Abstract
Editing of RNA changes the read-out of information from DNA by altering the nucleotide sequence of a transcript. One type of RNA editing found in all metazoans uses double-stranded RNA (dsRNA) as a substrate and results in the deamination of adenosine to give inosine, which is translated as guanosine. Editing thus allows variant proteins to be produced from a single pre-mRNA. A mechanism by which dsRNA substrates form is through pairing of intronic and exonic sequences before the removal of noncoding sequences by splicing. Here we report that the RNA editing enzyme, human dsRNA adenosine deaminase (DRADA1, or ADAR1) contains a domain (Zα) that binds specifically to the left-handed Z-DNA conformation with high affinity (KD = 4 nM). As formation of Z-DNA in vivo occurs 5′ to, or behind, a moving RNA polymerase during transcription, recognition of Z-DNA by DRADA1 provides a plausible mechanism by which DRADA1 can be targeted to a nascent RNA so that editing occurs before splicing. Analysis of sequences related to Zα has allowed identification of motifs common to this class of nucleic acid binding domain.
Keywords: RNA editing, negative supercoiling, B-Z junction, transcription
A well characterized editing mechanism affecting double-stranded RNA (dsRNA) involves the deamination of adenosine to produce inosine, which is translated as guanosine (1). This activity has been reported to be widespread throughout metazoa. The first example showing the physiological relevance of dsRNA editing in mammals was the replacement of a glutamine (CAG) by arginine (CGG) in the ion channel of a glutamate-responsive neuroreceptor (GluR). This change decreased the Ca2+ permeability of the receptor (2–4). Subsequently, other examples of GluR RNA editing also have been identified (5, 6), as well as editing of the serotonin-2C receptor (7) and the 4f-rnp gene from Drosophila (8). So far, two types of enzymes have been reported that are capable of performing dsRNA editing in vitro, DRADA1 (the prototype of the ADAR1 family that includes dsRAD1 and dsRAD2) and RED1 (the prototype of the ADAR2 family that includes DRADA2) (9–15). A third protein, RED2, which has strong sequence homology to RED1, has as yet no known in vitro or in vivo substrate (17). RED1 was cloned using low-stringency hybridization with probes prepared from DRADA1. Both DRADA1 and RED-1 are present in all tissues tested, suggesting that dsRNA editing is a widespread process. However, these enzymes show differences in editing specificity when transiently coexpressed with RNA substrates in vivo (13, 14). DRADA1 and RED1 are similar to each other in their catalytic and dsRNA binding motifs (9, 10, 13, 18), but differ in that the N terminus of DRADA1 contains domains absent from RED1. The possibility therefore exists that this difference in structure determines how RED1 and DRADA1 are used within cells.
METHODS
Identification of the Zα Domain.
The Z-DNA binding domain (Zα) initially was mapped to the N terminus of DRADA1 by testing baculovirus-expressed protein and showing that band shift activity required the presence of residues 1–296. This region of DRADA1 was then expressed as a C-terminal glutathione S-transferase fusion in Escherichia coli using a pGEX-5X1 cloning vector (Pharmacia) and shown to retain Z-DNA binding activity. The Zα binding domain was mapped further both by deletion and by PCR amplification of selected portions of DRADA1 cDNA. After expression in E. coli, glutathione S-transferase-fusion proteins were purified by affinity chromatography using glutathione-agarose (Sigma), and cleaved with factor Xa (New England Biolabs) to produce Zα peptide. The peptide was additionally purified by Mono S ion exchange chromatography (Pharmacia).
Z-DNA specific binding by the Zα domain of DRADA1 was demonstrated in a bandshift assay using nondenaturing 6% polyacrylamide gel electrophoresis (19). All assays were performed in a final volume of 20 μl and contained purified Zα (5 ng), 1 μg of sonicated salmon sperm DNA, 10 mM MgCl2, 25 mM NaCl, 25 mM Tris⋅HCl (pH 7.4), and 100 pg of radiolabeled Z-DNA probe (19). Specificity of Zα-binding to the probe was tested by competition with unlabeled B-form poly(dC-dG) (Pharmacia) or unlabeled, chemically brominated Z-form poly(dC-dG) (20). In addition, unlabeled supercoiled plasmid pDHg16 (21) containing a (dC-dG)13 insert, which adopts the Z-DNA conformation under the conditions used, also was used as competitor. Competition also was performed using the parental plasmid pDPL6 (21) that has no Z-DNA forming insert under these conditions.
Circular Dichroism (CD).
