Abstract
Carbon catabolite repression (CCR) of several Bacillus subtilis catabolic genes is mediated by ATP-dependent phosphorylation of histidine-containing protein (HPr), a phosphocarrier protein of the phosphoenolpyruvate (PEP): sugar phosphotransferase system. In this study, we report the discovery of a new B. subtilis gene encoding a HPr-like protein, Crh (for catabolite repression HPr), composed of 85 amino acids. Crh exhibits 45% sequence identity with HPr, but the active site His-15 of HPr is replaced with a glutamine in Crh. Crh is therefore not phosphorylated by PEP and enzyme I, but is phosphorylated by ATP and the HPr kinase in the presence of fructose-1,6-bisphosphate. We determined Ser-46 as the site of phosphorylation in Crh by carrying out mass spectrometry with peptides obtained by tryptic digestion or CNBr cleavage. In a B. subtilis ptsH1 mutant strain, synthesis of β-xylosidase, inositol dehydrogenase, and levanase was only partially relieved from CCR. Additional disruption of the crh gene caused almost complete relief from CCR. In a ptsH1 crh1 mutant, producing HPr and Crh in which Ser-46 is replaced with a nonphosphorylatable alanyl residue, expression of β-xylosidase was also completely relieved from glucose repression. These results suggest that CCR of certain catabolic operons requires, in addition to CcpA, ATP-dependent phosphorylation of Crh, and HPr at Ser-46.
Keywords: protein phosphorylation, Gram-positive bacteria
The bacterial phosphoenolpyruvate (PEP): sugar phosphotransferase system (PTS) catalyzes the transport and concomitant phosphorylation of carbohydrates via a protein phosphorylation chain including PEP-dependent phosphorylation of His-15 in histidine-containing protein (HPr) by enzyme I (EI). P-His-HPr phosphorylates the sugar-specific EIIAs. In Gram-positive bacteria, the PTS regulates also induction and carbon catabolite repression (CCR) of numerous catabolic genes (1). The central regulatory protein involved in these various functions is HPr. In Gram-positive bacteria, this small phosphoryl transfer protein can be phosphorylated at a regulatory serine (Ser-46) by ATP and the HPr kinase (2, 3), in addition to phosphorylation at the catalytic His-15 by PEP and EI (4, 5). PEP-dependent and ATP-dependent phosphorylation of HPr interfere with each other—i.e., P-His-HPr is a poor substrate for the HPr kinase and P-Ser-HPr is a poor substrate for EI (6, 7). ATP-dependent phosphorylation of HPr is stimulated by glycolytic intermediates such as fructose-1,6-bisphosphate (FBP) in Enterococcus faecalis (6) and in Streptococcus pyogenes (7). It has been reported that FBP is also implicated in CCR of the Bacillus subtilis gnt and iol operons (8, 9), and a potential role of phosphorylation of HPr at Ser-46 in CCR has therefore been investigated (10). The gnt operon contains the genes gntRKPZ encoding the repressor GntR, gluconate kinase, gluconate permease, and a gluconate-6-P-dehydrogenase (11), whereas the iol operon is composed of 10 genes encoding enzymes presumably implicated in inositol metabolism, including iolG encoding inositol dehydrogenase (12). In a B. subtilis strain carrying the ptsH1 mutation (Ser-46 of HPr is replaced with a nonphosphorylatable alanine), several catabolic genes were completely or partially relieved from CCR. These catabolic genes were also relieved from CCR in a ccpA mutant strain, defective in the key regulator of CCR in B. subtilis (13). The trans-acting protein CcpA, a member of the LacI–GalR family of repressors (14), mediates CCR by binding to the cis-active operator sequence cre (catabolite responsive element) (15). An interaction of P-Ser-HPr with CcpA has been demonstrated in vitro, and this interaction is strengthened by FBP (16). Fujita et al. (17) have shown that the protein complex formed between CcpA and P-Ser-HPr interacts specifically with the gnt cre.
In this communication, we report the discovery of a novel B. subtilis gene encoding the HPr-like protein Crh (for catabolite repression HPr). Because only the ATP-dependent but not the PEP-dependent site of phosphorylation is conserved in Crh, this HPr-like protein appears to carry out exclusively regulatory functions.
MATERIALS AND METHODS
Bacterial Strains, Growth Conditions, and Transformation Procedures.
The B. subtilis strains used for enzyme assays are listed in Table 1. Most of them were constructed by transforming QB5081 containing a translational levD′–′lacZ fusion (18) or by using DNA from QB5081 to transfer the translational levD′–′lacZ fusion into a ccpA mutant (QB7115). B. subtilis strains were grown in CSK medium (19) in the presence of 0.1% xylose, 0.5% inositol, or 0.2% fructose to induce synthesis of β-xylosidase, inositol dehydrogenase, or expression of the translational levD′–′lacZ fusion, respectively. To study the repressive effect of carbon sources, 1% glucose, fructose, or glycerol was added. Standard procedures were used to transform Escherichia coli and B. subtilis cells (20, 21).
Table 1.
