Abstract
We have purified and characterized a novel 60-kDa protein that binds to centromeric K-type repeat DNA from Schizosaccharomyces pombe. This protein was initially purified by its ability to bind to the autonomously replicating sequence 3002 DNA. Cloning of the gene encoding this protein revealed that it possesses significant homology to the mammalian centromere DNA-binding protein CENP-B and S. pombe Abp1, and this gene was designated as cbh+ (CENP-B homologue). Cbh protein specifically interacts in vitro with the K-type repeat DNA, which is essential for centromere function. The Cbh-binding consensus sequence was determined by DNase I footprinting assays as PyPuATATPyPuTA, featuring an inverted repeat of the first four nucleotides. Based on its binding activity to centromeric DNA and homology to centromere proteins, we suggest that this protein may be a functional homologue of the mammalian CENP-B in S. pombe.
Centromeres are distinct chromosomal structures in eukaryotic cells that direct chromosome segregation during cell division. The centromere is the site of kinetochore formation that mediates spindle microtubule attachment and the regulation of chromosome movement. It also plays an important role in checkpoint regulation during mitosis (reviewed in refs. 1–3).
Centromeric DNA in human cells contains a large number of tandemly arranged repetitive sequences composed mostly of α-satellite DNA (4, 5). Although these repeats are common to all human chromosomes, their role in kinetochore assembly or chromosome segregation has not been determined because of the large size and complexity of centromeres. Despite this complexity, several centromeric proteins such as CENP-A, CENP-B, and CENP-C have been identified and characterized in human cells through the use of sera from patients with the autoimmune CREST (calcinosis, Raynaud’s phenomenon, esophageal dysmotility, sclerodactyly, and telangiectasia) syndrome of scleroderma (6–10). CENP-B, the best characterized centromeric protein in mammalian cells, is located in the centromere region beneath the kinetochore and binds to a 17-bp consensus sequence, the CENP-B box in α-satellite DNA (11–14). The association of CENP-B with centromeric DNA appears to be essential for the maintenance of centromeric heterochromatin structure.
Functional centromeric DNAs have been isolated and characterized in two organisms, Saccharomyces cerevisiae (15, 16) and Schizosaccharomyces pombe (17–19). In S. cerevisiae, functional centromeres, which were identified by their ability to confer mitotic and meiotic stability to minichromosomes, are relatively short, spanning about 125 bp, and contain three conserved DNA elements: CDEI, CDEII, and CDEIII (20, 21). The 25-bp CDEIII is the essential element for centromere function and is the site at which the CBF3 protein complex binds (22).
The centromeres of S. pombe are much more complex than those of S. cerevisiae and appear to more closely resemble centromeres of higher eukaryotes. The centromeres of the three chromosomes of S. pombe vary in size from 40 to 100 kb and are characterized by a complex organization of several classes of specific repeated DNA elements (18, 19). All centromeres contain a 4- to 7-kb central core DNA sequence embedded within a domain containing several inverted and direct repeats (23–25). This organization of repeats in each centromere differs among chromosomes and strains of S. pombe, but the basic inverted repeat configuration appears to be conserved (24). Among these elements, the central core containing several functionally redundant domains and the K-type repeats are essential for centromeric function (26).
Despite the identification and functional analysis of centromeric DNA elements in S. pombe, little is known about the proteins interacting with these DNA elements. The gene product of the swi6 gene, whose mutation represses centromere silencing and elevates chromosome loss (27), has been localized to heterochromatin DNA regions including the centromere (28). Also, Abp1 protein, which was initially identified as an autonomously replicating sequence-binding protein (29), is capable of binding to central core DNA of S. pombe centromere in vitro and is required for the proper segregation of chromosomes (30). Here we report the purification and biochemical characterization of a putative centromere-binding protein from S. pombe. This protein was initially purified from S. pombe based on its binding activity to ars3002 DNA, one of the autonomously replicating DNA sequences (ARS) located near the ura4 gene on chromosome III. The cloning of the gene encoding this protein showed that this protein possesses significant homology to centromere-binding proteins such as mammalian CENP-B and S. pombe Abp1. To determine the centromeric DNA-binding activity of this protein, we have examined the interaction of this protein with centromeric DNA fragments and also determined the consensus sequence to which this protein binds. The results of this study suggest a possible role for this protein in centromeric functions.
MATERIALS AND METHODS
Gel Mobility Shift and DNase I Footprinting Assays.
DNA-binding assays were carried out in reaction mixtures (20 μl) containing 25 mM Hepes⋅KOH (pH 7.5), 5 mM MgCl2, 2 mM DTT, 0.1 mg/ml BSA, 5% glycerol, 2 μg poly(dI-dC), 4–40 fmol of 32P-labeled DNA (5,000 to 15,000 cpm/fmol), and indicated amounts of purified protein. After incubation at 30°C for 20 min, loading buffer [2 μl, 0.5% bromophenol blue/0.5% xylenecyanol/40% (vol/vol) glycerol] was added, and the reaction mixtures were electrophoresed through a 3.5% polyacrylamide gel in 0.5× TBE (45 mM Tris/45 mM boric acid/1 mM EDTA) for 3 hr at 10 V/cm. After electrophoresis, gels were dried and subjected to autoradiography. The protein–DNA complexes formed were quantitated by phosphoimager analysis (Molecular Probes).