Experiments were performed using an Aviv model 62DS spectrometer. The DNA polymer stock, (Pharmacia, average length 2,900 base pairs) was dissolved in H2O and used without further purification. All measurements were performed at 30°C with 1 ml of 25 mM NaCl/50 mM Tris⋅HCl/0.1 mM Na2EDTA, pH 7.4, containing DNA at a final concentration of 46 μM. Samples were allowed to equilibrate for 15 min after addition of peptide before spectra were measured. The baseline was corrected against buffer.
Determination of Binding Constant.
A BIAcore instrument (Pharmacia Biosensor) was used according to the manufacturer’s specification. Three hundred response units of biotinylated poly(dC-dG), stabilized in the Z-DNA conformation by chemical bromination (20), was immobilized on a streptavidin-coated chip (SA5). All measurements were performed at 25°C in PBS buffer (1 mM KH2PO4/10 mM Na2HPO4/137 mM NaCl/1 mM KCl/1 mM Na2EDTA/0.05% Tween 20, pH 7.4) with a continuous flow rate of 20 μl/min. The association rate (kon) was determined by maintaining flow at each protein concentration for a 180-s period. The dissociation rate (koff) was measured by changing the flow to buffer for an additional 200 s. These values were used to calculate the equilibrium constant KD for Zα and for the Fab of the Z-DNA specific Z22 mAb (22).
Secondary Structure Analysis.
Sequences were analyzed by various sequence-structure programs located on the Internet at http://www.cse.ucsc.edu/research/compbio/sam.html (Hidden Markov Model, training, alignment and database searching/scoring); http://www.sdsc.edu/meme/meme/website (motif analysis), http://blocks.fhcrc.org (blockmaker and lama analysis); http://bonsai.lif.icnet.uk/bmm/dsc/dsc read align.html (dsc structure prediction); and http://www.embl-heidelberg.de/predictprotein/ppDoPred.html (phd structure prediction).
RED1.
RED1 was obtained from J. Yang (Harvard University, Boston) who had purified it from a baculovirus expression system.
RESULTS
We have shown previously that the chicken homologue of DRADA1 binds Z-DNA in vitro with high affinity (23). Subsequently, we have expressed different regions of human DRADA1 in E. coli and mapped a tight Z-DNA binding site (Zα) to a domain encompassed by amino acids 121–197 (Fig. 1C). Binding of Zα to Z-DNA was demonstrated using a band-shift assay that uses an oligonucleotide (dC-dG)n modified by incorporation of 5-bromodeoxycytosine, which causes the probe to adopt a Z-DNA conformation under low salt conditions in the presence of Mg2+ (19). As shown in Fig. 1, binding of Zα to the probe is competed by unlabeled poly(dC-dG) stabilized in the Z-form by chemical bromination (Fig 1A, lanes 4–6), but not by the unmodified B-form of the polymer (Fig. 1A, lanes 1–3). Furthermore, competition is observed with an unlabeled plasmid, pDHg16, that contains an insert maintained in the Z-conformation by negative supercoiling (Fig. 1B, lanes 4–6), but not with the parental plasmid pDPL6 that lacks this insert (Fig. 1B, lanes 1–3). These results indicate that Zα has specificity for Z-DNA, and not some other feature of the probe, such as a bromine atom. RED1, which does not have a domain equivalent to Zα, fails to bind to the probe used in this assay (data not shown). Another Z-DNA binding region, Zβ, has been identified in DRADA1 (Fig. 1C) and will be discussed elsewhere.
Figure 1.
Demonstration of Z-DNA specific binding by the Zα domain of DRADA1 in a bandshift assay using nondenaturing 6% polyacrylamide gel electrophoresis (19). All assays were performed in a final volume of 20 μl and contained purified Zα (5 ng), 1 μg of sonicated salmon sperm DNA, 10 mM MgCl2, 25 mM NaCl, 25 mM Tris⋅HCl (pH 7.4), and 100 pg of radiolabeled Z-DNA probe. (A) Specificity of Zα-binding to the probe was tested by competition with unlabeled B-form poly(dC-dG) (Pharmacia) (lanes 1–3) or unlabeled, chemically brominated Z-form poly(dC-dG) (20) (lanes 4–6) that were titrated in 5-fold dilution steps, starting at 35 ng (lanes 3 and 6). Lanes without added competitor are marked + while those without added Zα are labeled −. The two band shifts (arrows) arise from one or two complexes of Zα bound to the probe. (B) Unlabeled supercoiled plasmid pDHg16(21) also was used as competitor at bacterial superhelical density (lanes 4–6). This plasmid contains a (dC-dG)13 insert, which adopts the Z-DNA conformation under these conditions. Competition was compared with that of the parental plasmid pDPL6 (21), which has no Z-DNA forming insert under these conditions (lanes 1–3). Titrations were performed in 5-fold steps, starting at 500 ng of plasmid DNA (lanes 3 and 6). (C) A diagram of DRADA1 showing the locations of Zα (residues 121–197), Zβ, dsRNA binding motifs (DRBM 1–3), and the catalytic domain. Zβ is a second Z-DNA binding domain present in DRADA1 that currently is being analyzed.