Genotype of strains used in the enzyme assays
| Strain | Genotype* |
|---|---|
| QB5081 | trpC2 amyE::(pΔB levD′–′lacZ cat) |
| QB5224 | trpC2 ptsH1 amyE::(PΔB levD′–′lacZ cat) |
| QB7098 | trpC2 crh::aphA3 amyE::(PΔB levD′–′lacZ cat) |
| QB7103 | trpC2 ptsH1 crh::aphA3 amyE::(PΔB levD′–′lacZ cat) |
| QB7108 | trpC2 ptsH1 crh::aphA3 amyE::(pspac crh cat) |
| QB7112 | trpC2 ptsH1 crh::aphA3 amyE::(pspac crh1 cat) |
| QB7115 | trpC2 ccpA::spc amyE::(PΔB levD′–′lacZ cat) |
cat, pC194 chloramphenicol acetyltransferase gene; spc, spectinomycine resistance gene from Staphylococcus aureus; aphA3, Enterococcus faecalis kanamycin resistance gene (25).
Northern Blot Analysis.
Total RNA from B. subtilis 168 was prepared using the RNeasy Midi kit (Quiagen, Chatsworth, CA). RNA was fractionated on formaldehyde-agarose gels (1% agarose/20 mM Mops, pH 7.0/5 mM sodium acetate/1 mM EDTA/1.9% formaldehyde) and transferred to nylon membranes (Hybond-N; Amersham). DNA probes were prepared from specific PCR products, 32P-labeled by random priming (Prime-a-Gene labeling system; Promega) with [α-32P]dATP. Prehybridization (2 h) and hybridization (at least 12 h) were performed at 63°C. After washing, membranes were exposed to a Phosphor screen and analyzed on a Storm 860 (Molecular Dynamics).
Plasmid Constructions.
A 285-bp fragment containing the crh gene was amplified by PCR using chromosomal DNA of the ΔptsH strain GM273 (10) and two specific primers containing a NdeI site or PstI site, respectively. The NdeI–PstI fragment was cloned into the expression vector pT7–5 (6× His) (22). A modified Crh, carrying a polyhistidine fused to the C terminus, was expressed from the resulting plasmid pAG1 after transformation in the E. coli strain BL21(DE3) (Novagen) producing T7 RNA polymerase. Plasmids pAG2 and pAG3, used for overproducing HPr and EI with a polyhistidine sequence at the N terminus, are derivatives of pQE30 (Quiagen). The ptsH and ptsI genes from B. subtilis were obtained by PCR using appropriate synthetic oligonucleotides and plasmid pTS20 as template (23). pAG2 and pAG3 were obtained by cloning a BamHI–HindIII fragment containing the ptsH gene or a BamHI–SalI fragment containing the ptsI gene into pQE30.
By using appropriate oligonucleotides, a 1-kb DNA fragment containing the crh gene with an EcoRI site at the 5′ end and a BamHI site at the 3′ end was amplified by PCR and cloned into the integrative plasmid pJH101 (24) to give plasmid pRC12. A 1.5-kb SmaI–StuI DNA fragment containing the kanamycin resistance gene aphA3 (25) was inserted at the unique AgeI restriction site of pRC12 made blunt by using the Klenow fragment of DNA polymerase I. The resulting plasmid pRC14 was linearized with PstI and used to transform B. subtilis 168 or QB5081 leading to the disruption of the chromosomal copy of the crh gene in these strains by integration of the kanamycin resistance cassette. The BamHI–EcoRI fragment of pRC12 was cloned into M13 mp19 to give M13 mp19crh. The Muta-Gene M13 in vitro mutagenesis kit (Bio-Rad) was used to obtain the crh1 (crhS46A) allele. A 434-bp PvuII–BstBI DNA fragment containing the promoterless crh gene or crh1 allele was inserted in pDR67 (26) cut with SmaI and ClaI to give plasmid pRC16 or pRC17, respectively. Plasmid pDR67 carries the pC194 chloramphenicol acetyltransferase gene cat and a spac promoter between two fragments of the B. subtilis amyE gene. The crh and crh1 alleles were integrated downstream of the spac promoter allowing induction of their expression with 0.5 mM isopropyl β-d-thiogalactoside after integration in monocopy at the amyE locus of the B. subtilis genome.
Protein Purifications.
E. coli BL21(DE3) was transformed with plasmid pAG1, whereas E. coli M15 containing the pREP4 plasmid (Quiagen) was transformed with plasmids pAG2 or pAG3. Transformants were grown at 37°C in 2× TY medium until the culture reached an OD595 of 0.7. Expression of the genes incorporated in plasmids pAG1, pAG2, or pAG3 was induced by adding isopropyl β-d-thiogalactoside (TEBU; 5 Prime → 3 Prime, Inc.) to a final concentration of 1 mM. Incubation was continued for a further 4 h. Protein purification was carried out as described by Cortay et al. (22). Fractions containing Crh(His)6, HPr(His)6, or EI(His)6 were pooled and desalted on a Sephadex G25 column (Pharmacia) run with 20 mM ammonium bicarbonate. Protein concentration was determined spectrophotometrically (Bio-Rad protein assay). Protein solutions were stored at −80°C.
