Abstract
The replication system of bacteriophage T4 uses a trimeric ring-shaped processivity clamp (gp45) to tether the replication polymerase (gp43) to the template-primer DNA. This ring is placed onto the DNA by an ATPase-driven clamp-loading complex (gp44/62) where it then transfers, in closed form, to the polymerase. It generally has been assumed that one of the functions of the loading machinery is to open the clamp to place it around the DNA. However, the mechanism by which this occurs has not been fully defined. In this study we design and characterize a double-mutant gp45 protein that contains pairs of cysteine residues located at each monomer-monomer interface of the trimeric clamp. This mutant protein is functionally equivalent to wild-type gp45. However, when all three monomer-monomer interfaces are tethered by covalent crosslinks formed (reversibly or irreversibly) between the cysteine pairs these closed clamps can no longer be loaded onto the DNA nor onto the polymerase, effectively eliminating processive strand-displacement DNA synthesis. Analysis of the individual steps of the clamp-loading process shows that the ATPase-dependent interactions between the clamp and the clamp loader that precede DNA binding are hyperstimulated by the covalently crosslinked ring, suggesting that binding of the closed ring induces a futile, ATP-driven, ring-opening cycle. These findings and others permit further characterization and ordering of the steps involved in the T4 clamp-loading process.
Keywords: replication complex, processivity clamp, ATP hydrolysis, subunit crosslinking
The sliding processivity clamp of the DNA replication machinery is a ring-shaped protein complex that plays essential roles in the regulation of DNA replication, recombination, and repair in both prokaryotic and eukaryotic organisms (1–3). One role of the sliding clamp in DNA replication is to assist the polymerases that are responsible for copying the genome of the organism. These replication polymerases are often poorly processive in the high-salt environment of the cell and undergo repeated association and dissociation events during the extension of a nascent DNA chain in vitro (4). Left unchecked, this inefficiency would blunt the intrinsic kinetic proficiency of the polymerase that is required to complete genomic replication within the time constraints imposed by the cell cycle. The sliding clamp protein provides an elegantly simple solution to this problem. By encircling the DNA like a bead on a string, the sliding clamp is able to tether its cognate polymerase to the nucleic acid and transform a nearly distributive enzyme into a remarkably processive one, while still permitting the control of processivity that is required in discontinuous lagging strand synthesis (1).
Although sliding clamps have been identified in many different organisms and viruses, the best studied are those of bacteriophage T4, Escherichia coli, and yeast. In T4, the sliding clamp is the gp45 replication accessory protein and exists as a 25-kDa monomer that self-assembles into trimers (5, 6). Several published reports had suggested that gp45 is a protein ring (4, 7, 8), and this notion was confirmed by the x-ray structure, which revealed that gp45 crystallizes as an asymmetric torus (I. Moarefi and J. Kuriyan, personal communication). However, the question of how this ring is actually placed around the DNA remained. Various lines of evidence show that the DNA is not “broken” in the ring-loading process; thus the gp45 ring itself must be opened to permit loading onto the DNA. Yet direct evidence of ring opening during loading of the T4 clamp has been lacking.
The current view is that loading of the gp45 clamp onto the DNA replication fork is orchestrated by a coordinated series of interactions involving significant conformational rearrangements within the clamp-loading complex (6, 10–13). The placement of the gp45 clamp on the DNA depends on the action of the gp44/62 clamp-loader subassembly, which consists of four gp44 monomers and one gp62 monomer bound together so tightly that the complex can be dissociated only in the presence of denaturants (6). The gp44/62 clamp loader is also an ATPase and couples ATP hydrolysis with the loading of gp45 onto DNA. Indeed, the rate of ATP hydrolysis by gp44/62 is stimulated nearly 30-fold by the gp45 clamp and an additional 30-fold on adding template-primer DNA (14, 15). In the presence of ATP, gp44/62 and gp45 associate with a Kd of ≈10 nM (13), poising the resultant complex to interact with DNA. Gp45 acts as a specificity factor (14) to target the clamp-loading complex to the primer-template DNA (p-t DNA) junction. After loading the complex dissociates, leaving the gp45 clamp on the DNA where it then transfers to DNA polymerase to form the highly processive holoenzyme complex (12, 13, 16–18).
To better understand the role of the monomer-monomer interfaces of gp45 in clamp loading and to attempt to determine the point in the loading cycle at which clamp opening actually occurs, we describe here the design and construction of the gp45 double mutant gp45-K82C;S166C. Residues K82C and S166C face each other across the monomer-monomer interfaces of the trimer at a separation of ≈10 Å. Because the gp45-K82C;S166C double mutant does not contain any cysteine residues other than the two created by the above site-specific mutations, thiol-specific crosslinkers can be used to chemically seal the gp45 ring at each interface. Gp45-K82C;S166C can be efficiently crosslinked, either irreversibly with bismaleimidohexane (BMH) or reversibly with 1,4-di-[3′-(2′-pyridyldithio)propionamido]-butane (DPDPB) (see Materials and Methods) to form a covalently closed ring.