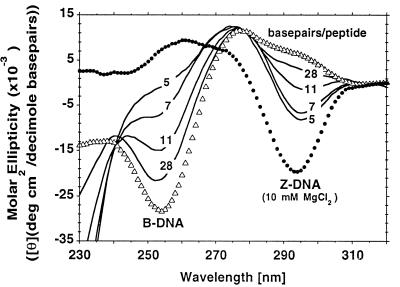
The structural specificity of Zα was examined further by using CD to follow the conversion of B-form poly(m5dC-dG) to the Z-DNA conformation induced by Zα. Poly(m5dC-dG) has been shown previously to form Z-DNA readily under physiological conditions in the presence of metal ions, polyamines, and synthetic peptides (24, 25). Formation of Z-DNA is indicated by the loss of a negative peak at 258 nm and the appearance of a new one at 295 nm. Such a change in the poly(m5dC-dG) CD spectrum is indeed caused by Zα (Fig. 2), indicating that Zα stabilizes the Z-DNA conformation of DNA. This effect was maximal under the conditions used at a molar ratio of one Zα peptide per 5 base pairs of DNA. The isosbestic point of the Zα-induced transition differs from that obtained with MgCl2, which also is shown in Fig. 2. The trough at 295 nm is also less negative. These results suggest that Zα may stabilize a left-handed conformation with a CD spectrum that differs slightly from the Mg2+-stabilized Z-DNA, and indicates that the transition from B-form poly(m5dC-dG) occurs in a single step. Such findings are similar to those found with an anti-Z-DNA antibody in which the spectrum of the protein-DNA complex differs slightly from that of metal-ion stabilized Z-DNA (26). In both cases, the protein traps the DNA polymer in a left-handed conformation.
Figure 2.
CD spectra of poly(m5dC-dG) in the presence of increasing amounts of Zα peptide. The Zα peptide alone had no observable effect on spectra in the region of 250 to 320 nm, so spectra with DNA were not corrected for the presence of peptide. The spectra measured using Zα therefore contain a large negative peak present below 245 nm that is attributable to the peptide. Molar ellipticities were calculated using base pairs of DNA. Numbers next to each spectra (solid lines) reflect the molar ratio of base pairs to peptide. The spectra obtained with DNA alone (i.e., no added MgCl2) (B form) and DNA in the presence of 10 mM MgCl2 (Z form) are also shown for comparison.
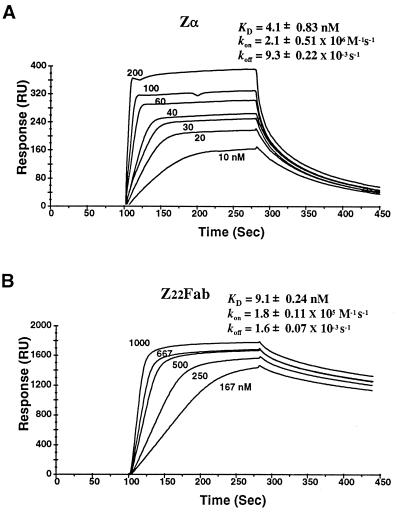
The affinity of Zα for Z-DNA was further assessed by using a BIAcore instrument to measure, under physiological conditions, the direct interaction between peptide and poly(dC-dG) stabilized in the Z-DNA conformation by chemical bromination. For comparison, binding to the same polymer of an Fab prepared from the anti-Z-DNA mAb Z22 was measured (Fig. 3) (22). The equilibrium dissociation constant between Zα and Z-DNA was 4 nM and that of the Fab was 9 nM. The latter result is in agreement with previous measurements (27). However, the rates of association and dissociation of the two molecules were markedly different. Whereas the Fab demonstrated slow on and slow off kinetics, Zα showed the opposite with fast rates of association and dissociation. This result suggests that Zα is optimized for rapid interaction with Z-DNA. No interaction between Zα and random sequence B-DNA was measurable using this approach.
Figure 3.