The HPr kinase was partially purified from the B. subtilis strain GM273 (10). Cells were grown overnight at 37°C in 100 ml Luria–Bertani medium. Bacteria were harvested by low-speed centrifugation, resuspended in 1 ml of a buffer containing 100 mM Tris⋅HCl (pH 7.4), 5 mM MgCl2, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1 mg/ml lysozyme, and 200 units/ml of benzonase (Merck) and disrupted by sonication. After centrifugation, the crude extract was applied onto a DEAE-5PW Protein PAK glass column (0.8 × 7.5 cm; Waters) equilibrated with 50 mM Tris⋅HCl (pH 7.4). Proteins were eluted with a linear gradient of 0–500 mM NaCl.
Protein Phosphorylation Assay.
Phosphorylation of Crh(His)6 or HPr(His)6 by PEP and EI(His)6 was carried out in a total volume of 15 μl. A total of 1.5 μg of Crh(His)6 or HPr(His)6 was incubated for 10 min at 37°C with 2 μg of EI(His)6 in 50 mM Tris⋅HCl (pH 7.4), 15 mM MgCl2, and 1 μM [32P]PEP (0.5 μCi; 1 Ci = 37 GBq). [32P]PEP was prepared from [γ-32P]ATP as described by Roossien et al. (27). Phosphorylation of Crh(His)6 or HPr(His)6 by the HPr kinase was carried out using various FBP concentrations. A typical assay contained, in a total volume of 20 μl, 1.5 μg of Crh(His)6 or HPr(His)6 which was incubated for 10 min at 37°C with 5 to 16 μl of partially purified HPr kinase in 20 mM Tris⋅HCl (pH 7.4), 10 mM MgCl2, 1 mM DTT, 20 mM FBP, and 50 μM [γ-32P]ATP (4 μCi). The phosphorylation reaction was stopped by adding an equal volume of sample buffer (28) to the assay mixtures before applying them onto a SDS/PAGE. In the case of ATP-dependent phosphorylation, gels were treated after electrophoresis for 5 min with boiling 16% trichloroacetic acid before they were dried and exposed to autoradiography (Biomax MR; Kodak).
Determination of the Phosphorylation Site in Crh.
Crh(His)6 (500 μg) was phosphorylated as described above in a total volume of 1 ml and subsequently purified on a Ni-NTA column. A total of 100 μg of phosphorylated or unphosphorylated protein was lyophilized and resuspended in 100 μl of 70% formic acid containing CNBr (20 μg/ml). The samples were incubated in the dark for 20 h at room temperature, lyophilized, and dissolved in 100 μl 20 mM ammonium bicarbonate. Tryptic fragments were obtained by incubating 100 μg of unphosphorylated or phosphorylated protein for 7 h at 37°C in 100 μl of 20 mM Tris⋅HCl (pH 8) containing 20 μg of trypsin. Matrix-assisted laser desorption ionization mass spectra (MALDI-MS) of peptides derived from phosphorylated or unphosphorylated Crh(His)6 were recorded on a RETOF (time of flight) instrument from Perseptive Biosystems (Framingham, MA) as described by Charrier et al. (29).
Enzyme Assays.
β-Galactosidase activities were measured after growth of B. subtilis strains in CSK medium using the method of Miller (30). One unit of β-galactosidase activity is defined as the amount of enzyme which produces 1 nmol of o-nitrophenol per min at 28°C. β-Xylosidase activities were determined as previously described using p-nitrophenyl β-d-xylopyranoside as substrate (31). Inositol dehydrogenase activities were determined as described (9) by measuring the formation of NADH from NAD plus inositol at 37°C.
RESULTS
The crh Gene of B. subtilis Encodes a HPr-Like Protein.
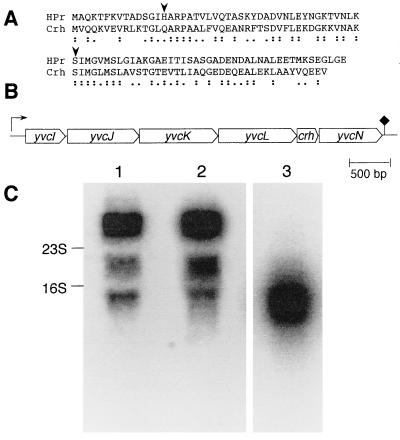
Within the B. subtilis genome sequencing program, a gene present on a 115-kb DNA fragment located between cysB (292°) and hisA (298°) on the B. subtilis genome (32) and encoding a protein composed of 85 amino acids has been identified. This protein exhibits 45% sequence identity and 67% similarity to HPr (Fig. 1A). However, the active site His-15 of HPr was replaced with a glutamine, whereas the ATP-dependent site of phosphorylation of HPr, which has been demonstrated to be implicated in CCR, was conserved. The protein encoded by the above gene was therefore called Crh (for catabolite repression HPr). The B. subtilis crh gene appears to be part of a putative operon composed of six ORFs (Fig. 1B). The function of the proteins encoded by these ORFs has not been identified. The expression of the crh gene was studied in vivo by carrying out Northern blot analysis (Fig. 1C). Annealing with two probes, corresponding to the putative ORFs crh (lane 1) and yvcJ (lane 2), respectively, allowed the detection of a transcript consisting of 4,500 nt, which corresponds to the length of the putative operon containing crh and the five additional ORFs. This result indicates that crh is expressed in vivo in B. subtilis.