We show here that these chemically sealed gp45 clamps stimulate ATPase activity comparably to (or more than) wild-type (wt) gp45 in the clamp-loading pathway, up to the point of interaction of the complex with p-t DNA cofactor. Here, relative to the level reached with wt gp45, the ATPase reaction is strongly inhibited, and strand-displacement synthesis is comparably reduced. However, the DNA-dependent step in the clamp-loading process can be reactivated by adding DTT to the reversibly crosslinked gp45-DPDPB clamps, which cleaves the disulfide bonds in the DPDPB crosslinker and restores the ability of the gp45 interfaces to open. These results and others are used to gain further insight into the events of the T4 clamp-loading process.
Materials and Methods
Materials.
Radiolabeled [γ-32P]ATP was obtained from New England Nuclear, phosphoenolpyruvate, NADH, ATP, ADP, rabbit muscle pyruvate kinase, and rabbit muscle lactate dehydrogenase from Sigma, dNTPs from Amersham Pharmacia, the BMH and DPDPB crosslinkers from Pierce, and SYPRO orange protein stain from Molecular Probes. The overexpression vector containing the gp43 exo− polymerase gene was kindly provided by Nancy Nossal (National Institutes of Health, Bethesda, MD). All T4 replication proteins were expressed and purified as described (15).
Site-Directed Mutagenesis, Expression, and Purification of gp45-K82C;S166C and gp45-S166C.
The gp45-K82C;S166C and gp45-S166C genes were constructed by PCR mutagenesis as described (13), and the primary structures of the mutants were confirmed by DNA sequencing. The mutant plasmids were transformed into BL21(DE3) cells and expressed and purified as described (13). Purified gp45-K82C;S166C and gp45-S166C proteins were stored at −20°C in 20 mM Hepes (pH 7.5), 50 mM KOAc, 0.2 mM EDTA, 0.5 mM DTT, and 50% glycerol.
Crosslinking of gp45-K82C;S166C with BMH and DPDPB.
Various concentrations of BMH or DPDPB were reacted with 38-μM concentrations (trimers) of the gp45-K82C;S166C double mutant in 100-μl vol of 1× HMK buffer [50 mM Hepes, pH 7.4/50 mM KOAc/6 mM Mg(OAc)2] for 75 min at 22°C, and unreacted crosslinker was removed by passage through BioSpin P6 columns equilibrated with 1× HMK buffer. Protein concentrations were determined by Bradford assay using gp45 as the protein standard or by measuring the absorbance of the flow-through at 280 nm. Glycerol was added to a final concentration of 30%, and the crosslinked proteins were stored at −20°C until used. The efficiency of crosslinking was determined by resolving the gp45-BMH and gp45-DPDPB proteins on SDS/PAGE gels, staining with SYPRO orange (Molecular Probes), and analyzing the intensities of each band in the blue fluorescence mode of a Storm 860 PhosphorImager (Molecular Dynamics). Quantitative cleavage of the DPDPB crosslinker was achieved by incubating the gp45-DPDP protein with 30 mM DTT for 10 min at 37°C.
Spectrophotometric ATPase Assay.
This assay, which measures the loss of NADH absorbance at 340 nm (ɛM = 6,250 cm−1⋅M−1) as ADP is converted to ATP through the coupled action of pyruvate kinase and lactate dehydrogenase in the presence of phosphoenolpyruvate (PEP), has been used by us (14) and others (19) to measure the kinetics of the ATPase of the gp44/62 complex. Reactions contained 3 mM PEP, 500 μM NADH, 6 units lactate dehydrogenase, 6 units pyruvate kinase, 350 nM gp44/62, 350 nM gp45, 350 nM 50/30/32-mer template-primer-flap DNA and, in some cases, 350 nM gp43 exo− in 25 mM Tris⋅OAc (pH 7.5), 150 mM KOAc, 10 mM Mg(OAc)2, and 10 mM 2-mercaptoethanol. Reactions without DTT (Table 1) were performed in the same assay buffer. Under the conditions of the assay the regeneration of ATP is not rate-limiting, and the steady-state concentration of ADP is essentially zero.
Table 1.
Stimulation of the gp44/62 clamp holder loader ATPase by protein and nucleic acid cofactors
| Sliding clamp | +gp44/62 | +DNA | +SA | +gp exo− |
|---|---|---|---|---|
| gp45 | 1.5 ± 0.5 | 44 ± 5 | 40 ± 5 | 4.3 ± 2 |
| (1.2 ± 0.3) | (43 ± 3) | (39 ± 4) | (4.0) | |
| gp45-K82C;S166C | 2.7 ± 0.6 (2.5 ± 0.8) | 33 ± 3 (33 ± 3) | 28 ± 4 (27 ± 4) | 3.7 ± 2 (2.9 ± 2) |
| gp45-BMH | 4.8 ± 0.4 | 9.0 ± 0.4 | 7.0 ± 0.2 | 4.7 ± 0.4 |
| (4.4 ± 1) | (9.7) | (7.8) | (5.4) | |
| gp45-DPDPB | 3.1 ± 0.6 (3.3 ± 0.6) | 7.7 ± 1 (30 ± 3) | 6.1 ± 0.7 (26 ± 5) | 3.7 ± 0.1 (3.9 ± 3) |
All values are in units of μmol ATP hydrolyzed per min under the reaction conditions described in Materials and Methods and represent the average of 2-4 experiments when SDs are given. DNA refers to the 30/50/32 pt-flap construct used in these assays. SA, strepavidin. Values in parentheses were measured in the presence of DTT.
