Abstract
Transforming growth factor β (TGF-β) regulates a variety of physiologic processes, including growth inhibition, differentiation, and induction of apoptosis. Some TGF-β-initiated signals are conveyed through Smad3; TGF-β binding to its receptors induces phosphorylation of Smad3, which then migrates to the nucleus where it functions as a transcription factor. We describe here the association of Smad3 with the nuclear protooncogene protein SnoN. Overexpression of SnoN represses transcriptional activation by Smad3. Activation of TGF-β signaling leads to rapid degradation of SnoN and, to a lesser extent, of the related Ski protein, and this degradation is likely mediated by cellular proteasomes. These results demonstrate the existence of a cascade of the TGF-β signaling pathway, which, upon TGF-β stimulation, leads to the destruction of protooncoproteins that antagonize the activation of the TGF-β signaling.
Transforming growth factor β (TGF-β) is a member of a large family of cytokines that affect a variety of biological processes, including cell growth regulation, embryonic pattern formation, immune regulation, and apoptosis (1–5). Signaling by TGF-β is initiated by the binding of TGF-β to its heteromeric complex of type I and II cell surface receptors (6, 7). Each receptor is a transmembrane protein that possesses a cytoplasmic serine/threonine kinase domain (8, 9). Binding of TGF-β to the ectodomain of the type II receptor induces heterooligomerization between the type II and the type I receptors. Once the two receptor subunits are in close proximity, the constitutively active type II receptor kinase phosphorylates the GS domain of the type I receptor kinase; this phosphorylation in turn activates the type I receptor kinase, which dispatches most, if not all, of the downstream signals from the receptor complex (10–13).
Genetic analysis in Drosophila and Caenorhabditis elegans has indicated that Smad proteins are the intracellular mediators of TGF-β signaling (14, 15). Once activated, the type I TGF-β receptor kinase phosphorylates a Smad2 or Smad3 protein at an SSXS motif present at the C terminus of both proteins (16, 17), resulting in the oligomerization of Smad2 or -3 with their common partner, Smad4 (18, 19). The resulting heteromeric Smad protein complexes then migrate to the nucleus, where they regulate expression of a large number of target genes, most of which remain to be identified (20).
Several nuclear proteins have been found to interact with Smad proteins. Such interactions either influence Smad function directly or, alternatively, affect the functions of its binding partners. Thus, p300, the AP-1 complex, TFE3, FAST-2, vitamin D nuclear receptor, Gli3, and Evi-1 have all been reported to bind to Smad3 and thereby participate in the regulation of natural or artificial reporter constructs known to be inducible by TGF-β/activin (21–29).
Recently, we reported that the Ski oncoprotein can also bind Smad3 in vivo in a ligand-dependent manner (30). The resulting complexes are able to bind to DNA, but the ability of these complexes to function as activators of TGF-β-responsive promoters is blocked. Overexpression of Ski in mink lung epithelial cells causes the cells to acquire resistance to growth inhibition by TGF-β. This resistance cannot be traced to any effects of Ski on the ability of TGF-β to induce expression of the p15INK4B gene, whose product is a potent inhibitor of the CDK4 and CDK6 G1 cyclin-dependent kinases. However, in the presence of overexpressed Ski, myc expression, which is normally suppressed by TGF-β treatment, remains high. This suggests a plausible mechanism by which Ski confers resistance to TGF-β-mediated growth suppression.
We report here on the interaction of SnoN, a Ski-related protein, with Smad3. In addition to influencing the binding of Ski and SnoN to the Smad3/4 transcription complex, we find a second functional interaction between the TGF-β receptor and the Ski and SnoN proteins that involves their metabolic stability. These results extend our understanding of the molecular mechanisms mediating the intracellular signaling of TGF-β and should also lead to a clearer mechanistic view of the oncogenic activity of SnoN and Ski.
Materials and Methods
Expression of Smad3 and SnoN.