Association and dissociation constants for interactions between Zα and Z-DNA polymer, compared with that of mAb Z22 Fab (22). The binding was determined using surface plasmon resonance (BIAcore, Pharmacia Biosensor) and are labeled in RU, or response units, which measure the mass bound to the surface of a sensor chip. The equilibrium constant KD for Zα (A) and Z22 Fab (B) are shown. The association rate (kon) was measured over a 180-s period, and the dissociation rate (koff) was over a 200-s time frame. Neither protein gave measurable association when a biotinylated 400-bp mixed sequence B-DNA fragment was attached to the sensor chip.
Sequences that show strong similarity to Zα were identified by a blast search (28) of GenBank. In total, eight sequences from DRADA1-related proteins, two E3L proteins from vaccinia, and variola virus and a murine expressed sequence tag were found. A further search with an Hidden Markov Model (29) trained using these sequences failed to identify any more related domains (240,346 sequences searched, 0.01 significance level). Fig. 4A shows an HMM-generated multiple alignment of the Z-DNA binding domains (29). In addition to the Zα domain, the human, rat, bovine, and Xenopus genes all have another Z-DNA binding domain, Zβ, that differs from Zα in the amino terminus. E3L is of interest because it contains a dsRNA binding site with similarity to the dsRNA binding motifs of DRADA1, but it has no deaminase domain and is not an editing enzyme. The sequences in Fig. 4A were analyzed as a group, using position-specific scoring matrices, to identify conserved sequence and structural elements. Multiple alignment using meme (30), a program that creates letter probability matrices for each sequence position, identified three conserved consensus motifs, as indicated in Fig. 4B. In addition, secondary structure elements were analyzed using the dsc program (31), which uses sequence variation at each position to refine structural predictions, and the phd program, which uses a neural net strategy to achieve the same outcome (32). Three regions of strong helix formation are predicted by both programs, and these are labeled helix A, B, and C in Fig. 4C. Two of these helical regions incorporate motif I and motif II identified by the meme program. The carboxy terminal motif III identified by meme is predicted to be unstructured by dsc and to contain two strands of a β-sheet by the phd program. Another conceptually different method also was used to examine the potential of these sequences to adopt particular structures. This approach uses two programs, blockmaker (33) and lama (34), that attempt to identify structural and functional properties of proteins through sensitive detection of sequence similarities. blockmaker searches protein databases for stretches of highly conserved sequence, called blocks, and records these in the blocks database that can be assessed through the Internet address given in Methods. lama scores the relationship between blocks, allowing comparison of newly found blocks with those already known. The output of lama is a Z-score that is derived by Pearson correlation analysis of amino acid usage within blocks. A Z-score of 8.3 or greater almost certainly establishes that blocks are related, as all scores generated with blocks created by randomizing the blocks database fall below this value. Scores of 8.3 or greater allow predictions based on the properties of a related block to be made with great confidence. Scores between 5.6 and 8.3 are suggestive of a relationship between blocks. Indeed, scores in this range are found when blocks belonging to different protein families with a common structural fold are compared (34). The predictive value of scores between 5.6 and 8.3 is increased when a particular block shows relationships to numerous other blocks with the same structural fold. blockmaker was used to create a block (ZA_Block, Fig. 4D.) from the aligned sequences shown in Fig. 4A. To maintain the predicted alignment of residues and because blockmaker does not compensate for insertion of residues, the analysis was performed using Zβ sequences with the insertion between motif I and motif II removed. The block was tested against the blocks database (33) with lama. Significant similarity was found with BL01051A (the helix–turn–helix ICLR family, alignment score 39, Z-score 8.1 in the top 0e−00 percentile of scores), BL00043 (the helix–turn–helix GNTR family, alignment score 29,Z-score 7.9 in the top 0.0e−00 percentile of scores), BL0042B (the helix–turn–helix CRP family, alignment score 25, Z-score 7.3 in the top 7.5e−03 percentile of scores), BL00894A (the helix–turn–helix DEOR family, alignment score 23, Z-score 6.9 in the top 2.5e−02 percentile of scores) and with BL00356 (the helix–turn–helix LACI family, alignment score 21, Z-score 6.7 in the top 3.5e−02 percentile of scores). When the analysis was performed with a block made without using Zβ sequences, the Z-scoe was 7.6 with BL01051A and 7.2 with BL00043. The alignment between the cobbler, or composite sequence, of each of the above blocks is shown in Fig. 4D. The regions of similarity map to helix B and helix C predicted by the dsc and phd programs, and also to the helix–turn–helix region of BL00043, BL01051A, BL00894A, BL00356, and BL01128A. Taken together these predicted relationships suggest that the Zα family contains a type of helix–turn–helix binding domain, and that the three α-helical turns of helix C are involved in the recognition of Z-DNA. Helix–turn–helix previously have been shown to bind non-B-DNA structures such as bends and RNA (35–37). The presence of a β-sheet structure in motif III, as predicted by the phd program, would place Zα in the winged-helix class of transcription proteins (35, 38–43) that play important roles in embryogenesis, tumorigenesis, and the maintenance of differentiated cell states (44). It will be of interest to see whether such predictions are borne out once the structure of the Zα is solved.