For DNase I footprinting assays, the DNA-binding reaction was carried out using the same conditions described above except that reactions contained 2% polyvinyl alcohol. Reaction mixtures were digested with bovine pancreatic DNase I (Worthington) at 25°C for 1 min followed by electrophoresis through 6% polyacrylamide gels containing 8 M urea.
Preparation of Substrates and Competitors for Gel Mobility Shift and DNase I Footprinting Assays.
A 361-bp fragment of ars3002 DNA (nucleotide 3373–3733, obtained through accession number Z27236 in the GenBank database, which has been renumbered nucleotide 1–361 in this study) was used as the substrate in the gel mobility shift assay routinely employed for the purification of the protein. This DNA was synthesized by PCR using pYJ7 plasmid as a template and subcloned into BamHI and HindIII sites of pBluescriptII KS(+) (Stratagene). The NotI- and XbaI-digested DNA fragments containing ars3002 were labeled with [α-32P]dCTP [3,000 Ci/mmol (1 Ci = 37 GBq), Amersham] by Klenow.
For competition assays, several DNA fragments derived from the central core DNA sequence of centromere 2 (cc2) or the K"-repeat DNA of centromere 1 (described in Fig. 4A) were synthesized by PCR from the cc2 or K"-repeat DNAs subcloned in pBluescript KS(−) (kindly provided by L. Clarke, University of California, Santa Barbara) using the Pfu polymerase (Stratagene). The PCR primers used in this study were designed based on the nucleotide sequence of centromere regions that were obtained from the GenBank database under the following accession numbers: X03806 and X13763 (K" repeat DNA sequence), and X66739 and X66741 (cc2 DNA sequence).
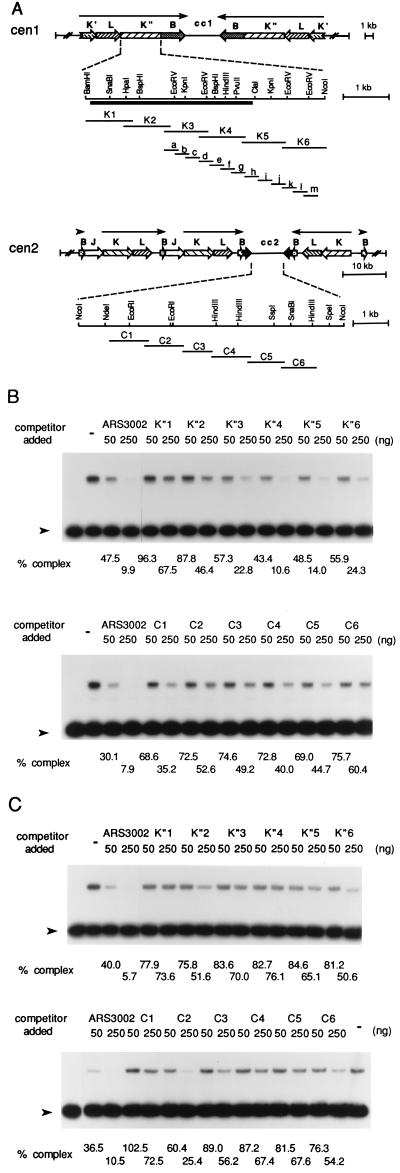
Figure 4.
Preparation of centromeric DNA fragments and determination of the binding activity of the Cbh or Abp1 protein to centromeric DNA. (A) Structures of centromeres in chromosome I (cen1) and chromosome II (cen2) and the positions of centromere DNA fragments generated by PCR in this study. The solid bar indicates the highly conserved region in all K-type repeats of S. pombe centromeres. (B and C) Competition assays using centromere DNA fragments. Indicated amounts of unlabeled ars3002 DNA or centromere DNA fragments were incubated with 40 fmol (10 ng) of 32P-labeled ars3002 DNA (361 bp) and 10 ng of Cbh (B) or Abp1 (C) protein. The percentage of complex formed (amount of complex with competitor × 100/amount of complex without competitor) was determined by phosphoimager analysis and is presented at the bottom of each lane. Lane – indicates reactions carried out in the absence of competitor.
For DNase I footprinting assays, the K"-repeat d (K-d) fragment and two ars3002 DNA fragments (ars3002-A, nucleotide 1–210; ars3002-B, nucleotide 179–361) were synthesized by PCR and subcloned into BamHI and HindIII sites of pBluescriptII KS(+). The XhoI- and SacI-digested or XbaI- and KpnI-digested DNA fragments were labeled after extension with [α-32P]dCTP (3,000 Ci/mmol) by Klenow.