N-terminally hemagglutinin (HA)-tagged SnoN was cloned in both the pEXL (22) and the pCI-Neo vectors (Promega). BOSC cells were transfected by the calcium phosphate precipitation method (32) and 36 hr later were labeled for 4 hr in methionine-free medium (GIBCO/BRL) containing 150 μCi/ml [35S]methionine (ICN). Cells were lysed in 0.4 ml of buffer C [150 mM NaCl/1% Nonidet P-40/50 mM Tris⋅HCl (pH 7.5)/50 mM NaF/50 mM β-glycerophosphate/1 mM Na3VO3/1 mM DTT/5 mM EDTA/1× protease inhibitor mixture (Roche Molecular Biochemicals)/10% glycerol] with NaCl adjusted to 400 mM. The mixture was then centrifuged at 100,000 × g for 15 min, the lysate was diluted with buffer C without NaCl, and appropriate Abs (5 μg) were added. The lysates were incubated at 4°C for 3 hr with 10 μl of protein-A beads prebound with 1 μl of rabbit anti-mouse Ab (Upstate Biotechnology, Lake Placid, NY). The immunoprecipitates were eluted from beads by boiling for 5 min at 100°C in 30 μl of 2× SDS sample buffer [100 mM Tris⋅HCl (pH 6.8)/200 mM DTT/4% SDS/0.2% bromphenol blue/20% glycerol] and resolved by electrophoresis through a 10% SDS/PAGE gel, followed by autoradiography.
HA-tagged SnoN was also cloned in the pMX-IRES-CD2 vector (X.L., unpublished data). The pMX-HA-SnoN-IRES-CD2 construct was transfected into BOSC cells by calcium phosphate precipitation (32). Forty-eight hours after transfection, virus supernatants were collected. A total of 5 × 105 Mv1Lu L20 cells were infected with viral supernatant in the presence of 4 μg/ml Polybrene for 4 hr, and fresh medium was added afterward. Forty-eight hours after the infection, cells were sorted by FACStar cell sorter (Becton Dickinson) according to their cell surface CD2 level, which was detected by Cy3-labeled anti-CD2 Ab (PharMingen) and used as populations. Cells expressing Flag-tagged Smad3 were generated essentially as described (16).
In Vitro Translation.
A 1-μg sample of plasmid expressing HA-hSki and HA-hSnoN was in vitro-translated with the TNT Coupled Reticulocyte Lysate System (Promega). The glutathione S-transferase (GST) fusion proteins were mixed with in vitro-translated proteins in 300 μl of buffer C for 3 hr at 4°C, collected on glutathione-Sepharose beads, and the bound proteins were separated by SDS/PAGE, visualized through fluorography.
Luciferase Reporter Gene Assay.
Twenty four hours prior to transfection, Mv1Lu or HepG2 cells were seeded at 1 × 105 or 5 × 104 cells per well in a 12-well plate and grown for 24 hr. DEAE-dextran-mediated transfections (10) or calcium phosphate precipitation were performed with 0.75 μg of p3TP-Lux together with a total of 1 μg of the vector alone or expressing the indicated Smad3 and SnoN. A total of 0.5 μg of pCH110 or pSVβ (CLONTECH) encoding β-galactosidase was included in each sample as a control for the efficiency of transfection. After 20 hr of incubation in the absence or presence of 100 pM TGF-β1, luciferase activity was determined by using the luciferase assay system (Promega), and β-galactosidase activity was measured with the luminescent β-gal detection kit (CLONTECH).
Measurement of Protein Stability.
Cells were incubated in methionine-free medium for 30 min, and then changed into the same but fresh medium containing 1 mCi of [35S]methionine for 2 hr. They were then rinsed twice with PBS−, and once with chase medium (DMEM containing 100 μg/ml additional methionine and cysteine), followed by incubation in the chase medium for the indicated time. Alternatively, cells were treated with cycloheximide at 10 μg/ml concomitant with or without TGF-β treatment for the indicated period of time. Cells were then lysed with buffer C and processed as described above.
Results
Ligand-Dependent Association of the SnoN Protein with Smad3.
We have recently characterized in detail the functional role of Ski in the TGF-β signal transduction pathway. In vivo, Ski interacts physically with Smad3 in a TGF-β-dependent manner. Binding of Ski to Smad3 blocks transcriptional activation of TGF-β-inducible promoters regulated by Smad3, while leaving intact the DNA-binding activities of Smad3. Our studies did not address the function of SnoN, the only other protein known to have clear relatedness to Ski. To explore the functional similarities and differences between the two proteins, we began by assessing the binding activity of SnoN to Smad3 and the consequences of this binding on Smad3-regulated transcription, using assays similar to those described previously (30).