The Polymerase Holoenzyme Strand-Displacement Assay.
Strand-displacement assays were performed essentially as described (17), except that the gp43 exo− polymerase, which carries the D219A mutation that deletes the 3′-5′ exonuclease activity of the gp43 (20) was used, and a 50-fold excess of unlabeled “trap” (30-mer/50-mer p-t-flap) DNA was added to prevent any rebinding of the labeled DNA substrate. The extent of strand displacement was linear with increasing concentrations of input gp45. Essentially no strand displacement occurred in the absence of gp45, and quantitative displacement was observed at stoichiometric gp45 concentrations.
Results
Design and Characterization of the gp45-K82C;S166C Double Mutant.
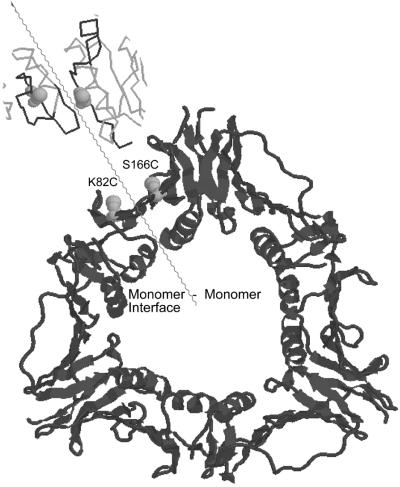
The gp45-K82C mutant of gp45 has been described (13).§ This gp45 variant was designed to take advantage of the lack of cysteines in wt gp45 to create a site-specific target for labeling with thiol-reactive reporter groups. As described (12, 13, 16, 17), the gp45-K82C mutant protein has been labeled with both fluorophores and crosslinkers and used to monitor protein–protein and protein–DNA interactions during the clamp- loading process. Residue 82, which was modified in creating the gp45-K82C mutant, is positioned very close to the monomer-monomer interface of the gp45 trimer. Thus the gp45-K82C DNA sequence was used as the starting point for PCR mutagenesis. A second cysteine was inserted at residue S166, which is located ≈10 Å away from position 82 across the monomer-monomer interface (I. Moarefi and J. Kuriyan, personal communication), to form the gp45-K82C;S166C double mutant (see Fig. 1). Each trimer of the double mutant gp45-K82C;S166C thus contains six cysteine residues.
Figure 1.
Ribbon diagram of the gp45 sliding clamp, showing the locations of the K82C and S166C residues relative to the monomer-monomer interface within the trimer. The structure was rendered from the gp45 Protein Databank coordinate file (I. Moarefi and J. Kuriyan, personal communication) by using the program raswin. The central figure is a ribbon structure showing the front face of gp45. The wavy line marks the binding interface between two adjacent monomers. The labels K82C and S166C mark the positions of these two residues on opposite sides of the interface in two separate monomers. The backbone structure shown in the upper left represents a close-up of the interface as observed in a top view of the edge of the ring.
Equilibrium ultracentrifugation had shown that gp45 exists as a trimer in solution (6, 14, 21). We used the same technique (F.D., unpublished work) to show that gp45-K82C;S166C, at a concentration of 33 μM (expressed as gp45 monomers) and in a buffer containing 30 mM Tris⋅OAc (pH 7.6), 160 mM KOAc, and 6 mM Mg(OAc)2, exists as a homogeneous population of trimers with a weight average molecular mass of 7.5 × 104 (± 2%) g/mol (the calculated molecular mass of the gp45-K82C;S166C monomer is 24,849 g/mol). Thus the double-mutant protein also exists totally as trimers under the solution conditions of this study, demonstrating that the K82C;S166C mutations do not interfere with trimer ring formation.
Crosslinking with BMH.
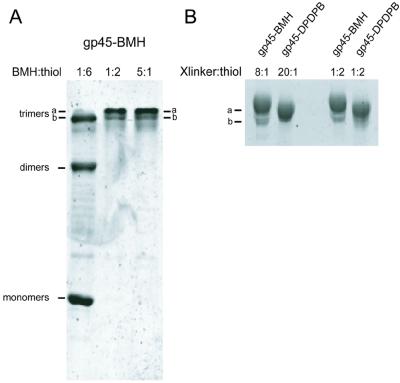
Initial crosslinking studies were performed with the irreversible homobifunctional crosslinker BMH, which contains two maleimide groups separated by a 16-Å polymethylene linker. Fig. 2 shows that the addition of a molar excess of BMH to a solution of gp45-K82C;S166C results in the efficient conversion of the denatured protein to a species that migrates more slowly on an SDS/PAGE gel, at a position shown previously (16) to correspond to that of a crosslinked gp45 trimer. No crosslinked dimers were formed, and PhosphorImager analysis of the SYPRO orange-stained bands showed that at least 90% of the input double-mutant gp45 subunits had been crosslinked into trimers.
Figure 2.