Purification of ars3002 Binding Protein.
The DNA-binding activity was measured by the gel mobility shift assay as described above using 4 fmol of the 361-bp ars3002 DNA fragment as the substrate. All steps were carried out at 4°C, and all buffers contained 0.5 mM phenylmethanesulfonyl fluoride, 1 mM benzamidine⋅HCl, 2 μg/ml leupeptin, 2 μg/ml pepstatin A, 2 μg/ml antipain, 2 μg/ml aprotinin, and 1 mM DTT.
Exponentially growing S. pombe 972h− cells (200 g) were resuspended in an equal volume of buffer A (50 mM Tris⋅HCl, pH 8.0/0.5 M NaCl/5 mM MgCl2/1 mM EDTA/1 mM EGTA/10% glycerol) and disrupted using the DynoMill (Impandex, Glen Mills, Clifton, NJ). Saturated ammonium sulfate solution (saturated at 4°C) was added to 7.5% saturation, and cell debris and insoluble materials were removed by centrifugation at 15,000 × g for 30 min. The supernatant was adjusted to 70% ammonium sulfate with solid ammonium sulfate (0.389 g/ml), and after centrifugation the pellet was resuspended in 200 ml of buffer B (25 mM Hepes⋅KOH, pH 7.5/5 mM MgCl2/1 mM EDTA/1 mM EGTA/0.02% Nonidet P-40/10% glycerol). After dialysis against two changes of 4 liters of buffer B + 0.1 M NaCl for 12 hr, the solution was loaded onto an SP-Sepharose (Pharmacia) column (2.5 × 17 cm) equilibrated with buffer B + 0.1 M NaCl. The column was washed with 500 ml of buffer B + 0.1 M NaCl, and protein was eluted with a 2-liter linear gradient of NaCl (0.1–0.5 M) in buffer B. The active fractions (peaking at 0.2 M NaCl) were pooled (180 ml) and dialyzed against 4 liters of buffer B + 0.05 M NaCl for 8 hr and loaded onto a Q-Sepharose (Pharmacia) column (1.5 × 7.5 cm) equilibrated with buffer B + 0.05 M NaCl. After washing with 200 ml of buffer B + 0.05 M NaCl, the column was eluted with a 400-ml linear gradient of NaCl (0.05–0.25 M) in buffer B. Active fractions (peaking at 0.13 M NaCl) were pooled (50 ml), diluted to 0.1 M NaCl with buffer B, and loaded onto a double-stranded DNA cellulose (Sigma) column (1.0 × 4.0 cm) equilibrated with buffer B + 0.1 M NaCl. This column was washed with 30 ml of buffer B + 0.1 M NaCl and eluted with a 100-ml linear gradient of NaCl (0.1–0.4 M) in buffer B. Active fractions (peaking at 0.2 M NaCl) were pooled (12 ml), diluted to 0.1 M NaCl with buffer B, and loaded onto a sequence-specific DNA affinity column (0.7 × 2.0 cm) equilibrated with buffer B + 0.1 M NaCl. For the preparation of the sequence-specific DNA affinity column, two complementary oligonucleotides (5′-Biotin-TATATACTAACAAGGGATTGTAATAATATTATATAAAAAGTCTATTACC-3′ and 5′-GGTAATAGACTTTTTATATAATATTATTACAATCCCTTGTTAGTATATA-3′) were chemically synthesized, annealed, and conjugated to streptavidin agarose beads (GIBCO/BRL). The protein was eluted with a 20-ml linear gradient of NaCl (0.1–0.4 M) in buffer B. The active fractions (peaking at 0.2 M NaCl) were pooled (2.5 ml) and then concentrated by centrifugation in a centricon 30 (Amicon) to a volume of 0.25 ml. A portion of this fraction (0.2 ml) was applied to a 15–35% glycerol gradient in buffer B + 0.3 M NaCl. After centrifugation at 45,000 rpm for 18 hr in an SW 50.1 rotor at 4°C, fractions (0.18 ml) were collected from the bottom of the tube. Active fractions (fractions 10–14) were pooled and used in all experiments described below. This fraction was also used for peptide sequencing.
Purification of Abp1.
The breakage of S. pombe 972h− cells (200 g) and ammonium sulfate fractionation were carried out as described above. Abp1 protein was purified from this fraction as described (29), with the following changes. The protein was eluted with linear gradient of NaCl (0.1–0.5 M) at SP-Sepharose chromatography step, and only a single step of DNA-specific affinity chromatography was performed.
Cloning of the cbh Gene.
