage-induced focus formation of Fbh1. (A) fbh1Δ cells expressing NLS-GFP-Fbh1 (wild type) and NLS-GFP-Fbh1-fb were treated as in Figure 3 with MMS (0.1%) for 2 h at 30°C and observed by fluorescence microscopy. The upper and lower panels show GFP and DAPI images, respectively. The scale bar indicates 10 μm. (B) Quantitative analysis of NLS-GFP-Fbh1 foci. Cells with Fbh1 foci were counted and divided by the total number of cells. At least 100 cells were counted per strain. The result represents the average of three independent measurements.
An important unresolved issue is the identification of physiological substrates of SCFFbh1 that presumably regulate the HR pathway. A recent study has shown that hFBH1 has a short half-life when it is expressed in S. cerevisiae, and its degradation depends on the presence of a functional F-box and yeast SCF components [17], suggesting that one candidate for a SCFhFBH1 substrate is hFBH1 itself. The rapid turnover of hFBH1 might contribute to the tight regulation of hFBH1 helicase activity. However, if SCFhFBH1 complex formation is also necessary for its nuclear transport and recruitment to DNA damage sites, the alteration in subcellular localization caused by the F-box mutation might affect hFBH1 stability in budding yeast cells. In addition, many F-box proteins identified to date target more than one substrate for degradation [28-30]. Since our present data suggest that Fbh1 functions as an SCFFbh1 ubiquitin ligase complex for HR repair, the most plausible candidates as SCFFbh1 substrates are HR proteins, including Rhp51. For example, SCFFbh1 might contribute to regulate Rhp51-dependent HR by promoting the ubiquitination of Rhp51 or other recombination proteins. Future studies will be needed to determine whether Fbh1 is a physiological SCFFbh1 substrate and to identify other SCFFbh1 substrates, which would provide a means to conduct a more detailed analysis of its function in HR repair.
Conclusion
In this study, we characterized the in vivo function of Fbh1, focusing on the role of its F-box and helicase domains. Our results show that the assembly of SCFFbh1 mediated by the F-box domain of Fbh1 is required for its recruitment to both the nucleus and DNA damage sites, whereas the helicase domain is involved in controlling the action of Rhp51. Thus, Fbh1 is tightly regulated by SCF components, which might prevent it from functioning inappropriately in HR.
Methods
S. pombe strains and plasmids
All yeast strains used in this study are listed in Table 1. S. pombe cells were grown in YES or EMM medium, and standard genetic and molecular procedures were employed as previously described [31]. An fbh1 cDNA clone was constructed by PCR from total S. pombe cDNA with the primers CSF (5'-GGCGGATCCCATATGAGTGCTCAACATTTACA-3') and CSR (5'-GGCGGATCCCTACTGATCATGTACAGCAAA-3'). The fbh1 cDNA fragment was cloned into the BamHI site of pUC119 to produce pUCcfbh1. An fbh1 genomic DNA was obtained from a genomic library [11], and a BamHI-KpnI fragment containing the fbh1 coding region was cloned into pUC119 to produce pUCgfbh1. The fbh1-fb and fbh1-hl mutant genes were constructed by PCR-mediated site-directed mutagenesis of pUCcfbh1 and pUCgfbh1. All mutant clones were sequenced to ensure that only the desired mutation had been introduced. BamHI-KpnI fragments of the fbh1 mutants were introduced into the vector pU19, which carries the ura4+ gene for directing gene replacement. The resulting plasmids were digested with AgeI and integrated into the S. pombe genome. Transformed strains were then plated onto EMM2 plates containing 5-fluoroorotic acid to select ura- cells.
Table 1.