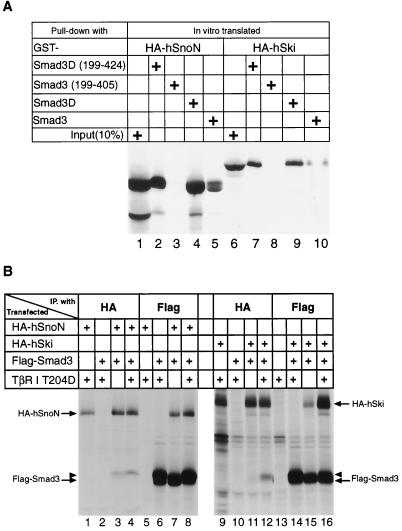
First, we characterized the relative abilities of SnoN and Ski to bind in vitro to wild-type and mutant versions of Smad3. As previously described, we generated a mutant version of Smad3, termed Smad3D, in which we converted the three C-terminal serines into aspartic acid residues to mimic the physiologic phosphorylation of Smad3 by the activated TGF-β type I receptor kinase (30). We also generated a truncated form of Smad3D that lacks the N-terminal 198 amino acids, as well as a mutant that contains only the middle portion (amino acids 199–405) of Smad3. These proteins were expressed as recombinant GST fusion proteins and were mixed with the full-length human SnoN and Ski proteins that were tagged with an influenza virus HA antigen, and synthesized by in vitro translation in the presence of [35S]methionine. Upon binding to the SnoN and Ski protein in vitro, the GST fusion proteins were retrieved on glutathione-Sepharose beads, and the bound proteins were analyzed by SDS/PAGE.
As shown in Fig. 1A, the in vitro-translated SnoN and Ski proteins behaved identically. Each bound quite strongly to the Smad3D protein or its C-terminal MH2-containing domain, but more weakly to the wild-type, unmodified version of Smad3, and did not bind at all the middle portion of Smad3, which served as a negative control. These results show that SnoN and Ski exhibit similar affinity in vitro for the Smad3D protein, which, as mentioned, mimics the phosphorylated form of Smad3 found in cells after exposure to TGF-β.
Figure 1.
In vitro and in vivo protein–protein interaction between Smad3 and SnoN. (A) In vitro binding of Ski and SnoN proteins to GST-Smad3 recombinant fusion proteins. In vitro-translated HA-tagged human Ski and SnoN proteins were exposed to the various GST-Smad3 recombinant fusion proteins and captured on glutathione beads; 10% of the translation products were directly immunoprecipitated with anti-HA Ab. Proteins bound to the beads were separated by SDS/PAGE and visualized by fluorography. (B) In vivo association of SnoN and Ski with Smad3 protein. HA-SnoN, HA-Ski, and Flag-Smad3 together with the constitutively active type I TGF-β receptor were ectopically expressed in BOSC cells through transient transfection. Lysates prepared from metabolically labeled cells 36 hr after transfection were subjected to immunoprecipitation with anti-HA or anti-Flag Ab. The SnoN and Ski proteins that were associated with Smad3 protein as well as the Smad3 protein associated with SnoN and Ski proteins were retrieved on protein G beads, separated by SDS/PAGE, and visualized through fluorography. The arrowheads denote the slowly migrating Smad3, which most likely is the form phosphorylated by the activated type I TGF-β receptor kinase.
To further assess possible in vivo interactions of Smad3 with SnoN, through transient transfection, we ectopically expressed an HA-tagged SnoN protein (HA-SnoN) with a Flag-tagged Smad3 protein (Flag-Smad3) in BOSC23 cells, derived from a human embryonic kidney cell line transformed by the adenovirus E1A protein (32). These cells were labeled metabolically for 4 hr with [35S]methionine 40 hr posttransfection. Immunoprecipitation of resulting cell lysates with Ab directed against the HA tag at the N terminus of SnoN revealed the expected expression of the protein (Fig. 1B, lane 1), which was absent in a control transfection in which only N-terminally Flag-tagged Smad3 was expressed by transient transfection (Fig. 1B, lane 2). Similarly, immunoprecipitation with the anti-Flag Ab yielded only the expected N-terminally Flag-tagged Smad3 protein in cells ectopically expressing this protein (Fig. 1B, lane 6), but not in control cells where only HA-tagged SnoN was ectopically expressed (Fig. 1B, lane 5). However, the coexpression of HA-SnoN with Flag-Smad3 resulted in complex formation between these two proteins, as demonstrated by the presence of Smad3 in immunoprecipitates from lysates treated with the anti-HA Ab directed against the tagged SnoN protein (Fig. 1B, lane 3). In a reciprocal fashion, complex formation between SnoN and Smad3 could also be demonstrated by the coimmunoprecipitated SnoN protein observed when the lysate was treated with the anti-Flag Ab directed against the tagged Smad3 protein (Fig. 1B, lanes 7). Thus, SnoN and Smad3 can form physical complexes in vivo when both proteins are overexpressed. Such association can occur even in the absence of TGF-β treatment of cells.