BMH and DPDPB efficiently crosslink gp45 monomers into trimers. (A) The lane labeled 1:6 shows the SDS/PAGE pattern of crosslinked products after reaction of 19 μM BMH with 19.3 μM gp45-K82C;S166C (containing 116 μM protein thiols); the lane labeled 1:2 shows the products formed after reaction of 58 μM BMH with 19.3 μM gp45-K82C;S166C (116 μM protein thiols); and the lane labeled 5:1 shows the products formed after reaction of 470 μM BMH with 15.7 μM gp45-K82C;S166C (94 μM protein thiols). Note that when the crosslinker/thiol ratio is 1:2 for these homobifunctional crosslinkers there is exactly one thiol-reactive species for each protein thiol. (B) The mobility of gp45-BMH and gp45-DPDPB in SDS/PAGE is unaffected by the addition of excess crosslinker.
Close inspection of the gel band corresponding to BMH-crosslinked gp45-K82C;S166C (henceforth referred to as gp45-BMH) revealed that this trimer band actual consists of two sub-bands (labeled a and b in Fig. 2). Quantification of these sub-bands (lanes labeled BMH:thiol 1:2 and 5:1) showed that roughly 75% of the gp45-BMH was contained in the slower moving a sub-band, and the remaining 25% in the b sub-band. We show (Fig. 2B) that the relative intensities of these bands do not change, even when the input ratio BMH crosslinker to protein thiol was reduced from 8:1 to a stoichiometric level of 1:2 (one crosslinker per two thiols). However, when the BMH/thiol ratio was further reduced to 1:6, a distribution of the protein into monomer, dimer, and trimer bands was observed (Fig. 2A, lane labeled 1:6). The only trimer band observed at this substoichiometric crosslinking ratio comigrated with sub-band b.
Each gel band in the 1:6 crosslinking reactions was quantified by PhosphorImager analysis after staining with SYPRO orange. Of the total staining intensity, 43% of the signal was in the monomer band, 30% in the dimer band, and the remaining 27% in the trimer band. These results were compared with the expected statistical distribution of gp45 subunits into monomers and crosslinked dimers and trimers, assuming that each protein thiol is equally likely to be covalently modified by BMH and that BMH binds “cooperatively” to each apposed pair of cysteine residues (i.e., no BMH monoadducts remain). A calculated distribution of 44.4% monomers, 29.6% dimers, 22.2% “open” trimers (crosslinked at only two of the three interfaces), and 3.7% “closed” trimers (crosslinked at all three interfaces) was obtained, corresponding very closely to the experimental distribution.
This good correlation supports the conclusion that the trimer sub-band b (Fig. 2A, lane labeled 1:6) contains two BMH crosslinks and thus corresponds to open trimers that have not been covalently closed at all three interfaces. Furthermore, band b observed in the 1:2 and 5:1 BMH:thiol lanes of Fig. 2A comigrates exactly with the trimers obtained in the 1:6 crosslinking reaction. Thus we propose that sub-band b, like the single trimer species observed with the 1:6 crosslinking reaction, corresponds to an open trimer and that sub-band a, (the dominant product of the crosslinking reaction at BMH/thiol ratios of 1:2 or greater), corresponds to a closed trimer that has been covalently sealed by three BMH crosslinkers.
Although covalent ring closure does occur in the presence of excess BMH, the existence of an unclosed trimer fraction (sub-band b), which remains in the reactions performed at higher BMH/thiol ratios, shows that the crosslinking of the third trimer interface is somewhat disfavored. This finding may reflect a reduced accessibility of the cysteines to BMH at the last interface or result because formation of the first two crosslinks may separate the remaining target thiol and maleimide group to a distance that does not permit crosslinking. This latter possibility is consistent with a recent study that suggests that in solution (in contrast to the situation in the crystal) the dominant gp45 species may be the open trimer (21). In this situation the unreacted maleimide of a covalently attached BMH mono-adduct might crosslink with a nearby amine group rather than the target thiol, although the rate of reaction with amines at the pH of the experiment is roughly 1,000 times slower than the reaction rate with thiols (data not shown).
Crosslinking with DPDPB.
We also have sealed the gp45 trimer ring with DPDPB, which is a reversible homobifunctional 20-Å crosslinker capped by pyridyl disulfide groups. This permits us to use DTT to reverse covalent ring closure and return the clamp to a state with functionally normal interfaces. Gp45-K82C;S166C monomers were efficiently converted to crosslinked trimers on reaction with DPDPB, thereby creating gp45-DPDPB. At 1:2 and 20:1 ratios of DPDPB/thiol, gp45-DPDPB is also fully trimeric, moving as a single electrophoretic species on SDS/PAGE gels with a mobility midway between those of sub-bands a and b seen with gp45-BMH (we note that DPDPB contains only thiol crosslinkers, and thus cannot interact with amines). The incubation of gp45-DPDPB with 30 mM DTT for 10 min at 37°C quantitatively converts these crosslinked trimers back to monomers (data not shown), whereas gp45-BMH is unaffected by DTT.
Effects of Crosslinked Clamps on the ATPase Activity of the Clamp Loader.