Figure 1.
(A) Alignment of the amino acid sequences of HPr and Crh from B. subtilis. The Crh amino acid sequence was derived from the DNA sequence of the crh gene (Gene Bank accession number Z94043). Identical residues are indicated by colons and conservative exchanges by single dots. The phosphorylatable His-15 and Ser-46 in HPr are indicated by arrows. (B) Organization of the operon containing crh and five additional ORFs. The direction of translation is indicated by thick arrows, the putative promoter is indicated by an arrow, and the presumed transcription terminator is indicated by a rhomb. (C) Northern blot analysis. Two PCR products corresponding to the whole crh (258 bp) (lane 1) or yvcJ (417 bp) (lane 2) were 32P labeled and used as probes for Northern blot analysis. The presented autoradiogram was obtained after 10 days exposure to the hybridized membrane. With both probes, a 4.5-kb transcript was detected that is in agreement with the length of the putative operon containing crh. The two other bands correspond to the commonly observed compression artifacts triggered by 16S and 23S rRNAs. The positions of the 16S and 23S rRNAs are indicated by arrows (1,541 and 2,904 bases, respectively). In a control experiment (lane 3), the membrane was hybridized with a probe specific for yvcE, a 1,422-bp-long ORF located about 2,000 bp upstream of the crh-containing operon. The result of a 48-h exposure is shown.
In Vitro Phosphorylation.
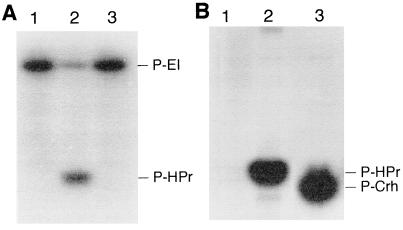
To analyze its biochemical properties and its potential phosphorylation, large amounts of Crh were produced after cloning the crh gene into the expression vector pT7–5 (6× His) (22), allowing the fusion of a polyhistidine sequence to the C terminus of Crh. After overproduction and purification of Crh(His)6, we tested whether it can be phosphorylated by PEP and ATP, similar to its homologue, the phosphocarrier protein HPr. In contrast to HPr(His)6, Crh(His)6, lacking His-15, was not phosphorylated by [32P]PEP in the presence of purified EI(His)6 (Fig. 2A, lane 3), but both proteins were phosphorylated by [γ-32P]ATP using a crude extract of B. subtilis strain GM273 carrying a deletion of ptsH (Fig. 2B, lanes 2 and 3). The phosphate bond in P-Crh(His)6 was resistant to treatment with boiling 16% trichloroacetic acid, suggesting phosphorylation at Ser, Thr, or Tyr (33).
Figure 2.
In vitro phosphorylation of HPr(His)6 and Crh(His)6. [32P]PEP and purified EI (A) or [γ-32P]ATP and a crude extract of B. subtilis GM273 (B) were incubated together with HPr(His)6 (lane 2) or Crh(His)6 (lane 3); lane 1, control without HPr(His)6 or Crh(His)6. Proteins were separated by SDS/PAGE as described. In the case of ATP-dependent phosphorylation (B), the gel was treated for 10 min with boiling 16% trichloroacetic acid prior to autoradiography.
Phosphorylation by Partially Purified HPr Kinase.
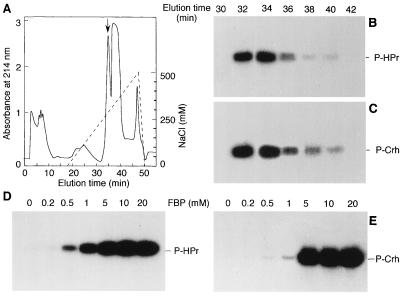
To test whether HPr kinase can phosphorylate Crh, we partially purified this enzyme from B. subtilis on a DEAE column. Proteins were eluted with a linear gradient of 0–500 mM NaCl (Fig. 3A). HPr kinase and Crh kinase activity were tested by incubating an aliquot of every other fraction with [γ-32P]ATP and Crh(His)6 or HPr(His)6 as described in Materials and Methods. HPr kinase from B. subtilis eluted at around 250 mM NaCl, similar to HPr kinase from E. faecalis (6) and S. pyogenes (7). HPr kinase from B. subtilis was found to phosphorylate both HPr and Crh (Fig. 3 B and C). In the absence of HPr or Crh in the assay mixture, no radioactive band comigrating with HPr or Crh was observed (data not shown). No additional Crh-specific protein kinase could be detected in the fractions. Because FBP stimulates the ATP-dependent phosphorylation of HPr in E. faecalis (6) and in S. pyogenes (7), we tested the effect of FBP on Crh and HPr phosphorylation in B. subtilis. Phosphorylation of both proteins was stimulated by FBP, and the maximal rate was obtained with 5 mM FBP (Fig. 3 D and E). However, at FBP concentrations lower than 5 mM, Crh(His)6 was less phosphorylated than HPr(His)6.