Four distinct peptide sequences were obtained from the trypsin-digested peptides of purified protein (glycerol gradient fraction). Based on the sequences obtained, degenerate oligonucleotides were synthesized and used as primers for PCR synthesis with S. pombe cDNA as a template. A PCR product of about 0.6 kb in length was cloned into PCRII with the TA cloning kit (Invitrogen), sequenced, and used as a probe for the screening of the cDNA library constructed in λZAPII (kindly supplied by D. Young, Calgary University, Canada). Two clones containing the complete open reading frame, including all of the tryptic sequences obtained from the purified protein, were obtained, and the entire nucleotide sequence was determined by the dideoxy chain termination method using Sequenase 2.0 (United States Biochemical).
RESULTS
Purification and Characterization of a Novel ars3002 Binding Protein in S. pombe Cells.
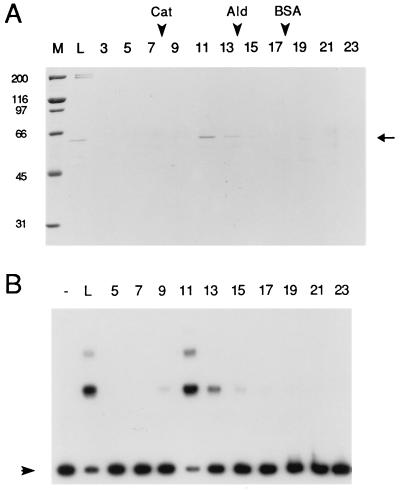
For the purification of ars binding protein from S. pombe cells, the 361-bp core sequence of ars3002 was used as substrate in the gel mobility shift assay (31). A novel DNA-binding activity that interacted with this DNA fragment was identified from S. pombe extracts, and this activity was purified approximately 7,500-fold with a yield of about 2% using the procedure described in Materials and Methods (Table 1). The purified protein was sedimented in a glycerol gradient, and the resulting fractions were analyzed by the gel mobility shift assay and SDS/PAGE. The ars3002 binding activity cosedimented with a 60-kDa protein possessing a sedimentation coefficient of 8.5 (Fig. 1).
Table 1.
Summary of purification
| Fraction | Amount of protein, mg | Total activity, units* | Specific activity, units/μg |
|---|---|---|---|
| Ammonium sulfate | 6,000 | 108,000 | 0.02 |
| SP-Sepharose | 268 | 32,160 | 0.12 |
| Q-Sepharose | 62.6 | 23,162 | 0.37 |
| ds-DNA Cellulose | 1.1 | 8,085 | 7.7 |
| DNA affinity column | 0.059 | 3,570 | 60.5 |
| Glycerol gradient | 0.018 | 2,700 | 151.0 |
One unit of activity was defined as the amount of protein that converted 50% of the substrate to complexes in the gel mobility shift assay described in Materials and Methods.
Figure 1.
SDS/PAGE and ars3002 binding assay of fractions obtained after glycerol gradient sedimentation. Glycerol gradient (15–35%) sedimentation was performed as described in Materials and Methods. (A) Glycerol gradient fractions (15 μl) were subjected to 9% SDS/PAGE and stained with Coomassie blue. The positions of size-marker proteins in the glycerol gradient are indicated by arrowheads: Cat, catalase (11.3 S); Ald, aldolase (7.3 S); BSA (4.3 S). (B) The glycerol gradient fractions (0.5 μl) were subjected to the gel mobility shift assay using 4 fmol of 32P-labeled ars3002 DNA as the substrate. Arrowheads indicate the position of free ars3002 DNA. Lanes: M, size marker; L, material loaded onto the glycerol gradient; 3–23, glycerol gradient fractions; –, buffer only.
To determine the binding specificity of this protein to ars3002 DNA, competition experiments were carried out using several synthetic DNA polymers as competitors. The addition of most synthetic DNA polymers to binding reactions did not affect the binding activity of the protein to ars3002 DNA (data not shown). Only a large excess of the synthetic copolymer poly(dA-dT)⋅poly(dA-dT) showed weak competition activity. The addition of a 100-fold excess of poly(dA-dT)⋅poly(dA-dT) to labeled ars3002 DNA inhibited the reaction about 50% in the mobility shift assay (data not shown). These results suggest that this protein binds to ars3002 DNA in a sequence-specific manner and that the binding sequence might include an alternating AT sequence motif.
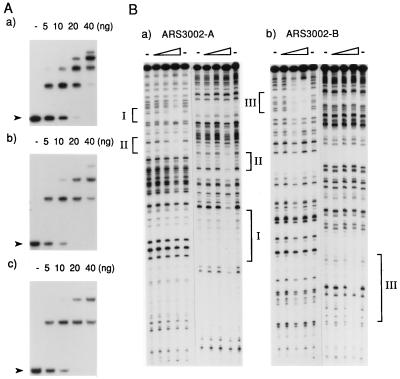
When the binding reaction was carried out with increasing levels of purified protein, multiple bands were detected in the gel mobility shift assay (Fig. 2A), suggesting that this protein was capable of binding to ars3002 DNA at multiple sites or it was multimerized. To determine the binding sites in ars3002, two DNA fragments from ars3002 were prepared for both gel mobility and DNase I footprinting analyses. One fragment spanned nucleotide 1–210 of ars3002 (ars3002-A) and the second spanned nucleotide 179–361 (ars3002-B). Both DNA fragments were bound by the protein as efficiently as the full-length ars3002 DNA (Fig. 2A). The DNase I footprinting assay showed that ars3002 DNA contained three protein-binding sites that were located between nucleotide 82 and 98, 125 and 141, and 342 and 358 (Fig. 2B).