We also wished to investigate the influence of TGF-β signaling on this association. To do so, we ectopically expressed in these BOSC cells a constitutively active type I TGF-β receptor that carries a threonine-to-aspartic acid substitution at its residue 204 (TβRI T204D) (33). This constitutively active type I receptor kinase yields phosphorylation of the C-terminal serine residues of Smad3 (data not shown) and resulted in an increase in the level of a slowly migrating species of Smad3 seen in lanes 6 and 8 (arrowheads) of Fig. 1B. As is also shown in these lanes, in the presence of the constitutively active receptor, complex formation between SnoN and Smad3 was modestly increased, as indicated by the coimmunoprecipitated SnoN with Smad3 or vice versa (compare SnoN between lanes 7 and 8 in the anti-Flag precipitation; Smad3 between lanes 3 and 4 in the anti-HA precipitation).
An apparently contrasting result was observed when we conducted transient transfection experiments using HA-tagged Ski instead of the SnoN protein. Consistent with previously reported results (30), there was a much more robust TGF-β signaling-dependent complex formation between Smad3 and Ski (Fig. 1B, compare Ski between lanes 15 and 16 in the anti-Flag precipitation and Smad3 between lanes 11 and 12 in the anti-HA precipitation). We concluded that, in an overexpression system, Smad3 and SnoN can form physical complexes, the levels of which can be enhanced by the activation of the TGF-β signal transduction pathway. However, unlike the behavior of Ski: Smad3 complexes, the formation of SnoN:Smad3 complexes did not appear to be strongly induced by activation of TGF-β signaling.
Transcriptional Repression by the SnoN Protein of a TGF-β-Responsive Gene.
One of the consequences of TGF-β signaling in the cell is the transcriptional activation of specific genes, among them the plasminogen activator inhibitor-1 (PAI-1) gene (22, 34, 35). In addition, artificially constructed promoters containing the Smad3/4 binding consensus sequence [Smad binding element (SBE): GTCTAGAC] (36) are also activated transcriptionally in response to TGF-β treatment.
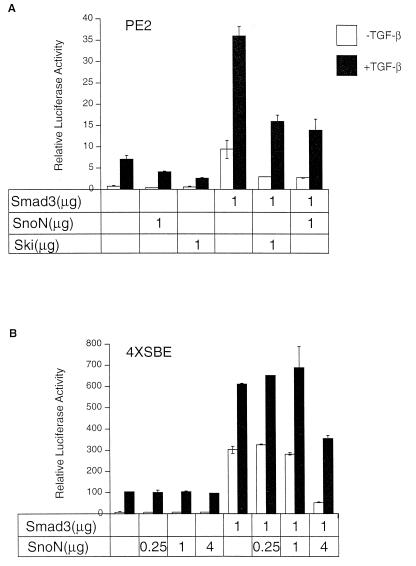
To investigate the functional consequences of the binding of SnoN to Smad3, we measured effects of ectopic expression of these two proteins on the activity of PE2, a portion of the natural PAI-1 promoter responsive to the activation of TGF-β signaling (22) and on the activity of the 4XSBE promoter, which contains four SBE sites in tandem; each promoter drove expression of a luciferase reporter construct. When these chimeric genes are introduced into TGF-β-responsive cells, their expression is known to be greatly increased after TGF-β treatment (22, 36). Moreover, ectopic expression of Smad3/4 is known to further increase the responsiveness of the PE2 promoter to TGF-β-mediated activation (22).
As seen in Fig. 2A, when the PE2 promoter construct was introduced into HepG2 cells, which express receptors for and are responsive to TGF-β (37), introduction of a Smad3 expression construct enhanced the ability of TGF-β to activate the PE2 promoter. In contrast, such activation was reduced by the coexpression of either SnoN protein or Ski in these cells.
Figure 2.
Transcriptional repression by SnoN of the PE2 and 4XSBE (SBE: GTCTAGAG) promoters. (A) HepG2 cells were transfected with PE2-luciferase construct together with 1 μg of Smad3 and SnoN expression plasmids. After incubation for 20 hr in the absence or presence of 100 pM TGF-β, luciferase as well as β-galactosidase activity was determined. (B) HepG2 cells were transfected with 4XSBE-luciferase construct promoter together with 1 μg of Smad3 and 0.25, 1, or 4 μg of SnoN expression plasmids. Luciferase and β-galactosidase activities were determined as in A. In both A and B, relative luciferase activities, whose values are the averages of the duplicate samples, were plotted after normalization with the β-galactosidase, and representative results from three separate experiments are presented. The filled bars represent cells that had been treated with TGF-β, and the open bars represent cells that had not been treated.