To determine the functional effects of crosslinking the gp45 monomer-monomer interfaces in gp45-BMH and gp45-DPDPB, we performed a series of ATPase assays designed to measure the rates of ATP hydrolysis by gp44/62 at different steps in the clamp-loading pathway. The basal rate of ATP hydrolysis by gp44/62 is too low to be measured accurately. However, the addition of either gp45 or p-t DNA (14) dramatically stimulates ATP hydrolysis, as documented for the double-mutant gp45 clamps in Table 1 and Fig. 3. We note that the addition of p-t DNA to a reaction containing gp44/62 and the double-mutant gp45 increases the ATPase rate 30-fold, reflecting the loading of the gp45 clamp onto the DNA. Streptavidin (SA) can be bound to the biotinylated template DNA of the p-t-flap 30/50/32-mer DNA construct (22) and prevents the loaded gp45 clamp from sliding off over the 5′ end of the template (the clamp is blocked at the 3′ end by the partially annealed 32-mer flap DNA). This blockage results in free gp45 being returned to solution at a much slower rate, with this step becoming at least partially rate-limiting in the ATPase cycle under these conditions (19). Table 1 shows that the ATPase rate decreases by 10–20% on adding SA to a reaction containing gp44/62, gp45, and the p-t-flap DNA construct.
Figure 3.
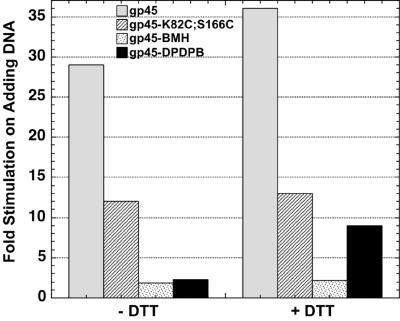
Pretreatment of gp45-DPDPB with DTT dramatically enhances the DNA-dependent stimulation of the gp44/62 ATPase activity. The values of fold stimulation on adding DNA were calculated by dividing the value for the ATPase rate in the presence of DNA by the value obtained in the absence of this cofactor (data from Table 1).
The addition of the gp43 exo− polymerase to the above reaction results in the assembly of the gp43-gp45 polymerase holoenzyme on the DNA construct (23), with the loaded gp45 clamp binding to the gp43 polymerase to create a tightly associated DNA-bound complex characterized by a dissociation time of minutes (24), meaning that the gp45 clamp is trapped within holoenzyme for a time that is long compared with the rate of ATP hydrolysis. Thus these polymerase-bound clamps are not available to stimulate the gp44/62 ATPase in solution. We find that the ATPase rate is reduced ≈10-fold on adding gp43 exo− polymerase to the double-mutant gp45 protein in both the absence and the presence of DTT (Table 1). In addition, we note that the rate of ATP hydrolysis in the presence of gp45-K82C;S166C and gp44/62 is nearly double that measured with wt gp45. However, after adding p-t DNA the rate of ATP hydrolysis with the gp45-K82C;S166C mutant is only 75% of that observed with wt gp45 (Table 1). As a consequence the DNA-dependent stimulation of the gp44/62 ATPase rate by gp45-K82C;S166C is ≈2.5-fold less than that achieved by adding DNA to wt gp45 (Fig. 3).
In the absence of p-t DNA, the ATPase-stimulatory effect of gp45-BMH is greater than that of wt gp45 and gp45-K82C;S166C. In fact gp45-BMH hyperstimulates gp44/62 relative to gp45-K82C;S166C, inducing ATP hydrolysis at a rate that is 1.8-fold higher than that seen with gp45-K82C;S166C and 3.2-fold higher than that with wt gp45. This finding confirms that the ATP cycle of the clamp loader (ATP binding and hydrolysis) is related to ring opening; the mechanistic significance of this hyperstimulation is considered further in Discussion. However, unlike the major further stimulation of the ATPase activity seen with both wt gp45 and gp45-K82C;S166C on adding p-t DNA, the ATPase of the complexes containing the fully crosslinked rings is only modestly further enhanced by this cofactor. Furthermore, interaction with gp43 exo− polymerase decreases the ATPase rate only slightly, returning the rate to the level measured with gp45-BMH and gp44/62 alone. These results suggest that fully crosslinked gp45 clamps can neither be loaded onto DNA nor serve to tether the DNA polymerase.
The simplest explanation for this set of results is that gp45 clamps must open to encircle the DNA and that the formation of covalent crosslinks across all three monomer-monomer interfaces prohibits such opening. In support of this conclusion we have demonstrated (data not shown) that gp45-BMH, crosslinked under conditions that create a significant population of monomers, dimers, and open trimers (but few if any closed trimers), stimulates the ATPase activity of gp44/62 to levels comparable to those seen with wt gp45, both with and without added p-t DNA (P.P., unpublished results). Thus, tethering gp45 subunits at one or two interfaces of the trimer ring does not inhibit clamp loading, whereas gp45-BMH that has been crosslinked exclusively to trimers is unable to stimulate the gp44/62 ATPase to a level characteristic of clamp assembly on DNA. We conclude that DNA-dependent ATPase stimulation, and (by extension) clamp loading onto DNA, requires only one free (i.e., untethered) monomer-monomer interface.
Consistent with this conclusion, we also have shown that the ATPase-stimulating properties of gp45-DPDPB are similar to those of gp45-BMH (Table 1), with hyperstimulation of ATPase activity again being observed in the absence of the DNA cofactor. Adding DNA induces a marginal (2-fold) further rate increase (Fig. 3). Thus both gp45-BMH and gp45-DPDPB are stimulated by DNA to only one-sixth the level reached with the noncrosslinked gp45-K82C;S166C (Fig. 3). As expected, preincubation of gp45-DPDPB with DTT restores the DNA-dependent stimulation of the gp44/62 ATPase to the level of the uncrosslinked gp45-K82C;S166C clamp (Table 1 and Fig. 3), whereas preincubating wt gp45, gp45-K82C;S166C, or gp45-BMH with DTT had no effect.