Figure 2.
Properties of Cbh protein-ars3002 DNA complexes. (A) The indicated amounts of purified protein were used in the gel mobility shift assay with 4 fmol of the 361-bp ars3002 (a), 210-bp ars3002-A fragment (b), or 182-bp ars3002-B fragment (c) DNA as substrate. Arrowheads indicate the positions of free DNA in each experiment. (B) DNase I footprinting assay for the determination of binding sites in ars3002. Three levels of protein (15, 30, and 60 ng) were used in the DNase I footprinting assay with 4 fmol of 32P-labeled ars3002-A (a) or ars3002-B (b) fragments. (a and b) Left portion shows the T-rich strand, and the right shows the complementary strand. The regions protected by the Cbh protein are indicated as I (nucleotide 82–98), II (nucleotide 125–141), and III (nucleotide 342–358). Lanes indicated by the symbol – were reactions carried out without protein.
Cloning and Amino Acid Sequence Analysis.
The cDNA clone containing the complete coding region of this protein was obtained as described in Materials and Methods. This cDNA clone contained one open reading frame encoding a putative protein of 514 amino acids with a predicted molecular mass of 59.9 kDa (Fig. 3), which was identical to the size of the purified protein estimated by SDS/PAGE analysis.
Figure 3.
The amino acid sequence of Cbh protein (spcbh) aligned with the human CENP-B (hcenp-b) and the S. pombe Abp1 (spabp1). Boxed regions indicate exact matches between these sequences.
The amino acid sequence of this protein showed significant homology to the human and mouse CENP-B proteins, which bind to the CENP-B box (C/TTTCGTTGGAARCGGGA) present in the highly repeated α-satellite DNA of the centromere region (9, 11). Based on this homology, this gene was named cbh+ (CENP-B homologue). Also, the sequence of Cbh is similar to that of S. pombe Abp1, which was initially identified as an ARS-binding protein (29) and later shown to play a role in chromosome segregation (30). The amino acid sequence showed about 25% identity and 46% homology to human CENP-B and 40% identity and 67% homology to Abp1, and included several short, highly conserved amino acid regions, as shown in Fig. 3.
The Cbh Protein Binds to the S. pombe Centromere K-Type Repeat DNA in Vitro.
The homology of the Cbh protein to mammalian CENP-B suggests that this protein may play a role in centromere functions. To test this possibility, the centromeric DNA-binding activity of this protein was determined using the gel mobility shift assay. For this purpose, the central core sequence of centromere 2 (cc2) and the centromere 1 K"-repeat DNA were examined as possible DNA substrates because these two DNA elements are essential for the function of centromeres in S. pombe (26).
Several DNA fragments of about 1 kb derived from the K"-repeat and cc2 regions were generated by PCR (designated as K1 to K6 for the K" repeat and C1 to C6 for the cc2 region as shown in Fig. 4A), and the binding of the Cbh protein to these DNA fragments was analyzed using a competition assay in which ars3002 DNA was employed as the labeled substrate. As shown in Fig. 4B, DNA fragments derived from regions of the cc2 DNA did not effectively block the binding of Cbh to labeled ars3002 DNA. In contrast, the K"-repeat DNA fragments K4 and K5 were effective competitors, comparable to ars3002 DNA in preventing complex formation with the labeled ars3002 DNA. K3 and K6 DNA fragments also showed significant competition, suggesting that the Cbh protein binds to these K"-repeat DNA regions as well. This centromere-binding specificity of Cbh protein was quite distinct from that of Abp1 protein. When the same competition experiments were performed with Abp1 protein, only C2 fragment showed significant competition (Fig. 4C). These results showed that these two proteins have distinct binding specificity to centromere DNA even though they share strong amino acid sequence homology.