Figure 4.
Sequence and structural analysis of the Zα and related domains. Human Zα and Zβ (hza, hzb, and HSU10439A), rat Zα and Zβ (rza, rzb, and RNU18942), bovine Zα and Zβ (bza, bzb, and this paper), two Zα-related sequences (xa1 and xa2) and two Zβ related sequences (xb1 and xb2) present in xenopus dsRAD1 and dsRAD2 (XLU88065 and XLU88066, respectively), the vaccinia E3L protein (S64006), and the variola equivalent (var, VVCGAA), as well as a mouse expressed sequence tag (AA204007) with relationship to Zα are shown. An HMM-generated alignment of sequences (29) is presented in A, with dots indicating gaps and blanks inserted to aid in viewing the data. Residues conserved in all sequences are shown in bold. (B) Common sequence motifs present in this group of sequences. These motifs were extracted using the meme program (30) and are presented as a multilevel consensus sequence. (C) Structural prediction for Zα made using the dsc program (31), which incorporate sequence variation at each position to improve accuracy. H is used to indicate predicted regions of helix, E for β-sheet residues, and C for coiled or loop elements. An alternative prediction made using the phd program also is shown (32). (D) Alignment between the cobbler (or composite) sequence of the blocks (33) that characterize the ICLR helix–turn–helix family (HTH_ICLR, BL01051A), the GNTR helix–turn–helix family (HTH_GNTR, BL00043), the CRP helix–turn–helix family (HTH_CRP, BL0042B), the DEOR helix–turn–helix family (HTH_DEOR, BL00894A), the LACI helix–turn–helix family (HTH_LACI), and the Zα group of related sequences (ZA_BLOCK). The ZA_BLOCK was generated after removal of the inserted R and P residues present in helix C of Zβ-related sequences, a move necessary to maintain alignment of other residues during analysis. The Z-scores indicate a high probability that the relationships revealed by lama (34) are true positive results. For the HTH families, sequences corresponding to the DNA binding region are capitalized.
Five of the 14 residues in motif II have positively charged side chains, suggesting that this region is a site of interaction with DNA. Lysines at either end potentially could anchor motif II to the phosphate backbone of DNA. Asparagine and leucine are highly conserved residues, making it likely that these amino acids are involved in direct DNA contacts. Indeed, both residues would lie on the same face of the helix if motif II were α-helical. The proline and tryptophan in motif III is completely conserved, raising the possibility that one or the other amino acid also makes DNA contacts. Tryptophan, for example, could bind to Z-DNA in a nonintercalative mode similar to that seen in the cocrystal of proflavin and Z-DNA (45). Intercalation of tryptophan between a dG-dC step has been reported for the human ETS1-DNA complex (38) (discussed in ref. 43), whereas intercalation of proline is found in an IHF-DNA complex (46). Alternatively, the conserved proline and tryptophan may be required for correct protein folding. The variation in the amino terminus between Zα, Zβ, and E3L may allow specific interaction with other proteins, and thus affect binding specificity or function of these domains.
DISCUSSION
The potential for involvement of Z-DNA in biological processes has been much discussed since the x-ray crystallographic description of this structure (47). The demonstration that Z-DNA could be stabilized by negative supercoiling brought this conformation into the realm of the biologist (48, 49). Subsequently, it was shown that formation of Z-DNA occurs in vivo and in agarose-embedded, permeabilized, metabolically active nuclei as a result of transcription-induced supercoiling in the underwound region 5′ or behind a moving RNA polymerase (50–52). The level of unrestrained supercoiling present in vivo nevertheless is limited by the relaxing action of topoisomerases and the accommodation of negative supercoils into nucleoprotein structures. Due to the transient nature of Z-DNA in vivo, direct experimental demonstration of the involvement of Z-DNA in biological processes has been difficult. The indirect approach of finding Z-DNA binding proteins also has been beset by methodological problems (53, 54). The data presented here shows that a natural protein exists that is specific for Z-DNA. The nuclear location of this protein and the high affinity for Z-DNA make it unlikely that this finding is adventitious, underscoring the possibility that this non-B-DNA structure is exploited by nature in regulation of biological processes.