Effects of Crosslinked Clamps on Strand-Displacement DNA Synthesis.
The strand-displacement synthesis assay was designed to discriminate between polymerase molecules that are tethered to the DNA template by a sliding clamp and those that are untethered. Binding the clamp to the polymerase forms a highly processive holoenzyme that can displace a partially annealed DNA oligomer (the flap DNA) from the DNA template during DNA synthesis (25). An unassisted polymerase is poorly processive without the benefit of a sliding clamp and dissociates before the flap DNA can be fully displaced. The fraction of gp43 exo− polymerase that is blocked by the flap DNA in the absence of a clamp corresponds directly to the fraction of primer and flap DNA that is annealed to the template strand (data not shown).
In the absence of DTT the strand-displacement activity of gp45-K82C;S166C is slightly lower than, but still very comparable to, that of wt gp45 (Table 2). However, both gp45-BMH and gp45-DPDPB are much less effective in this assay, with activity levels corresponding to only one-sixth those seen with gp45-K82C;S166C. This finding shows that fully crosslinked clamps assemble onto the polymerase holoenzyme complexes very inefficiently; i.e., only to an extent comparable to their stimulation of the DNA-dependent ATPase assay (Table 1). Pretreatment of gp45-DPDPB with DTT restores strand-displacement activity to the level seen with gp45-K82C;S166C, again suggesting that the covalent crosslinks at the interfaces of the gp45-DPDPB trimers can be completely cleaved by DTT and that removing the DPDPB tethers restores ring opening and thus clamp loading onto DNA.
Table 2.
Functional activity of gp45 clamps in the polymerase holoenzyme strand displacement assay
| Sliding clamp | +DTT | % Activity | +DTT | % Activity |
|---|---|---|---|---|
| gp45 | 218 | 100 | 218 | 100 |
| gp45-K82C;S166C | 203 | 93 | 195 ± 7.8 | 89 |
| gp45-DPDPB | 35 ± 3 | 16 | 196 ± 5.3 | 90 |
| gp45-BMH | 36 | 17 | 42 ± 0.7 | 19 |
Values are presented in active wt gp45 concentration units (nM), with the dynamic range and linearity of the scale determined by calibration with a standard curve of 0, 55, 109, and 218 nM wt gp45. The relative active concentrations of the gp45 variants then were determined after back-extrapolation from the amount of strand displacement with each modified gp45. %Activity was calculated relative to 218 nM wt gp45, which was defined as 100% active. This parameter can be calculated in this way because the assay is linear over the entire (0–218 nM) gp45 concentration range examined. Data listed as +DTT were obtained from reactions in which all of the clamp-loader proteins, rather than just gp45-DPDPB, were incubated with DTT.
Discussion
Like gp45, the E. coli β-clamp and the eukaryotic PCNA trimer crystallize as protein rings, and thus it is not surprising that these three molecular clamps are structurally and functionally homologous. Yet, because the x-ray structures of all three clamps show that they crystallize primarily as closed protein rings in the absence of DNA (I. Moarefi and J. Kuriyan, personal communication; refs. 26 and 27), it is also unclear how the homologous clamp-loading machinery manipulates these clamps to achieve concatenation onto the DNA, though again it generally has been assumed that the process involves ring opening at one or more monomer-monomer interfaces.
To study this process several laboratories have attempted to modify the subunit interfaces within the clamp to destabilize the rings, but until recently (21, 28) no effort had been made to determine the consequences of sealing the clamps to form covalently closed rings. We have created such a construct for gp45 by crosslinking the subunit interfaces with both irreversible and reversible thiol-specific crosslinkers to create a trimer ring that is incapable of opening or dissociating into subunits. An unexpected finding with these crosslinked gp45 clamps is that they do not inhibit the DNA-independent interactions with the clamp loader that stimulate ATP hydrolysis. In fact these covalently sealed rings actually hyperstimulate ATP hydrolysis appreciably in the absence of DNA, relative to the level achieved with uncrosslinked gp45. However, the closed rings do not further stimulate the ATPase activity of the clamp-loader complex when the DNA cofactor is bound, nor do they increase the processivity of the T4 DNA polymerase in strand-displacement synthesis reactions.
In related studies O’Donnell and colleagues (28) recently have characterized a double mutant of the β-clamp of E. coli (β-R103C-I305C) that also can be sealed under oxidative conditions to create a disulfide linkage across the dimer interface. The β-dimer was not efficiently sealed at both interfaces and a mixture of uncrosslinked, singly crosslinked, and presumably doubly crosslinked clamps were obtained. Those studies showed that a single disulfide crosslink (creating an open β-dimer) does not abolish clamp loading. In addition a fraction of crosslinked β-dimers was identified that appeared to be closed at both interfaces and could not be loaded onto DNA. Consistent with those conclusions, our results show that fully crosslinked T4 clamps also cannot be loaded onto DNA, whereas partially crosslinked clamps (crosslinked to the dimer or open trimer level) can be loaded and are fully functional. Thus closed T4 rings can interact with the clamp-loader complex to stimulate (indeed, to hyperstimulate) the ATPase activity of the gp44/62 complex, but cannot be loaded onto the DNA. However these results do not tell us where the ring-opening process occurs in the clamp-loading process or the associated ATP binding and hydrolysis cycle; this information is derived from kinetic studies that will be described elsewhere (P.P., M. C. Young, G.J.L., and P.H.v.H., unpublished work).