Strains used in this study
| Strain | Genotype | Source |
| MP111 | h+ leu1-32 ura4-D18 | [11] |
| MPF1 | h+ fbh1::LEU2 leu1-32 ura4-D18 | [11] |
| 12521 | h+ fbh1::KanMX-leu1-32 ura4-D18 ade6-704 | Lab. stock |
| C11 | h+leu1-32 ura4-D18 fbh1-fb | This study |
| C12 | h+ leu1-32 ura4-D18 fbh1-hl | This study |
| C13 | h+leu1-32 ura4-D18 fbh1-fb/hl | This study |
| C100 | h+ ars1::pREP41-EGFP N-Fbh1 leu1-32 ura4-D18 ade6-704 fbh1::KanMX | This study |
| C101 | h+ars1::pREP41-EGFP N-Fbh1-fb leu1-32 ura4-D18 ade6-704 fbh1::KanMX | This study |
| C102 | h+ ars1::pREP41-EGFP N-Fbh1-hl leu1-32 ura4-D18 ade6-704 fbh1::KanMX | This study |
| C103 | h+ars1::pREP41-EGFP N-Fbh1-fb/hl leu1-32 ura4-D18 ade6-704 fbh1::KanMX | This study |
| B54 | smt-0 rhp51::his3+leu1-32 ura4-D18 his3-D1 arg3-1 | Y. Tsutsui |
| C109 | rhp51::his3+fbh1-hl leu1-32 ura4-D18 his3-D1 arg3-1 | This study |
| C15 | smt-0 rad22::arg3+ura4-D18 leu1-32 arg3-D1 | Lab. stock |
| C105 | mat1PD::LEU2 rad22::arg3+fbh1::KanMX ura4-D18 leu1-32 arg3-D1 | This study |
| C106 | smt-0 rad22::arg3+fbh1-fb ura4-D18 leu1-32 arg3-D1 | This study |
| C107 | mat1PD::LEU2 rad22::arg3+fbh1-hl ura4-D18 leu1-32 arg3-D1 | This study |
| C108 | smt-0 rad22::arg3+fbh1-fb/hl leu1-32 ura4-D18 his3-D1 arg3-1 | This study |
| skp1-94 ura4-D18 leu1-32 his3-D1 | T. Toda | |
| MPF25 | smt-0 fbh1::LEU2 rhp51::his3+ura4-D18 leu1-32 his3-D1 arg3-D1 | [11] |
Expression of the GFP-Fbh1 and YFP-Skp1 fusions in S. pombe
Wild type and mutant fbh1 cDNAs were cloned separately into the vector pREP41 EGFP N [32] to express enhanced green fluorescent protein (EGFP) fusion proteins under the control of the medium-strength nmt1 promoter. The resulting plasmids were linearized at the unique MluI site within the ars1 sequence of the plasmid pREP41 and then introduced into the ars1 locus of the fbh1Δ strain. To construct pREP41/NLS-GFP-Fbh1-fb, two complementary DNA oligonucleotides encoding a NLS sequence (PKKKRKV) from the SV40 large T antigen were synthesized and inserted at the N-terminus-encoding region. The skp1 cDNA was cloned into the plasmid pREP41 YFP N to express a yellow fluorescent protein-Skp1 fusion under the control of the nmt1 promoter. Cells harboring pREP41 YFP-Skp1 were grown in EMM2 medium with appropriate supplements and containing 0.1% MMS in the absence of thiamine for 2 h. Cells were fixed with 70% ethanol and observed by fluorescence microscopy. More than 100 individual cells were scored for each strain.
Spot assays
Logarithmically growing cells were harvested and resuspended in water. Five-fold serial dilutions of cultures of the indicated mutants were spotted onto YES plates containing the indicated concentration of chemical genotoxins. Plates were incubated at 30°C for 3–4 days. All spot assays were repeated at least twice to ensure that the results were reproducible.
Immunoprecipitation
The skp1 fragment was amplified by PCR from an S. pombe cDNA library and cloned into the vector pREP42 HA N, which encodes a triple C-terminal hemagglutinin (HA) tag [32]. The fbh1 mutant strains C100, C101, C102 and C103 were transformed with pREP42 HA N or pREP42 HA-Skp1. The transformants were grown in EMM2 medium with appropriate supplements in the absence of thiamine to express N-terminally HA-tagged skp1 from the thiamine-repressible nmt1 promoter. Mid-log-phase cells from a 50-ml culture were collected, washed with buffer A (50 mM Tris-HCl, pH 7.5, 15 mM EGTA, 100 mM NaCl, 0.1% (w/v) Triton ×-100, protease inhibitor cocktail for yeast (Sigma), 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) and resuspended in 500 μl buffer A. Cells were disrupted with the same volume of acid-washed glass beads using a Mini-BeadBeater-8 (BioSpec Products). The supernatant fraction was collected by centrifugation and used for immunoprecipitation. Fifty microliters of protein G-agarose (GE Healthcare) was added to absorb nonspecific Protein G binding protein. Twenty microliters of anti-HA antibody (12CA5, Roche) and 40 μl Protein G-agarose were used per 400 μl cell lysate, and the mixture was rotated for 2 h at 4°C. The beads were washed three times with buffer A, resuspended in 25 μl 5% SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer, and boiled for 5 min. After centrifugation, the supernatants were separated by SDS-PAGE and analyzed by western blotting with an ECL Advance Western blotting detection kit (GE Healthcare).