Similarly, as shown in Fig. 2B, in HepG2 cells, activation of the 4XSBE promoter in response to TGF-β was greatly increased by cointroduction of a Smad3 expression construct, and Smad3-mediated transcriptional activation was again attenuated by the expression of SnoN protein. The transcriptional block imposed by SnoN on this promoter is less dramatic than that seen on the PE2 promoter. Indeed, only when SnoN was expressed at a very high level could it block the transcriptional activation. In contrast, as we reported previously (30), Ski can effectively block TGF-β-induced transcriptional activation, even when expressed at a lower level. We note that the activity of the cytomegalovirus promoter, used as internal control in these experiments, was not affected by the presence of SnoN, indicating that the transcriptional repression of SnoN is not a result of a general, nonspecific inhibition of cellular transcriptional activity. These results indicate that the SnoN protein acts as a direct antagonist of Smad3 transcriptional activation. These data also suggest that the effects of SnoN on Smad3-mediated gene activation are dependent on the sequence context of the Smad3-binding site, which may vary greatly from one promoter to another.
Effects of TGF-β on the Metabolic Stability of SnoN.
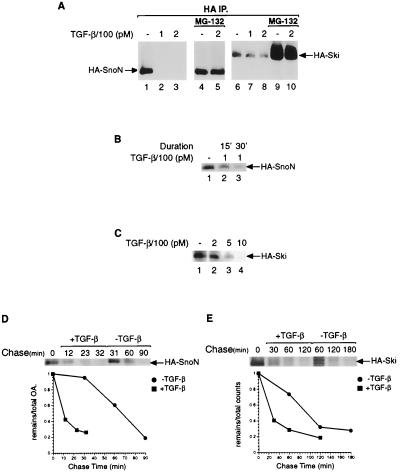
To examine more carefully the consequences of this interaction between Smad3 and SnoN, we generated a clone of mink lung epithelial cells that stably expressed HA-tagged SnoN. As shown in Fig. 3 A and B, HA-tagged SnoN rapidly disappeared in response to TGF-β in a manner dependent on the duration of the treatment: a 30-min exposure to TGF-β resulted in a more drastic reduction of SnoN levels than did a 15-min exposure (Fig. 3B, lanes 3 and 2, respectively).
Figure 3.
TGF-β causes degradation of SnoN and Ski proteins. (A) Mink lung epithelial cells stably expressing SnoN or Ski were pretreated with MG-132 or vehicle (DMSO) for 1 hr before treatment with the indicated concentrations of TGF-β for 30 min. Lysates were immunoprecipitated with the anti-HA Ab; the immunoprecipitates (IP) were collected on protein G beads, separated by SDS/PAGE, transferred to poly(vinylidene difluoride) (PVDF) membrane, and immunoblotted with anti-HA Ab. The amount of lysate used in lanes 4 and 5 was one-third that employed in lanes 1–3. (B) Same as A, except that mink lung epithelial cells stably expressing SnoN protein were treated for different time with TGF-β. (C) Same as A, except an increasing concentration of TGF-β was used with mink lung epithelial cells stably expressing Ski protein. (D and E) Measurement of stability of SnoN (D) and Ski protein (E). (D) Mink lung cells stably expressing HA-SnoN were treated with cycloheximide concomitant with TGF-β. At the indicated times, cells were lysed and the lysate was immunoprecipitated with the anti-HA Ab. The immunoprecipitates were collected on protein G beads, separated by SDS/PAGE, and transferred to PVDF membrane followed by immunoblotting with the anti-HA Ab. (E) Mink lung cells stably expressing Ski were labeled with [35S]methionine for 3 hr, followed by a chase in the presence of 100 μg/ml methionine and cysteine. Cells were lysed and processed as in D, except that the SDS/PAGE gel was dried and exposed to a phosphoimager plate (FUJIX, Tokyo).
To gauge quantitatively the stability of SnoN, we measured the half-life of SnoN protein in the absence or presence of TGF-β treatment. As seen in Fig. 3D, the half-life of SnoN changed from 60 min in mink lung epithelial cells that had not been treated with TGF-β to 10 min after the cells were treated with 100 pM TGF-β, consistent with the rapid decline in the steady-state levels of SnoN protein after TGF-β treatment as gauged by an immunoblot assay (Fig. 3 A and B).