The opening of the ring has been proposed to occur as a stepwise process in both the E. coli and the T4 systems, with the clamp loader binding the clamp and ATP, then opening the clamp, and finally binding to p-t DNA, which triggers a process that results in closure of the clamp around the DNA and dissociation of the clamp-loader complex (13, 29, 30). Clamp opening in E. coli now has been convincingly demonstrated to occur before DNA binding and without ATP hydrolysis (28, 31). A similar mechanism had been suggested for T4, except that ATP hydrolysis was proposed to precede clamp opening (13, 30). Recent kinetic studies in our laboratory now suggest, in accord with findings with the E. coli system, that the binding of the open clamp also precedes ATP hydrolysis in the T4 system.
A model that fits these results and is also analogous to the E. coli clamp-loading pathway suggests that the T4 clamp binds to the gp44/62 clamp loader as a closed ring, which, in the presence of bound ATP, drives a conformational change of the resulting complex that opens the ring. Alternatively, if significant populations of open T4 rings exist in solution as suggested by Alley et al. (21), the ring actually may be bound by the clamp loader as an initially open trimeric species. In either case conformational rearrangements are induced in the gp44/62-gp45-ATP complex that result in significant changes in protein–protein contacts, as shown by crosslinking studies (16). These conformational rearrangements are transient, and eventually ATP hydrolysis will occur, presumably reversing these changes within the complex and releasing the clamp back into solution. In this scenario a covalently closed clamp bound to the clamp loader could stimulate a futile ATPase cycle, because the clamp cannot be opened and thus a proper poised complex carrying an open clamp cannot be formed. This fact could explain the hyperstimulation of the gp44/62 ATPase that is induced by the binding of the permanently closed clamp, inasmuch as the sealed clamp may resemble the dissociation product of the accessory proteins complex and induce accelerated hydrolysis. In contrast, when p-t DNA binds to a poised complex carrying an open clamp the ring is efficiently loaded onto the DNA, aided perhaps by favorable charge-charge interactions between the DNA and the inside of the gp45 ring (I. Moarefi and J. Kuriyan, personal communication). This interaction then could drive the closing of the clamp and the ATP-hydrolysis-dependent release of the clamp loader back into solution.
This model has the virtue of explaining our experimental observations with these covalently closed rings and also suggests that the T4 clamp-loading mechanism may be quite similar to that proposed for the E. coli system by the O’Donnell group (28, 29, 31). Presteady-state kinetic experiments that support and document this model will be presented elsewhere (P.P., M. C. Young, G.J.L., and P.H.v.H., unpublished work).
Acknowledgments
We are indebted to Dr. Nancy Nossal for the gift of the gp43 exo− overexpression plasmid and to Dr. John Kuriyan for providing us with the Protein Databank coordinate file for the gp45 sliding clamp structure before publication. This research was supported in part by U.S. Public Health Service Research Grants GM-15792 and GM-29158 (to P.H.v.H.) and by American Cancer Society Postdoctoral Fellowship PF-4303 (to G.J.L.). P.H.v.H. is an American Cancer Society Research Professor of Chemistry.
Abbreviations
- wt
wild type
- p-t DNA
primer-template DNA
- BMH
bismaleimidohexane
- DPDPB
1,4-di-[3′-(2′-pyridyldithio)propionamido]butane
Footnotes
In the earlier literature, the gp45-K82C mutant was designated gp45-K81C. We here introduce a correction in the naming of this mutant, based on findings from our laboratory (32) and others (33) that demonstrate that the original published DNA and protein sequences of gp45 (9) contained errors at three of the 228 DNA codons or amino acid residues. One of the errors corresponds to the omission of an Asp residue at position 70, which, when corrected, requires that all subsequent residues be shifted one position forward. Thus, the mutant that previously was designated gp45-K81C now becomes gp45-K82C, and so forth.