The nature of the interaction of Zα with Z-DNA will await structural studies. The CD experiments show a difference between salt-induced and Zα-stabilized Z-DNA. This outcome may reflect the sensitivity of CD to changes in the close environment of Z-DNA, possibly due to binding of Zα to the convex outer surface of Z-DNA. Alternatively, Zα may induce changes to the helical parameters of Z-DNA. For example, Zα may interact with the minor groove of Z-DNA in a manner analogous to that seen with some transcriptional regulators that bend the DNA helix (38, 55, 56). Alternatively the B-Z junction may be recognized by Zα. At the B-Z junction, the change in helical direction is associated with an inversion of base pairs. The major groove of B-DNA comes to overlie the minor groove of Z-DNA whereas the minor groove of B-DNA merges with the major convexity of Z-DNA. This area of transition has the potential to create structurally unique shapes to which Zα might bind (see for example ref. 57). The junction also may be bent (58), providing sites where the intercalation of the carboxy tryptophan (or other amino acids) could occur. However, binding involving intercalation might be expected to involve slower kinetics than those shown in Fig. 3. It is of interest that extremely slow association and dissociation phases, suggestive of a second binding mode, are found when Zα dimers rather than monomers are studied (data not shown). Potentially Zα could bind solely to the major groove of B-DNA adjacent to a B-Z junction. This event is unlikely as the B-DNA form of dC-dG (Fig. 1.) does not affect binding of Zα to Z-DNA polymers.
An unresolved question in these studies is the role of Zα in the biology of the editing enzyme DRADA1. Use of introns to form dsRNA, as has been demonstrated for editing of glutamate-responsive neuroreceptor subunit RNAs (5, 6, 59–61), means that DRADA1 must act soon after transcription and before splicing of the nascent pre-mRNA. Targeting of DRADA1 to a transcription-dependent structure, such as Z-DNA would facilitate its localization to nascent transcripts, so that editing occurred before splicing. It may turn out, however, that Z-DNA binding domains of DRADA1 regulate transcriptional events other than editing.
Demonstration of an interaction between Zα and DNA within living cells may be possible using rapid UV-laser-induced crosslinking techniques (62). Potential Z-DNA-forming sequence motifs are present in many human genes (63). Binding of Zα to such segments could indicate that under appropriate levels of negative supercoiling, pre-mRNA from that gene is edited. Such studies might provide further insight into the possible regulation of DRADA1 by Z-DNA and provide insight into the evolution of this editing mechanism (64).
Acknowledgments
We would like to thank Ky Lowenhaupt and Imre Berger for their help in preparing part of the Zα peptide used in these studies, Jinghua Yang for the gift of RED1, Shmuel Pietrokovski for reviewing the data generated from sequence analysis, and Ron Hough for supplying prepublication data. We would like to thank Imre Berger and his associates A. Ortiz and L. Ribas de Pouplana for sharing the results of their calculations, based on data in the present paper, suggesting that Zα forms a helix–loop–helix structure, and also for raising the possibility that the tryptophan residue of Zα binds in a manner similar to proflavin in its cocrystal with Z-DNA. This work was supported by grants from the National Institutes of Health (2 R37 CA04186-37 and RO1 GM40536), the Human Frontier Science Program (RG-3/95B), the Director, Office of Energy Research, Office of Health and Environment Research, Division of the U.S. Department of Energy under Contract No. DE-AC03-76SF00098), and the National Science Foundation (MCB 9305271).