Figure 3.
HPr and Crh phosphorylation by partially purified HPr kinase and activation by FBP. A crude extract of B. subtilis GM273 was applied onto a DEAE-5PW Protein PAK glass column (0.8 × 7.5 cm; Waters). Proteins were eluted with a linear gradient of 0–500 mM NaCl (A). Every other fraction was tested for ATP-dependent HPr and Crh phosphorylation using [γ-32P]ATP. As can be seen from the autoradiogram, phosphorylation of HPr (B) and Crh (C) was detected in fractions 32–40, corresponding to the peak indicated by the arrow in A. To test the effect of FBP on HPr and Crh phosphorylation, 1 μg of HPr(His)6 (D) or Crh(His)6 (E) was incubated for 10 min at 37°C with a B. subtilis HPr kinase preparation, [γ-32P]ATP, MgCl2, and FBP at the indicated concentrations.
Determination of the Site of Phosphorylation.
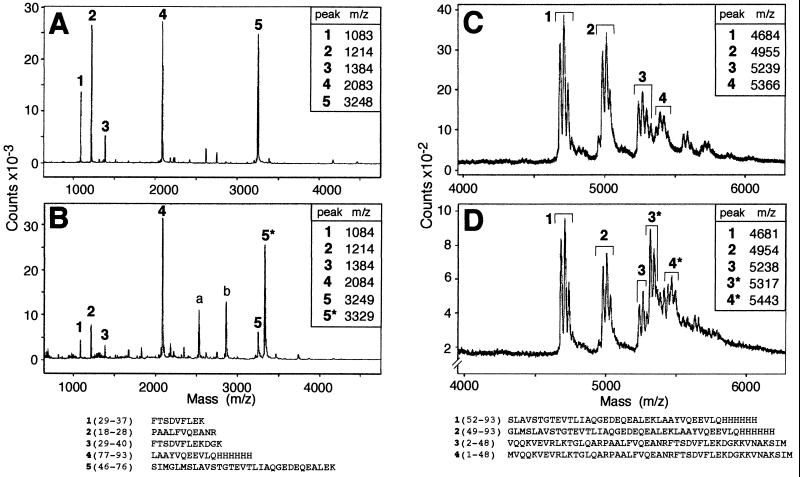
To determine which amino acid becomes phosphorylated in Crh, 100 μg of phosphorylated or unphosphorylated Crh(His)6 were either digested with trypsin or cleaved with CNBr. The obtained peptides were analyzed by MALDI-MS as described in Materials and Methods. The mass spectra obtained for tryptic digests of Crh(His)6 and partially phosphorylated Crh(His)6 are shown in Fig. 4 A and B. A strong peak exhibiting a molecular mass of 3248 was present in Fig. 4A, whereas only a small peak with a similar mass was observed in Fig. 4B. This peak corresponds to the tryptic peptide 5 which extends from amino acid Ser-46 to Lys-76 in the sequence of Crh and which has a calculated mass of 3,251. It differs from a peak exhibiting a mass of 3,329 and being present only in Fig. 4B by a mass of 80 Da, which corresponds to the mass of one phosphoryl group. This result establishes that the tryptic peptide 5 carries the phosphorylation site and that one of the three seryl or three threonyl residues present in this peptide represents the site of phosphorylation. To determine which amino acid becomes phosphorylated, 100 μg of phosphorylated or unphosphorylated Crh(His)6 was cleaved with CNBr. The mass spectra of the CNBr fragments are shown in Fig. 4 C [Crh(His)6] and D [partially phosphorylated Crh(His)6]. Each CNBr fragment gives at least a triplet, in which the first peak differs from the second and from the third by a mass of 28 or 56, respectively, corresponding to the mass of 1 or 2 formyl groups. The formyl groups were attached to the protein during the cleavage reaction, which was carried out in 70% formic acid. We observed a peak in Fig. 4C, and a smaller one in Fig. 4D, exhibiting a molecular mass of 5,239. This peak corresponds to peptide 3, extending from amino acid Val-2 to Met-48 in the sequence of Crh; it has a calculated mass of 5,240. This peak differs from the peak at 5,317 being present only in Fig. 4D by a mass of 78 Da, which corresponds to the mass of the phosphoryl group. This result indicates that peptide 3 extending from Val-2 to Met-48, contains the phosphorylation site. Because the tryptic and CNBr fragments carrying the phosphorylation site overlap only by three amino acids (Ser-46, Ile-47, and Met-48), Crh must be phosphorylated at Ser-46, as the other two amino acids cannot be phosphorylated.
Figure 4.
MALDI-MS of tryptic and CNBr cleavage products of Crh(His)6 or partially phosphorylated Crh(His)6. (A) Tryptic digests of Crh(His)6. (B) Tryptic digests of partially phosphorylated Crh(His)6. (C) CNBr cleavage of Crh(His)6. (D) CNBr cleavage of partially phosphorylated Crh(His)6. The sequences of the expected tryptic (Left) and CNBr fragments (Right) are listed, and peaks exhibiting the mass calculated for one of these fragments are labeled with the corresponding number. The two peaks exhibiting a mass of 2,530 and 2,861 (peaks a and b) in B, result probably from products present in the B. subtilis crude extract used as source for HPr kinase.