References
- 1.Kuriyan J, O’Donnell M. J Mol Biol. 1993;234:915–925. doi: 10.1006/jmbi.1993.1644. [DOI] [PubMed] [Google Scholar]
- 2.Kelman Z. Oncogene. 1997;14:629–640. doi: 10.1038/sj.onc.1200886. [DOI] [PubMed] [Google Scholar]
- 3.Jonsson Z O, Hindges R, Hubscher U. EMBO J. 1998;17:2412–2425. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newport J W, Kowalczykowski S C, Lonberg N, Paul L S, von Hippel P H. In: Mechanistic Studies of DNA Replication and Genetic Recombination: ICN-UCLA Symposia on Molecular and Cellular Biology. Alberts B, editor. XIX. New York: Academic; 1981. pp. 485–505. [Google Scholar]
- 5.Nossal N G, Alberts B M. In: The Mechanism of DNA Replication Catalyzed by Purified Bacteriophage T4 DNA Replication Proteins. Mathews C K, Kutter E M, Mosig G, Berget P B, editors. Washington, DC: Am. Soc. Microbiol.; 1983. pp. 71–81. [Google Scholar]
- 6.Jarvis T C, Paul L S, von Hippel P H. J Biol Chem. 1989;264:12709–12716. [PubMed] [Google Scholar]
- 7.Gogol E P, Young M C, Kubasek W L, Jarvis T C, von Hippel P H. J Mol Biol. 1992;224:395–412. doi: 10.1016/0022-2836(92)91003-8. [DOI] [PubMed] [Google Scholar]
- 8.Tinker R L, Kassavetis G A, Geiduschek E P. EMBO J. 1994;13:5330–5337. doi: 10.1002/j.1460-2075.1994.tb06867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spicer E K, Noble J A, Nossal N G, Konigsberg W A, Williams K R. J Biol Chem. 1982;257:8972–8979. [PubMed] [Google Scholar]
- 10.Nossal N G. FASEB J. 1992;6:871–878. doi: 10.1096/fasebj.6.3.1310946. [DOI] [PubMed] [Google Scholar]
- 11.Sexton D J, Carver T E, Berdis A J, Benkovic S J. J Biol Chem. 1996;271:28045–28051. doi: 10.1074/jbc.271.45.28045. [DOI] [PubMed] [Google Scholar]
- 12.Latham G J, Bacheller D J, Pietroni P, von Hippel P H. J Biol Chem. 1997;272:31677–31684. doi: 10.1074/jbc.272.50.31677. [DOI] [PubMed] [Google Scholar]
- 13.Latham G J, Pietroni P, Dong F, Young M C, von Hippel P H. J Mol Biol. 1996;264:426–439. doi: 10.1006/jmbi.1996.0651. [DOI] [PubMed] [Google Scholar]
- 14.Jarvis T C, Paul L S, Hockensmith J W, von Hippel P H. J Biol Chem. 1989;264:12717–12729. [PubMed] [Google Scholar]
- 15.Young M, Weitzel S E, von Hippel P H. J Mol Biol. 1996;264:440–452. doi: 10.1006/jmbi.1996.0652. [DOI] [PubMed] [Google Scholar]
- 16.Pietroni P, Young M C, Latham G J, von Hippel P H. J Biol Chem. 1997;272:31666–31676. doi: 10.1074/jbc.272.50.31666. [DOI] [PubMed] [Google Scholar]
- 17.Latham G J, Bacheller D J, Pietroni P, von Hippel P H. J Biol Chem. 1997;272:31685–31692. doi: 10.1074/jbc.272.50.31685. [DOI] [PubMed] [Google Scholar]
- 18.Sexton D J, Berdis A J, Benkovic S J. Curr Opin Chem Biol. 1997;1:316–322. doi: 10.1016/s1367-5931(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 19.Berdis A J, Benkovic S J. Biochemistry. 1996;35:9253–9265. doi: 10.1021/bi952569w. [DOI] [PubMed] [Google Scholar]
- 20.Frey M W, Nossal N G, Capson T L, Benkovic S J. Proc Natl Acad Sci USA. 1993;90:2579–2583. doi: 10.1073/pnas.90.7.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alley S C, Shier V K, Abel-Santos E, Sexton D J, Soumillion P, Benkovic S J. Biochemistry. 1999;38:7696–7709. doi: 10.1021/bi9827971. [DOI] [PubMed] [Google Scholar]
- 22.Kaboord B F, Benkovic S J. Proc Natl Acad Sci USA. 1993;90:10881–10885. doi: 10.1073/pnas.90.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaboord B F, Benkovic S J. Curr Biol. 1995;5:149–157. doi: 10.1016/s0960-9822(95)00036-4. [DOI] [PubMed] [Google Scholar]
- 24.Hacker K J, Alberts B M. J Biol Chem. 1994;269:24209–24220. [PubMed] [Google Scholar]
- 25.Reddy M K, Weitzel S E, von Hippel P H. Proc Natl Acad Sci USA. 1993;90:3211–3215. doi: 10.1073/pnas.90.8.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishna T S, Kong X-P, Gary S, Burgers P M, Kuriyan J. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 27.Kong X-P, Onrust R, O’Donnell M, Kuriyan J. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 28.Turner J, Hingorani M M, Kelman Z, O’Donnell M. EMBO J. 1999;18:771–783. doi: 10.1093/emboj/18.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertram J G, Bloom L B, Turner J, O’Donnell M, Beechem J M, Goodman M F. J Biol Chem. 1998;273:24564–24574. doi: 10.1074/jbc.273.38.24564. [DOI] [PubMed] [Google Scholar]
- 30.Sexton D J, Kaboord B F, Berdis A J, Carver T E, Benkovic S J. Biochemistry. 1998;37:7749–7756. doi: 10.1021/bi980088h. [DOI] [PubMed] [Google Scholar]
- 31.Hingorani M M, O’Donnell M. J Biol Chem. 1998;273:24550–24563. doi: 10.1074/jbc.273.38.24550. [DOI] [PubMed] [Google Scholar]
- 32.Latham G J. Res Commun Biochem Cell Mol Biol. 1998;2:139–147. [Google Scholar]
- 33.Yeh L S, Hsu T, Karam J D. J Bacteriol. 1998;180:2005–2013. doi: 10.1128/jb.180.8.2005-2013.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]