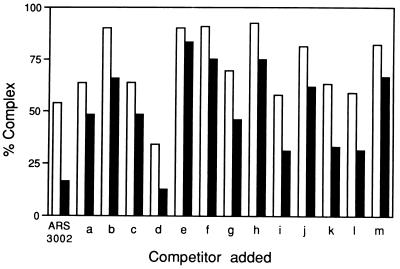
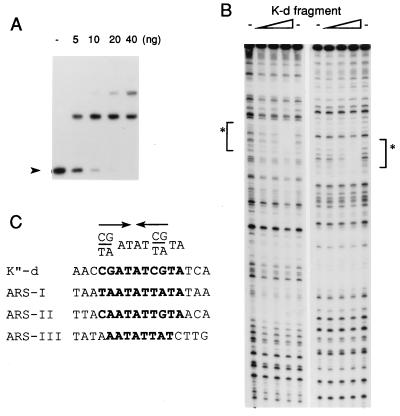
To further localize the binding region, smaller DNA fragments of about 300 bp were generated from these regions (designated as a to m in Fig. 4A), and the competition experiments were carried out as described above. With these DNA fragments, maximum competition was observed with fragment d, which blocked complex formation somewhat more effectively than ars3002. Fragments i, k, and l also showed significant competition (Fig. 5). These results suggest that Cbh protein is capable of binding to several sites in this K"-repeat DNA region. To determine the sequence that interacted with Cbh within the K-d fragment, DNase I footprinting assays were performed in which the K-d fragment was used as the labeled DNA substrate (Fig. 6). The Cbh protein bound to this DNA fragment efficiently in the gel mobility shift assay. Results from the DNase I footprinting assay showed that this DNA contained one Cbh binding site (nucleotide 2847–2862 in X03806). Comparison of the nucleotide sequences of the Cbh-binding site in the K-d fragment with those in ars3002 DNA (derived from the results presented in Fig. 2B) revealed a Cbh-binding consensus sequence PyPuATATPyPuTA, which featured an inverted repeat of the first four nucleotides (Fig. 6C).
Figure 5.
Competition assay using K-type repeat DNA fragments. Gel mobility shift assays were carried out after incubating K-type repeat DNA fragments (25 or 100 ng of fragments a to m described in Fig. 4A) with 40 fmol (10 ng) of 32P-labeled ars3002 DNA and 10 ng of the Cbh protein. After quantitation, the percentage of complex formed (amount of complex formed in the presence of competitor × 100/amount of complex formed in the absence of competitor) was determined, and the results are presented using a bar graph. The open and solid bars indicate the percentage of complex formed in the presence of 25 or 100 ng of competitor, respectively.
Figure 6.
Determination of the Cbh binding site in the K-type repeat DNA and the Cbh-binding consensus sequence. (A) Indicated amounts of the Cbh protein were used in the gel mobility shift assay with 4 fmol of 32P-labeled K-d DNA fragment. The position of free DNA is indicated by the arrowheads. (B) DNase I footprinting assay showing the binding site of the Cbh protein. Binding reactions were performed with 4 fmol of 32P-labeled K-d DNA fragment with 15, 30, or 60 ng of the Cbh protein. Left and Right show the results of the same experiments using complementary strands. Asterisks show the region protected by Cbh protein (nucleotide 2847–2862 in X03806). (C) The nucleotide sequences of Cbh-binding sites found in the K-d DNA fragment and the ars3002 DNA (Fig. 2B) and the consensus sequence. Arrows show the inverted repeat region in the consensus sequence.
DISCUSSION
In this study, we have purified and characterized a novel 60-kDa protein that is capable of binding to both K-type repeat and ars3002 DNAs of S. pombe. This protein, Cbh, was initially purified by its binding activity to ars3002 DNA. The ARS elements of S. pombe cells are larger as well as more complex than those of S. cerevisiae and usually lack a well defined boundary (31–33). However, the mapping of cis-acting elements by deletion and linker substitution analysis showed that ars3002 contains two essential and several stimulatory elements for ARS activity (34). In S. cerevisiae, ARS elements contain a single, essential element, the ARS consensus sequence (ACS) in domain A and two or three additional stimulatory elements in domain B (35, 36). The ACS is the recognition site of the origin-recognition complex (ORC), which plays an essential role in the initiation of DNA replication. The B1 element is required for efficient binding of ORC, and the binding of another protein, such as ABF1, to the B3 element is also required for the stimulation of some ARS elements (37, 38). The protein purified in this study, Cbh, may not be essential for ARS activity because this protein binds to three regions in ars3002 that are not essential for ARS activity (Fig. 3). In addition, Cbh did not bind efficiently to the essential element in ars1, another S. pombe ARS element that has been defined recently (39) (data not shown). However, the observed reduction of ARS activity upon deletion or substitution of a 55-bp region of ars3002 (nucleotide 94–148 in the 363-bp ars3002) that includes two sites to which Cbh binds (34) suggests that this protein may possess ARS stimulatory activity.