To assess whether the disappearance of SnoN protein was the result of degradation by cellular proteasomes, we pretreated the cells for 1 hr with MG-132, a proteasome inhibitor that is able to permeate the plasma membrane. In a subset of these cells, we continued this treatment for another 30 min in the presence of TGF-β; others received an additional 30 min of MG-132 treatment without addition of TGF-β. Treatment with MG-132 for a total of 90 min in the absence of TGF-β caused a 3-fold increase in the level of HA-SnoN (Fig. 3A, compare lanes 1 and 4; note that the sample in lane 4 is derived from one-third the number of cells as that in lane 1). This suggests that proteasomes are involved in the rapid turnover of SnoN protein, even in the absence of TGF-β. Pretreatment with MG-132 for 60 min before addition of 200 pM TGF-β completely prevented TGF-β-induced loss of HA-tagged-SnoN (Fig. 3A, compare lanes 3 and 5). These results demonstrate that inhibition of proteasome function leads to enhanced SnoN stability, preventing TGF-β-induced degradation of SnoN.
The related Ski protein also exhibited a rapid disappearance in response to TGF-β in a manner dependent on the concentration of TGF-β added (Fig. 3 A and C). As was the case with SnoN, the destruction of Ski could be completely prevented by pretreatment of cells with the MG-132 proteasome inhibitor (Fig. 3A, lanes 9 and 10). However, these two proteins behaved differently in that higher concentrations of TGF-β were required to cause the degradation of Ski (Fig. 3C). The normal half-life of Ski in mink lung epithelial cells is 100 min, longer than that of SnoN, and in response to treatment with 100 pM TGF-β, the half-life of Ski was shortened to 30 min (Fig. 3 D and E).
TGF-β-induced degradation is specific to SnoN and Ski, because under the identical conditions, the levels of Smad3 did not change (data not shown). Interestingly, in the presence of the MG-132 proteasome inhibitor, the complex between SnoN and the endogenous Smad3 could be readily detected when cells were activated by TGF-β, but was impossible to detect when cells were not treated with TGF-β (data not shown). This reinforced the notion that SnoN and Smad3 form a complex only in response to activation of TGF-β signaling. However, although a SnoN molecule may initially form a complex with Smad3 after TGF-β receptor activation, it may not succeed in blocking Smad3-mediated transcriptional activation subsequently, because activation of TGF-β signaling results in rapid degradation of SnoN (Fig. 3).
Discussion
We demonstrate here that the SnoN oncoprotein, like Ski, participates in modulating signaling by TGF-β. SnoN binds weakly to Smad3 in the absence of TGF-β treatment, and strongly to Smad3 after its phosphorylation by the ligand-activated TGF-β receptor kinase. This association reduces the subsequent ability of Smad3 to activate transcription of target genes, as demonstrated here through the ectopic expression of SnoN. The present work also reveals a second, countervailing effect of TGF-β signaling on SnoN: activation of the TGF-β receptor causes the SnoN and the related Ski molecules to be targeted for destruction, apparently by proteasomes. These findings reveal a connection between SnoN and Smad3 and shed light on the mechanism of both TGF-β signaling and the mechanism used by SnoN for cell transformation (38).
As reported recently, we identified the SnoN protein as a Smad3-binding protein by using an in vitro biochemical approach (30). We found that the Smad3D protein, which is a structural mimic of the receptor-phosphorylated form of Smad3, was able to bind a protein of 80 kDa in lysates prepared from mink lung epithelial cells, but was not able to do so when these cells were pretreated with TGF-β for 30 min. Paradoxically, once we identified this 80-kDa protein and analyzed its behavior through transient and stable expression, we found that Smad3:SnoN complexes behaved in an opposite fashion: their association was actually potentiated by TGF-β treatment of cells (Fig. 1, and data not shown). This discrepancy is resolved by the evidence presented here that TGF-β treatment rapidly leads to degradation of SnoN. Moreover, the rapid and apparently specific degradation of SnoN, as well as of Ski, in response to TGF-β treatment of the cell further supports the notion that these two proteins form integral components of the TGF-β signaling cascade. The degradation of SnoN most likely also occurs when cells express the constitutively active type I TGF-β receptor, which functionally mimics the consequences of physiologic activation of the TGF-β receptor through ligand binding. This would nicely explain the failure of robust complex formation between SnoN and Smad3 when these proteins are coexpressed together with the activated receptor (Fig. 1B), because the SnoN protein is rapidly and efficiently degraded in response to the activation of TGF-β signaling, leaving little of this protein available for binding to the Smad3 protein.