ABBREVIATIONS
- dsRNA
double-stranded RNA
- CD
circular dichroism
References
- 1.Bass B L. In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 383–418. [Google Scholar]
- 2.Verdoorn T A, Burnashev N, Monyer H, Seeburg P H, Sakmann B. Science. 1991;252:1715–1718. doi: 10.1126/science.1710829. [DOI] [PubMed] [Google Scholar]
- 3.Hume R I, Dingledine R, Heinemann S F. Science. 1991;253:1028–1031. doi: 10.1126/science.1653450. [DOI] [PubMed] [Google Scholar]
- 4.Sommer B, Kohler M, Sprengel R, Seeburg P H. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 5.Kohler M, Burnashev N, Sakmann B, Seeburg P H. Neuron. 1993;10:491–500. doi: 10.1016/0896-6273(93)90336-p. [DOI] [PubMed] [Google Scholar]
- 6.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger J R, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg P H. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 7.Burns C N, Chu H, Rueter S M, Hutchinson L K, Canton H, Sanders-Bush E, Emeson R. Nature (London) 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 8.Petschek J P, Mermer M J, Scheckelhoff A A S, Vaughn J C. J Mol Biol. 1996;259:885–890. doi: 10.1006/jmbi.1996.0365. [DOI] [PubMed] [Google Scholar]
- 9.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Proc Natl Acad Sci USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Connell M, Krause S, Higuchi M, Hsuan J J, Totty N F, Jenny A, Keller W. Mol Cell Biol. 1995;15:1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patterson J B, Samuel C E. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dabiri G A, Lai F, Drakas R A, Nishikura K. EMBO J. 1996;15:34–45. [PMC free article] [PubMed] [Google Scholar]
- 13.Melcher T, Maas S, Herb A, Sprengel R, Seeburg P H, Higuchi M. Nature (London) 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 14.Maas S, Melcher T, Herb A, Seeburg P, Keller W, Krause S, Higuchi M, O’Connell M A. J Biol Chem. 1996;271:12221–12226. doi: 10.1074/jbc.271.21.12221. [DOI] [PubMed] [Google Scholar]
- 15.Lai F, Chen C-X, Carter K C, Nishikura K. Mol Cell Biol. 1997;17:2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hough R F, Bass B L. J Biol Chem. 1994;269:9933–9939. [PubMed] [Google Scholar]
- 17.Melcher T, Maas S, Sprengel R, Higuchi M, Seeburg P H. J Biol Chem. 1996;271:31795–31798. doi: 10.1074/jbc.271.50.31795. [DOI] [PubMed] [Google Scholar]
- 18.Lai F, Drakas R, Nishikura K. J Biol Chem. 1995;270:17098–17105. doi: 10.1074/jbc.270.29.17098. [DOI] [PubMed] [Google Scholar]
- 19.Herbert A G, Rich A. Nucleic Acids Res. 1993;21:2669–2672. doi: 10.1093/nar/21.11.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moller A, Nordheim A, Kozolowski S A, Patel D J, Rich A. Biochemistry. 1984;23:54–62. doi: 10.1021/bi00296a009. [DOI] [PubMed] [Google Scholar]
- 21.Haniford D B, Pulleyblank D E. J Biomol Struct Dyn. 1983;1:593–609. doi: 10.1080/07391102.1983.10507467. [DOI] [PubMed] [Google Scholar]
- 22.Moller A, Gabriels J E, Lafer E M, Nordheim A, Rich A, Stollar B D. J Biol Chem. 1982;257:12081–12085. [PubMed] [Google Scholar]
- 23.Herbert A G, Lowenhaupt K, Spitzner J R, Rich A. Proc Natl Acad Sci USA. 1995;92:7550–7554. doi: 10.1073/pnas.92.16.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behe M, Felsenfeld G. Proc Natl Acad Sci USA. 1981;78:1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeuchi H, Hanamura N, Harada I. J Mol Biol. 1994;236:610–617. doi: 10.1006/jmbi.1994.1170. [DOI] [PubMed] [Google Scholar]
- 26.Lafer E M, Sousa R, Ali R, Rich A, Stollar B D. J Biol Chem. 1986;261:6438–6443. [PubMed] [Google Scholar]
- 27.Polymentis M, Stollar B D. J Immunol. 1995;154:2198–2208. [PubMed] [Google Scholar]
- 28.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 29.Krogh A, Brown M, Mian I S, Sjolander K, Haussler D. J Mol Biol. 1994;235:1501–1531. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- 30.Bailey T L, Elkan C. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA: AAAI Press; 1994. pp. 28–36. [PubMed] [Google Scholar]
- 31.King R D, Sternberg M J E. Protein Sci. 1996;5:2298–2310. doi: 10.1002/pro.5560051116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rost B, Sander C. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 33.Henikoff O, Henikoff J G. Genomics. 1994;19:97–107. doi: 10.1006/geno.1994.1018. [DOI] [PubMed] [Google Scholar]