The amino acid sequence of the protein Cbh shows significant homology to mammalian CENP-B. CENP-B, which was originally discovered as a centromeric autoantigen (7), binds to the CENP-B box of the highly repeated α-satellite DNA in the centromeric region and is thought to be essential for the heterochromatin structure of centromeres. This protein has a distinct domain structure; the N terminus of this protein is the DNA-binding domain and the dimerization domain is located at the C terminus (13, 40, 41). Although the functional domains of the Cbh protein have not been characterized as yet, the primary structure of this protein is similar to CENP-B. The basic N terminus is highly homologous to CENP-B (34% identity between amino acids 1 and 138), and an acidic domain is located at the C terminus. The N-terminal region of this protein also shows homology to other putative DNA-binding proteins such as PDC2, RAG3, and jerky (42, 43), suggesting that these proteins may constitute a family of proteins sharing a similar DNA-binding motif. The C terminus of the Cbh protein contains an acidic domain that presumably is required for its dimerization or multimerization. This 60-kDa protein sedimented to a position peaking between aldolase (138 kDa) and catalase (232 kDa) during glycerol gradient sedimentation. In the mobility shift assay, the interaction between Cbh and ars3002-B or the K-d DNA fragments yielded multiple bands, whereas the DNase I footprinting assay indicated the presence of only one Cbh-binding site in these DNA fragments (Figs. 2 and 6). These observations suggest that this protein may be dimerized or multimerized and that the C-terminal acidic domain might be required for the observed protein–protein interactions.
The centromeres of S. pombe contain special chromatin structures similar to centromeric heterochromatic regions found in mammalian cells (27, 44). This similarity suggests that a functional homologue of mammalian CENP-B might be required for the formation or maintenance of the centromeric heterochromatin structure in S. pombe. Based on the homology of the Cbh protein with CENP-B, this protein is a good candidate to be a functional homologue of mammalian CENP-B. This notion was supported by observations reported here that Cbh was capable of binding to K-type repeat DNA fragment (Figs. 4 and 5). Furthermore, the distribution of Cbh-binding sites appears to correlate with the centromeric function of these DNA elements. In minichromosome stability assays, deletion of the 2.1-kb KpnI-KpnI fragment, which contains the strong and one weak Cbh-binding sites, abolished centromeric function, and deletion of the 1.2-kb KpnI-NcoI fragment, which contains the two weak Cbh-binding sites, resulted in the reduction of minichromosome stability (26). In addition, the 2.1-kb KpnI-KpnI fragment region in the K-type repeat is highly conserved and repeated in all three centromeres of S. pombe (24, 25), suggesting that all three centromeres contain multiple sites capable of binding the Cbh protein.
The amino acid sequence of Cbh is also highly homologous to Abp1 (Fig. 3). Abp1 protein was initially identified and purified as an ARS-binding protein from S. pombe (29). However, this protein was thought to play a role in centromere function because this protein is capable of binding to central core DNA region in centromere 2 in vitro and the overexpression of this protein or null-mutation of this gene elevates mini-chromosome loss (Fig. 4C; ref. 30). In spite of the homology between these proteins, little is known about their functional relationship. A strain deleted of abp1 is viable but exhibits slow growth and elevated mini-chromosome instability (30), whereas cbh+ has been shown to be an essential gene strain (unpublished results). Furthermore, overexpression of the cbh+ gene did not suppress the phenotypes observed in the abp1-deleted strain (unpublished results). These differences might be due to the distinct DNA-binding specificity between these proteins. Abp1 preferentially binds to alternating AT sequences and interacted with portions of central core DNA fragments, whereas Cbh showed strong binding to K"-repeat DNA fragments (Fig. 4 B and C). These proteins, which have different DNA-binding specificity, might belong to a family of proteins with similar roles, such as formation of heterochromatin structure, and this will be addressed in future studies.
Acknowledgments
We thank Dr. D. Young of the Calgary University for providing the ZAPII S. pombe cDNA library and Dr. L. Clarke of the University of California, Santa Barbara, for providing vectors containing K-type repeats and cc2 DNA fragments. This work was supported by Grants GM34559 (J.H.) and GM49294 (J.A.H.) from the National Institutes of Health.
ABBREVIATIONS
- ARS
autonomously replicating sequence
- cc2
central core sequence of centromere 2
- K-d
K"-repeat d fragment
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U86754).