The related SnoN and Ski proteins, the only known members of this protooncogene family, differ significantly in their biological activities. When constitutively expressed in avian fibroblasts, Ski is able to cause anchorage-independent growth and morphological transformation (39). SnoN, in contrast, is able to achieve morphological transformation of avian fibroblasts only when expressed at very high levels (38). Overexpression of Ski in mink lung epithelial cells leads to repression of transcriptional activation by Smad3 and to resistance to the growth-inhibitory effect of TGF-β. The latter effect may be mediated by the continued expression of myc genes in spite of active TGF-β signaling in these cells (30). Unlike the effects observed with Ski, overexpression of SnoN in mink lung epithelial cells failed to render them resistant to TGF-β and did not cause any discernible effects on the transcriptional regulation of p15INK4B and myc genes (data not shown).
These very substantial differences between SnoN and Ski function can now be explained in terms of their respective metabolic stabilities. SnoN is readily degraded in response to the weak activation of TGF-β signaling, whereas Ski is degraded only when cells are stimulated with a very high level of TGF-β. As a direct consequence, only when SnoN is expressed at far higher levels than Ski can SnoN succeed in eliciting effects in a cell transformation assay or in an assay measuring resistance to the growth-inhibitory effects of TGF-β. These observations leave unresolved the normal physiologic roles of SnoN and Ski. We speculate that the antagonism between SnoN and Smad3 is responsible for the subtle modulation of TGF-β-induced transcription of certain genes in specific cell types at certain stages of development.
These observations suggest that the effects of TGF-β receptor activation can be profoundly affected by the presence of other proteins that may be constitutively present in specific cell types. Thus, cells that express SnoN or Ski may be relatively unresponsive to TGF-β stimulation. In certain cells, ligand activation of the TGF-β receptor may cause the rapid degradation of preexisting SnoN, resulting in the removal of an important obstacle to TGF-β-mediated transcriptional activation. In yet other cells, we speculate that SnoN may persist after TGF-β receptor activation, and thereby succeed in blocking an important component of the TGF-β signaling cascade involving the Smad3/4 transcription factors. Accordingly, the well-documented highly variable effects of TGF-β on various cell types may be attributable to the background of ancillary proteins, such as Ski and SnoN, that are capable of blunting and redirecting specific downstream effector pathways emanating from the TGF-β pathway.
Acknowledgments
We thank Maitreya Dunham for preparation of mink lung epithelial cells; Dr. Shunsuke Ishii for providing cDNAs encoding human Ski and SnoN; Dr. Bert Vogelstein for the 4XSBE-lux construct; and Drs. J. Massagué and R. Derynck for the cDNAs encoding human Smad3 and TβRI T204D. We also thank members of the Weinberg laboratory and Drs. Stefan Constantinescu and X. Hua of the Lodish laboratory for helpful discussions and comments. This work was supported in part by National Cancer Institute Grants R35CA39826 (to R.A.W.) and R01CA63260 (to H.F.L.). R.A.W. is an American Cancer Society Research Professor and a Daniel K. Ludwig Cancer Research Professor. Y.S. was supported by a Robert Steel Foundation for Pediatric Cancer Research postdoctoral fellowship, and X.L. was supported by a postdoctoral fellowship from the National Institutes of Health and U.S. Army Breast Cancer Research Program.