Preparation of yeast extracts and Western Blotting
Total protein extract was prepared from 5 × 106 cells from logarithmically growing culture as described previously [11]. Proteins were analyzed by SDS-PAGE, transferred to PVDF membranes, and probed with anti-Myc monoclonal antibody (Roche) or anti-a-tubulin antibody (Sigma). Detection was performed with HRP-conjugated secondary antibodies followed by treatment using the ECL advance Western blot detection kit (GE Healthcare).
Authors' contributions
CS performed experiments and drafted the manuscript. TM and HS participated in the experimental design and analyzed the data. TH performed experiments, participated in the experimental design, analyzed the data and finalized the manuscript. All authors read and approved the final manuscript.
Supplementary Material
The fbh1-fb allele is dominant for DNA repair. Wild type cells expressing GFP-wild type or mutant fbh1 under the control of the nmt1 promoter at the ars1 locus were incubated in EMM2 and spotted onto EMM2 plates containing MMS. The plates were incubated for 3 days at 30°C.
GFP-F-box domain mutant could not enter the nucleus. fbh1Δ cells expressing GFP-F-box (1–269 amino acids) were incubated in EMM2 without or with MMS (0.1%) treatment for 2 h at 30°C and observed by fluorescence microscopy. The upper and lower panels show GFP and DAPI images, respectively. The scale bar indicates 10 μm.
Acknowledgments
Acknowledgements
We thank Y. Tsutsui and T. Toda for strains. This work was supported by Grants-in-Aid for Scientific Research from The Ministry of Education, Science, Sports, and Culture of Japan, and CREST, JST.
Contributor Information
Chikako Sakaguchi, Email: sakaguchi@biken.osaka-u.ac.jp.
Takashi Morishita, Email: tmorishi@biken.osaka-u.ac.jp.
Hideo Shinagawa, Email: shinah@bioacademia.co.jp.
Takashi Hishida, Email: hishida@biken.osaka-u.ac.jp.
References
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Lambert S, Froget B, Carr AM. Arrested replication fork processing: interplay between checkpoints and recombination. DNA Repair (Amst) 2007;6:1042–1061. doi: 10.1016/j.dnarep.2007.02.024. [DOI] [PubMed] [Google Scholar]
- Game JC. The Saccharomyces repair genes at the end of the century. Mutat Res. 2000;451:277–293. doi: 10.1016/s0027-5107(00)00055-5. [DOI] [PubMed] [Google Scholar]
- Prado F, Cortes-Ledesma F, Huertas P, Aguilera A. Mitotic recombination in Saccharomyces cerevisiae. Curr Genet. 2003;42:185–198. doi: 10.1007/s00294-002-0346-3. [DOI] [PubMed] [Google Scholar]
- Dudas A, Chovanec M. DNA double-strand break repair by homologous recombination. Mutat Res. 2004;566:131–167. doi: 10.1016/j.mrrev.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Raji H, Hartsuiker E. Double-strand break repair and homologous recombination in Schizosaccharomyces pombe. Yeast. 2006;23:963–976. doi: 10.1002/yea.1414. [DOI] [PubMed] [Google Scholar]
- Haruta N, Kurokawa Y, Murayama Y, Akamatsu Y, Unzai S, Tsutsui Y, Iwasaki H. The Swi5-Sfr1 complex stimulates Rhp51/Rad51- and Dmc1-mediated DNA strand exchange in vitro. Nat Struct Mol Biol. 2006;13:823–830. doi: 10.1038/nsmb1136. [DOI] [PubMed] [Google Scholar]
- Akamatsu Y, Dziadkowiec D, Ikeguchi M, Shinagawa H, Iwasaki H. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc Natl Acad Sci USA. 2003;100:15770–15775. doi: 10.1073/pnas.2632890100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu Y, Tsutsui Y, Morishita T, Siddique MS, Kurokawa Y, Ikeguchi M, Yamao F, Arcangioli B, Iwasaki H. Fission yeast Swi5/Sfr1 and Rhp55/Rhp57 differentially regulate Rhp51-dependent recombination outcomes. Embo J. 2007;26:1352–1362. doi: 10.1038/sj.emboj.7601582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita T, Furukawa F, Sakaguchi C, Toda T, Carr AM, Iwasaki H, Shinagawa H. Role of the Schizosaccharomyces pombe F-Box DNA helicase in processing recombination intermediates. Mol Cell Biol. 2005;25:8074–8083. doi: 10.1128/MCB.25.18.8074-8083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Dixon J, Barr AR, Whitby MC. The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol Cell Biol. 2005;25:8084–8096. doi: 10.1128/MCB.25.18.8084-8096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Choi E, Lee SH, Lee C, Seo YS. A DNA helicase from Schizosaccharomyces pombe stimulated by single-stranded DNA-binding protein at low ATP concentration. J Biol Chem. 1997;272:18910–18919. doi: 10.1074/jbc.272.30.18910. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim JH, Lee SH, Kim DH, Kang HY, Bae SH, Pan ZQ, Seo YS. The novel human DNA helicase hFBH1 is an F-box protein. J Biol Chem. 2002;277:24530–24537. doi: 10.1074/jbc.M201612200. [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- Chiolo I, Saponaro M, Baryshnikova A, Kim JH, Seo YS, Liberi G. The human F-box DNA helicase FBH1 faces S. cerevisiae Srs2 and post-replication repair pathway roles. Mol Cell Biol. 2007;27:7439–7450. doi: 10.1128/MCB.00963-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/S0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/S0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/S0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Willems AR, Goh T, Taylor L, Chernushevich I, Shevchenko A, Tyers M. SCF ubiquitin protein ligases and phosphorylation-dependent proteolysis. Philos Trans R Soc Lond B Biol Sci. 1999;354:1533–1550. doi: 10.1098/rstb.1999.0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim J, Kim DH, Ryu GH, Bae SH, Seo YS. SCFhFBH1 can act as helicase and E3 ubiquitin ligase. Nucleic Acids Res. 2004;32:2287–2297. doi: 10.1093/nar/gkh534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/S0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- Winston JT, Koepp DM, Zhu C, Elledge SJ, Harper JW. A family of mammalian F-box proteins. Curr Biol. 1999;9:1180–1182. doi: 10.1016/S0960-9822(00)80021-4. [DOI] [PubMed] [Google Scholar]
- Cenciarelli C, Chiaur DS, Guardavaccaro D, Parks W, Vidal M, Pagano M. Identification of a family of human F-box proteins. Curr Biol. 1999;9:1177–1179. doi: 10.1016/S0960-9822(00)80020-2. [DOI] [PubMed] [Google Scholar]
- Hishida T, Iwasaki H, Yagi T, Shinagawa H. Role of Walker Motif A of RuvB protein in promoting branch migration of Holliday junctions. Walker motif a mutations affect ATP binding, ATP hydrolyzing, and DNA binding activities of RuvB. J Biol Chem. 1999;274:25335–25342. doi: 10.1074/jbc.274.36.25335. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Richardson WD, Markham AF, Smith AE. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Kominami K, Toda T. Fission yeast WD-repeat protein pop1 regulates genome ploidy through ubiquitin-proteasome-mediated degradation of the CDK inhibitor Rum1 and the S-phase initiator Cdc18. Genes Dev. 1997;11:1548–1560. doi: 10.1101/gad.11.12.1548. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- Chou S, Huang L, Liu H. Fus3-regulated Tec1 degradation through SCFCdc4 determines MAPK signaling specificity during mating in yeast. Cell. 2004;119:981–990. doi: 10.1016/j.cell.2004.11.053. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Craven RA, Griffiths DJ, Sheldrick KS, Randall RE, Hagan IM, Carr AM. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene. 1998;221:59–68. doi: 10.1016/S0378-1119(98)00434-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The fbh1-fb allele is dominant for DNA repair. Wild type cells expressing GFP-wild type or mutant fbh1 under the control of the nmt1 promoter at the ars1 locus were incubated in EMM2 and spotted onto EMM2 plates containing MMS. The plates were incubated for 3 days at 30°C.
GFP-F-box domain mutant could not enter the nucleus. fbh1Δ cells expressing GFP-F-box (1–269 amino acids) were incubated in EMM2 without or with MMS (0.1%) treatment for 2 h at 30°C and observed by fluorescence microscopy. The upper and lower panels show GFP and DAPI images, respectively. The scale bar indicates 10 μm.