References
- 1.Pluta A F, Mackay A M, Ainsztein A M, Goldberg I G, Earnshaw W C. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- 2.Earnshaw W C, Mackay A M. FASEB J. 1994;8:947–956. doi: 10.1096/fasebj.8.12.8088460. [DOI] [PubMed] [Google Scholar]
- 3.Willard H F. Trends Genet. 1990;6:410–416. doi: 10.1016/0168-9525(90)90302-m. [DOI] [PubMed] [Google Scholar]
- 4.Wevrick R, Willard H F. Proc Natl Acad Sci USA. 1989;86:9394–9398. doi: 10.1073/pnas.86.23.9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jabs E W, Goble C A, Cutting G R. Proc Natl Acad Sci USA. 1989;86:202–206. doi: 10.1073/pnas.86.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moroi Y, Peebles C, Fritzler M J, Steigerwald J, Tan E M. Proc Natl Acad Sci USA. 1980;77:1627–1631. doi: 10.1073/pnas.77.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Earnshaw W C, Rothfield N. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan K F, Hechenberger M, Masri K. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Earnshaw W C, Sullivan K F, Machlin P S, Cooke C A, Kaiser D A, Pollard T D, Rothfield N F, Cleveland D W. J Cell Biol. 1987;104:817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomkiel J E, Cooke C A, Saitoh H, Bernat R L, Earnshaw W C. J Cell Biol. 1994;125:531–545. doi: 10.1083/jcb.125.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. J Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muro Y, Matsumoto H, Yoda K, Nozaki N, Ohashi M, Okazaki T. J Cell Biol. 1992;116:585–596. doi: 10.1083/jcb.116.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoda K, Kitagawa K, Matsumoto H, Muro Y, Okazaki T. J Cell Biol. 1992;119:1413–1427. doi: 10.1083/jcb.119.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooke C A, Bernat R L, Earnshaw W C. J Cell Biol. 1990;110:1475–1488. doi: 10.1083/jcb.110.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke L, Carbon J. Annu Rev Genet. 1985;19:29–55. doi: 10.1146/annurev.ge.19.120185.000333. [DOI] [PubMed] [Google Scholar]
- 16.Cottarel G, Shero J H, Hieter P, Hegemann J H. Mol Cell Biol. 1989;9:3342–3349. doi: 10.1128/mcb.9.8.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke L, Amstutz H, Fishel B, Carbon J. Proc Natl Acad Sci USA. 1986;83:8253–8257. doi: 10.1073/pnas.83.21.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fishel B, Amstutz H, Baum M, Carbon J, Clarke L. Mol Cell Biol. 1988;8:754–763. doi: 10.1128/mcb.8.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M. Cell. 1989;57:739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]
- 20.Fitzgerald-Hayes M, Clarke L, Carbon J. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 21.Hieter P, Pridmore D, Hegemann J H, Thomas M, Davis R W, Phillippsen P. Cell. 1985;42:913–921. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- 22.Lechner J, Ortiz J. FEBS Lett. 1996;389:70–74. doi: 10.1016/0014-5793(96)00563-7. [DOI] [PubMed] [Google Scholar]
- 23.Hahnenberger K M, Carbon J, Clarke L. Mol Cell Biol. 1991;11:2206–2215. doi: 10.1128/mcb.11.4.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steiner N C, Hahnenberger K M, Clarke L. Mol Cell Biol. 1993;13:4578–4587. doi: 10.1128/mcb.13.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M. Mol Biol Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baum M, Ngan V K, Clarke L. Mol Biol Cell. 1994;5:747–761. doi: 10.1091/mbc.5.7.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allshire R C, Nimmo E R, Ekwall K, Javerzat J-P, Cranston G. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 28.Ekwall K, Javerzat J-P, Lorentz A, Schmidt H, Cranston G, Allshire R. Science. 1995;269:1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- 29.Murakami Y, Huberman J A, Hurwitz J. Proc Natl Acad Sci USA. 1996;93:502–507. doi: 10.1073/pnas.93.1.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halverson D, Baum M, Stryker J, Carbon J, Clarke L. J Cell Biol. 1997;136:487–500. doi: 10.1083/jcb.136.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu J, Carson D L, Dubey D D, Sharma K, Huberman J A. Chromosoma. 1994;103:414–422. doi: 10.1007/BF00362286. [DOI] [PubMed] [Google Scholar]
- 32.Caddle M S, Calos M P. Mol Cell Biol. 1994;14:1796–1805. doi: 10.1128/mcb.14.3.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maundrell K, Hutchison A, Shall S. EMBO J. 1988;7:2203–2209. doi: 10.1002/j.1460-2075.1988.tb03059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dubey D D, Kim S-M, Todorov I T, Huberman J A. Curr Biol. 1996;6:467–473. doi: 10.1016/s0960-9822(02)00514-6. [DOI] [PubMed] [Google Scholar]
- 35.Marahrens Y, Stillman B. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 36.Theis J F, Newlon C S. Mol Cell Biol. 1994;14:7652–7659. doi: 10.1128/mcb.14.11.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bell S P, Stillman B. Nature (London) 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 38.Rao H, Stillman B. Proc Natl Acad Sci USA. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clyne R K, Kelly T J. EMBO J. 1995;14:6348–6357. doi: 10.1002/j.1460-2075.1995.tb00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitagawa K, Matsumoto H, Ikeda M, Okazaki T. Mol Cell Biol. 1995;15:1602–1612. doi: 10.1128/mcb.15.3.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugimoto K, Hagishita Y, Himeno M. J Biol Chem. 1994;269:24271–24276. [PubMed] [Google Scholar]
- 42.Hohmann S. Mol Gen Genet. 1993;241:657–666. doi: 10.1007/BF00279908. [DOI] [PubMed] [Google Scholar]
- 43.Toth M, Grimsby J, Buzsaki G, Donovan G P. Nat Genet. 1995;11:71–75. doi: 10.1038/ng0995-71. [DOI] [PubMed] [Google Scholar]
- 44.Polizzi C M, Clarke L. J Cell Biol. 1991;112:191–201. doi: 10.1083/jcb.112.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]