Abbreviations
- TGF-β
transforming growth factor β
- HA
hemagglutinin
- GST
glutathione S-transferase
- SBE
Smad binding element
References
- 1.Hoodless P A, Wrana J L. Curr Top Microbiol Immunol. 1998;228:235–272. doi: 10.1007/978-3-642-80481-6_10. [DOI] [PubMed] [Google Scholar]
- 2.Kingsley D M. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 3.Massague J. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 4.Roberts A B, Flanders K C, Heine U I, Jakowlew S, Kondaiah P, Kim S J, Sporn M B. Philos Trans R Soc London B. 1990;327:145–154. doi: 10.1098/rstb.1990.0050. [DOI] [PubMed] [Google Scholar]
- 5.Massague J. Annu Rev Cell Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 6.Derynck R. Trends Biochem Sci. 1994;19:548–553. doi: 10.1016/0968-0004(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 7.Massague J, Weis-Garcia F. Cancer Surv. 1996;27:41–64. [PubMed] [Google Scholar]
- 8.Franzen P, ten Dijke P, Ichijo H, Yamashita H, Schulz P, Heldin C H, Miyazono K. Cell. 1993;75:681–692. doi: 10.1016/0092-8674(93)90489-d. [DOI] [PubMed] [Google Scholar]
- 9.Lin H Y, Wang X F, Ng-Eaton E, Weinberg R A, Lodish H F. Cell. 1992;68:775–785. doi: 10.1016/0092-8674(92)90152-3. [DOI] [PubMed] [Google Scholar]
- 10.Wrana J L, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang X F, Massague J. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 11.Wrana J L, Attisano L, Wieser R, Ventura F, Massague J. Nature (London) 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 12.Chen F, Weinberg R A. Proc Natl Acad Sci USA. 1995;92:1565–1569. doi: 10.1073/pnas.92.5.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huse M, Chen Y G, Massague J, Kuriyan J. Cell. 1999;96:425–436. doi: 10.1016/s0092-8674(00)80555-3. [DOI] [PubMed] [Google Scholar]
- 14.Raftery L A, Twombly V, Wharton K, Gelbart W M. Genetics. 1995;139:241–254. doi: 10.1093/genetics/139.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savage C, Das P, Finelli A L, Townsend S R, Sun C Y, Baird S E, Padgett R W. Proc Natl Acad Sci USA. 1996;93:790–794. doi: 10.1073/pnas.93.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Sun Y, Constantinescu S N, Karam E, Weinberg R A, Lodish H F. Proc Natl Acad Sci USA. 1997;94:10669–10674. doi: 10.1073/pnas.94.20.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macias-Silva M, Abdollah S, Hoodless P A, Pirone R, Attisano L, Wrana J L. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 18.Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Nature (London) 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 19.Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin C H, Miyazono K, ten Dijke P. EMBO J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heldin C H, Miyazono K, ten Dijke P. Nature (London) 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 21.Feng X H, Zhang Y, Wu R Y, Derynck R. Genes Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hua X, Liu X, Ansari D O, Lodish H F. Genes Dev. 1998;12:3084–3095. doi: 10.1101/gad.12.19.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janknecht R, Wells N J, Hunter T. Genes Dev. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurokawa M, Mitani K, Irie K, Matsuyama T, Takahashi T, Chiba S, Yazaki Y, Matsumoto K, Hirai H. Nature (London) 1998;394:92–96. doi: 10.1038/27945. [DOI] [PubMed] [Google Scholar]
- 25.Shen X, Hu P P, Liberati N T, Datto M B, Frederick J P, Wang X F. Mol Biol Cell. 1998;9:3309–3319. doi: 10.1091/mbc.9.12.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong C, Rougier-Chapman E M, Frederick J P, Datto M B, Liberati N T, Li J M, Wang X F. Mol Cell Biol. 1999;19:1821–1830. doi: 10.1128/mcb.19.3.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Feng X H, Derynck R. Nature (London) 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- 28.Yanagisawa J, Yanagi Y, Masuhiro Y, Suzawa M, Watanabe M, Kashiwagi K, Toriyabe T, Kawabata M, Miyazono K, Kato S. Science. 1999;283:1317–1321. doi: 10.1126/science.283.5406.1317. [DOI] [PubMed] [Google Scholar]
- 29.Liu F, Massague J, Ruiz i Altaba A. Nat Genet. 1998;20:325–326. doi: 10.1038/3793. [DOI] [PubMed] [Google Scholar]
- 30.Sun, Y., Liu, X., Ng-Eaton, E., Lane, W., Lodish, H. & Weinberg, R. (1999) Mol. Cell, in press. [DOI] [PubMed]
- 31.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 32.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wieser R, Wrana J L, Massague J. EMBO J. 1995;14:2199–2208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keeton M R, Curriden S A, van Zonneveld A J, Loskutoff D J. J Biol Chem. 1991;266:23048–23052. [PubMed] [Google Scholar]
- 35.Westerhausen D R, Jr, Hopkins W E, Billadello J J. J Biol Chem. 1991;266:1092–1100. [PubMed] [Google Scholar]
- 36.Zawel L, Dai J L, Buckhaults P, Zhou S, Kinzler K W, Vogelstein B, Kern S E. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 37.Carcamo J, Zentella A, Massague J. Mol Cell Biol. 1995;15:1573–1581. doi: 10.1128/mcb.15.3.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyer P L, Colmenares C, Stavnezer E, Hughes S H. Oncogene. 1993;8:457–466. [PubMed] [Google Scholar]
- 39.Colmenares C, Stavnezer E. Cell. 1989;59:293–303. doi: 10.1016/0092-8674(89)90291-2. [DOI] [PubMed] [Google Scholar]