- 34.Pietrokovski S. Nucleic Acids Res. 1996;24:3836–3845. doi: 10.1093/nar/24.19.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schultz S C, Schields G C, Steitz T A. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 36.Rivera-Pomar R, Niessing D, Schmidt-Orr U, Gehring W, Jackle H. Nature (London) 1996;379:746–749. doi: 10.1038/379746a0. [DOI] [PubMed] [Google Scholar]
- 37.Dubnau J, Struhl G. Nature (London) 1996;379:694–699. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- 38.Werner M H, Clore G M, Fisher C L, Fisher R J, Trinh L, Shiloach J, Gronenborn A M. Cell. 1995;83:761–771. doi: 10.1016/0092-8674(95)90189-2. [DOI] [PubMed] [Google Scholar]
- 39.Ramakrishnan V, Finch J T, Graziano V, Lee P L, Sweet R M. Nature (London) 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 40.Clark K L, Halay E D, Lai E, Burley S K. Nature (London) 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 41.Clubb R T, Omichinski J G, Savilahti H, Mizuuchi K, Gronenborn A M, Clore G M. Structure. 1994;2:1041–1048. doi: 10.1016/s0969-2126(94)00107-3. [DOI] [PubMed] [Google Scholar]
- 42.Fogh R H, Ottleben G, Ruterjans H, Schnarr M, Boelens R, Kaptein R. EMBO J. 1994;13:3936–3944. doi: 10.1002/j.1460-2075.1994.tb06709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kodandapani R, Pio F, Ni C-Z, Piccialli G, Klemsz M, Mckercher S, Maki R A, Ely K R. Nature (London) 1996;380:456–460. doi: 10.1038/380456a0. [DOI] [PubMed] [Google Scholar]
- 44.Kaufman E, Knochel W. Mech Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- 45.Westhof E, Hosur M V, Sundralingam M. Biochemistry. 1988;27:5742–5747. doi: 10.1021/bi00415a052. [DOI] [PubMed] [Google Scholar]
- 46.Rice P A, Yang S-W, Mizuuchi K, Nash H A. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 47.Wang A H-J, Quigley G J, Kolpak F J, Crawford J I, van Boom J H, van der Marel G, Rich A. Nature (London) 1979;282:680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 48.Klysik J, Stirdivant S M, Larson J E, Hart P A, Wells R D. Nature (London) 1981;290:672–677. doi: 10.1038/290672a0. [DOI] [PubMed] [Google Scholar]
- 49.Peck L J, Nordheim A, Rich A, Wang J C. Proc Natl Acad Sci USA. 1982;79:4560–4564. doi: 10.1073/pnas.79.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu L F, Wang J C. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rahmouni A R, Wells R D. Science. 1989;246:358–363. doi: 10.1126/science.2678475. [DOI] [PubMed] [Google Scholar]
- 52.Wittig B, Wolfl S, Dorbic T, Vahrson W, Rich A. EMBO J. 1992;11:4653–4663. doi: 10.1002/j.1460-2075.1992.tb05567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolfl S, Vahrson W, Herbert A G. In: DNA and Nucleoprotein Structure in Vivo. Saluz H P, Wiebauer K, editors. Austin, TX: Landes; 1995. pp. 137–159. [Google Scholar]
- 54.Herbert A, Rich A. J Biol Chem. 1996;271:11595–11598. doi: 10.1074/jbc.271.20.11595. [DOI] [PubMed] [Google Scholar]
- 55.Schumacher M A, Choi K Y, Zalkin H, Brennan R G. Science. 1994;266:763–770. doi: 10.1126/science.7973627. [DOI] [PubMed] [Google Scholar]
- 56.Werner M H, Huth J R, Gronenborn A, Clore G M. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- 57.Dai Z, Dauchez M, Thomas G, Peticolas W. J Biomol Struct Dyn. 1992;9:1155–1183. doi: 10.1080/07391102.1992.10507985. [DOI] [PubMed] [Google Scholar]
- 58.Liu M, Guo Q, Kallenbach N R, Sheardy R D. Biochemistry. 1992;1992:4712–4719. doi: 10.1021/bi00134a026. [DOI] [PubMed] [Google Scholar]
- 59.Higuchi M, Single F N, Kohler M, Sommer B, Sprengel R, Seeburg P H. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 60.Egebjerg J, Kukekov V, Heinemann S F. Proc Natl Acad Sci USA. 1994;91:10270–10274. doi: 10.1073/pnas.91.22.10270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Herb A, Higuchi M, Sprengel R, Seeburg P H. Proc Natl Acad Sci USA. 1996;93:1875–1880. doi: 10.1073/pnas.93.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wittig B, Dorbic T, Rich A. Proc Natl Acad Sci USA. 1991;88:2259–2263. doi: 10.1073/pnas.88.6.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schroth G P, Chou P J, Ho P S. J Biol Chem. 1992;267:11846–11855. [PubMed] [Google Scholar]
- 64.Herbert A G. Trends Genet. 1996;12:6–9. doi: 10.1016/0168-9525(96)81375-8. [DOI] [PubMed] [Google Scholar]