Effect of a crh Gene Disruption on CCR.
Phosphorylation of Crh at Ser-46 by the ATP-dependent HPr kinase suggested that Crh might also be implicated in CCR. To test this possibility, we inserted a kanamycin resistance cassette into the crh gene by transforming B. subtilis strain QB5081 (18) with plasmid pRC14, giving strain QB7098. The effect of the crh disruption on CCR of inositol dehydrogenase and β-xylosidase and of a translational levD′–′lacZ fusion was studied. These enzymes were chosen, because their CCR was not or only partially abolished in a ptsH1 mutant synthesizing HPrS46A (10, 34). Inositol dehydrogenase, β-galactosidase, and β-xylosidase activities were measured in the wild-type (QB5081), ptsH1 [QB5224 (34)], crh::aphA3, and ccpA::spc strains as well as in the double mutant ptsH1 crh::aphA3 under inducing or repressing (presence of fructose, glucose, or glycerol) conditions (Table 2). The three enzyme activities were less repressed in the ccpA mutant compared with the ptsH1 mutant. Disruption of the crh gene in a wild-type strain had almost no effect on CCR, whereas disruption of the crh gene in the ptsH1 mutant caused almost complete loss of the residual CCR. Enzyme activities similar to those found for the ccpA mutant strain were measured in the double mutant ptsH1 crh::aphA3 (Table 2). These results indicate that, in addition to CcpA, both HPr and Crh participate in CCR of certain genes in B. subtilis.
Table 2.
Effect of a crh gene disruption on CCR of inositol dehydrogenase, β-xylosidase, and of a levD′–′lacZ fusion
| System tested | Sugar added to CSK | Enzyme activity in units/mg of protein*
|
||||
|---|---|---|---|---|---|---|
| QB5081 | QB5224 | QB7098 | QB7103 ptsH1 | QB7115 | ||
| crh+ptsH+ | ptsH1 | crh::aphA3 | crh::aphA3 | ccpA::spc | ||
| β-Galactosidase (levD′–′lacZ fusion) | Fructose | 300 | 415 | 380 | 540 | 600 |
| Fructose + glucose | 20 | 110 | 35 | 450 | 290 | |
| Inositol dehydrogenase | Inositol | 280 | 330 | 280 | 340 | 390 |
| Inositol + fructose | 30 | 165 | 35 | 360 | 430 | |
| Inositol + glucose | 6 | 12 | 3 | 420 | 465 | |
| Inositol + glycerol | 45 | 340 | 50 | 670 | 480 | |
| β-Xylosidase | Xylose | 3,200 | 3,270 | 4,100 | 5,630 | 5,260 |
| Xylose + fructose | 230 | 856 | 245 | 4,870 | 3,090 | |
| Xylose + glucose | 30 | 80 | 35 | 2,360 | 3,550 | |
| Xylose + glycerol | 305 | 4,570 | 350 | 10,650 | 6,760 | |
Enzyme activities were calculated from two independent experiments for inositol dehydrogenase and from three independent experiments for the two other enzymes. Standard deviations did not exceed ±20% for β-xylosidase and ±12% for the two other enzymes.
Expression in trans of crh or crh1 in the ptsH1 crh::aphA3 Double Mutant.
To investigate whether phosphorylation of Crh at Ser-46 is necessary for its function in CCR, wild-type crh, and the crh1 allele, the latter encoding a mutant Crh in which Ser-46 is replaced with an alanine, were integrated at the amyE locus of the genome of the ptsH1 crh::aphA3 double mutant. Synthesis of wild-type Crh restored CCR sensitivity and a 55-fold lower β-xylosidase activity was measured in cells grown on xylose and glucose compared with cells grown only on xylose (Table 3). By contrast, glucose had only a weak repressive effect on β-xylosidase activity in cells synthesizing the two mutant proteins HPrS46A and CrhS46A, and enzyme activities closely similar to those measured in the ptsH1 crh::aphA3 double mutant were found (Tables 2 and 3). This result suggests that phosphorylation of Crh at Ser-46 is necessary for its participation in CCR of B. subtilis β-xylosidase.
Table 3.
Effect of the expression in trans of crh and the crh1 allele in the ptsH1 crh::aphA3 double mutant on CCR of β-xylosidase
| Strain and relevant genotype | β-Xylosidase activity in units/mg of protein*
|
||||
|---|---|---|---|---|---|
| Sugar added to CSK
| |||||
| None | Xylose | Xylose + fructose | Xylose + glucose | Xylose + glycerol | |
| QB7108 (ptsH1 crh+) | 60 | 2,250 | 720 | 40 | 2,350 |
| QB7112 (ptsH1 crh1) | 70 | 4,810 | 3,840 | 2,560 | 10,860 |
Enzyme activities were calculated from three independent experiments. Standard deviations did not exceed ±20%.
DISCUSSION
HPr of Gram-positive bacteria plays a central role in regulation of carbon metabolism similar to EIIAGlc in Gram-negative bacteria (35). Although protein phosphorylation is implicated in both cases, the mechanisms of CCR and inducer control are quite different. In Gram-positive bacteria, PEP-dependent phosphorylation of HPr at His-15 is not only necessary for PTS-catalyzed sugar transport and phosphorylation, but is also implicated in phosphorylation and regulation of transcriptional effectors (36, 37) and catabolic enzymes (29). By contrast, ATP-dependent phosphorylation of HPr at Ser-46 is involved in CCR (10) and inducer exclusion (35).
We here report the discovery of a B. subtilis gene, crh, which encodes a protein exhibiting 45% sequence identity to HPr. Because the protein Crh is missing the catalytic His-15, it cannot be phosphorylated by PEP and EI and is probably not involved in PTS-catalyzed sugar transport and phosphorylation. However, Ser-46, the site of ATP-dependent phosphorylation of HPr, and the sequence around Ser-46 are conserved in Crh. We could demonstrate that the metabolite-activated HPr kinase phosphorylates Crh at Ser-46. This result suggested that Crh might be involved in regulatory processes similar to P-Ser-HPr.
We could demonstrate that both proteins, HPr and Crh, participate in CCR of certain operons of B. subtilis. ptsH1 mutants, in which Ser-46 of HPr is replaced with an alanyl residue, exhibited reduced glucose repression compared with wild-type strains—i.e., only 3.5-fold repression of a levD′–′lacZ fusion—and 27- and 40-fold repression of inositol dehydrogenase and β-xylosidase synthesis, respectively. Disruption of the crh gene had little effect on CCR, whereas a ptsH1 crh::aphA3 double mutant was almost completely relieved from CCR of inositol dehydrogenase, β-xylosidase and the levD′–′lacZ fusion. Expression of the wild-type crh integrated at the amyE locus restored CCR of β-xylosidase synthesis in the double mutant, whereas expression of a crh allele encoding Crh in which Ser-46 is replaced with an alanyl residue had no effect on CCR. These results clearly demonstrate that both, P-Ser-HPr and P-Ser-Crh, are implicated in CCR of levanase, inositol dehydrogenase, and β-xylosidase of B. subtilis. Because a ccpA mutant exhibited a relief from CCR similar to the ptsH1 crh::aphA3 double mutant, it is likely that P-Ser-Crh exerts its effect on CCR via CcpA, similar to P-Ser-HPr. P-Ser-Crh might interact with CcpA and allows its binding to the cre sequences. Multiple cre sites have recently been described for the gnt operon of B. subtilis (38) and for the xyl operon of Bacillus megaterium (39). These novel cre sequences were not recognized by the CcpA/P-Ser-HPr complex, but were recognized by CcpA in the presence of glucose-6-P. However, the glucose-6-P-mediated interaction of CcpA with these novel cre sequences required either nonphysiological pH values or nonphysiological glucose-6-P concentrations. The presumed CcpA/P-Ser–Crh complex could possibly recognize these cre sequences in the presence of a metabolite different from FBP. It is interesting to note that the repressive effect of fructose or glycerol on inositol dehydrogenase and β-xylosidase synthesis in the ptsH1 mutant was much weaker compared with the repressive effect exerted by glucose, suggesting that Crh responds to a signal specific for glucose metabolism.
Because no effect on CCR could be observed in strains carrying a crh gene disruption, HPr seems to be able to substitute for Crh in CCR. β-Xylosidase synthesis in a ptsH1 mutant strain was repressed by glucose and fructose to a similar extent as in the wild-type strain. This repression must be due to Crh, because it was abolished when the crh disruption was introduced in the ptsH1 strain, but was restored when crh was integrated at the amyE site of the ptsH1 crh::aphA3 double mutant. The two related proteins, HPr and Crh, could possibly be used to allow B. subtilis cells to respond to different concentrations of glycolytic intermediates. We observed, for example, that Crh was less effectively phosphorylated compared with HPr when the FBP concentration was lower than 5 mM.
Similar to P-Ser-HPr, P-Ser-Crh could be involved in other regulatory mechanisms, such as inducer control (1). Although His-15 of HPr is replaced with a glutamine in Crh, the sequences around Gln-15 and His-15 in Crh and HPr are well conserved. B. subtilis HPr, in which His-15 had been replaced with an alanine, was reported to allosterically inhibit EI activity (40). Similarly, Crh could also act as allosteric inhibitor of EI.
Acknowledgments
We are grateful to G. Rapoport, in whose laboratory part of this work was carried out, for continuous encouragement and critical reading of the manuscript. We are thankful to F. Denizot and A. Klier for stimulating discussion, C. van Herrewege for his help with iconography, J. C. Cortay and D. Nègre for providing us with plasmid pT7–5 (6 × His), J. Bignon and L. Rousse for technical assistance, and C. Dugast for her help preparing the manuscript. We are also indebted to A. Guiseppi and J. Busuttil for DNA sequence analysis and Y. Quentin for computer analysis. This research was supported by the European Community Biotech Programme Contrat BIO2-CT-920137 (to J.D.) and a grant of the Actions Concertées Coordonneés (to J.H.), the Centre National de la Recherche Scientifique, the Institut Pasteur, and the Université Paris 7.
ABBREVIATIONS
- CCR
carbon catabolite repression
- HPr
histidine-containing protein
- PEP
phosphoenolpyruvate
- Crh
catabolite repression HPr
- EI
enzyme I
- PTS
phosphotransferase system
- FBP
fructose-1,6-bisphosphate
- cre
catabolite responsive element
- MALDI-MS
matrix-assisted laser desorption ionization mass spectra.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. Z94043).
References
- 1.Saier M H, Jr, Chauvaux S, Cook G M, Deutscher J, Paulsen I T, Reizer J, Ye J J. Microbiology. 1996;142:217–230. doi: 10.1099/13500872-142-2-217. [DOI] [PubMed] [Google Scholar]
- 2.Deutscher J, Saier M H., Jr Proc Natl Acad Sci USA. 1983;80:6790–6794. doi: 10.1073/pnas.80.22.6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deutscher J, Pevec B, Beyreuther K, Kiltz H-H, Hengstenberg W. Biochemistry. 1986;25:6543–6551. doi: 10.1021/bi00369a031. [DOI] [PubMed] [Google Scholar]
- 4.Beyreuther K, Raufuss H, Schrecker O, Hengstenberg W. FEBS Lett. 1982;138:102–103. [Google Scholar]
- 5.Weigel N, Powers D A, Roseman S. J Biol Chem. 1982;257:14499–14509. [PubMed] [Google Scholar]
- 6.Deutscher J, Engelmann R. FEMS Microbiol Lett. 1984;23:157–162. [Google Scholar]
- 7.Reizer J, Novotny M J, Hengstenberg W, Saier M H., Jr J Bacteriol. 1984;160:333–340. doi: 10.1128/jb.160.1.333-340.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowds B, Baxter L, McKillen M. Biochim Biophys Acta. 1978;541:18–34. [Google Scholar]
- 9.Nihashi J-I, Fujita Y. Biochim Biophys Acta. 1984;798:88–95. doi: 10.1016/0304-4165(84)90014-x. [DOI] [PubMed] [Google Scholar]
- 10.Deutscher J, Reizer J, Fischer C, Galinier A, Saier M H, Jr, Steinmetz M. J Bacteriol. 1994;176:3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reizer A, Deutscher J, Saier M H, Jr, Reizer J. Mol Microbiol. 1991;5:1081–1089. doi: 10.1111/j.1365-2958.1991.tb01880.x. [DOI] [PubMed] [Google Scholar]
- 12.Yoshida, K.-I., Aoyama, D., Ishio, I., Shibayama, T. & Fujita, Y. (1997) J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 13.Henkin T M. FEMS Microbiol Lett. 1996;135:9–15. doi: 10.1111/j.1574-6968.1996.tb07959.x. [DOI] [PubMed] [Google Scholar]
- 14.Weickert M J, Adhya S. J Biol Chem. 1992;267:15869–15874. [PubMed] [Google Scholar]
- 15.Weickert M J, Chambliss G H. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 17.Fujita Y, Miwa Y, Galinier A, Deutscher J. Mol Microbiol. 1995;17:953–960. doi: 10.1111/j.1365-2958.1995.mmi_17050953.x. [DOI] [PubMed] [Google Scholar]
- 18.Martin-Verstraete I, Débarbouillé M, Klier A, Rapoport G. J Mol Biol. 1992;226:85–99. doi: 10.1016/0022-2836(92)90126-5. [DOI] [PubMed] [Google Scholar]
- 19.Martin I, Débarbovillé M, Klier A, Rapoport G. J Bacteriol. 1989;171:1885–1892. doi: 10.1128/jb.171.4.1885-1892.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.Kunst F, Rapoport G. J Bacteriol. 1995;177:2403–2407. doi: 10.1128/jb.177.9.2403-2407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cortay J C, Nègre D, Scarabel M, Ramseier T, Vartak N B, Reizer J, Saier M H, Jr, Cozzone A J. J Biol Chem. 1994;269:14885–14891. [PubMed] [Google Scholar]
- 23.Gonzy-Tréboul G, Steinmetz M. J Bacteriol. 1987;169:2287–2290. doi: 10.1128/jb.169.5.2287-2290.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferrari F A, Ngyen A, Lang D, Hoch J A. J Bacteriol. 1983;154:1513–1515. doi: 10.1128/jb.154.3.1513-1515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trieu-Cuot P, Courvalin P. Gene. 1983;23:331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- 26.Ireton K, Rudner D Z, Siranosian K J, Grossman A D. Genes Dev. 1993;7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- 27.Roossien F F, Brink J, Robillard G T. Biochim Biophys Acta. 1983;760:185–187. doi: 10.1016/0304-4165(83)90141-1. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Charrier V, Buckley E, Parsonage D, Galinier A, Darbon E, Jaquinod M, Forest E, Deutscher J, Claiborne A. J Biol Chem. 1997;272:14166–14174. doi: 10.1074/jbc.272.22.14166. [DOI] [PubMed] [Google Scholar]
- 30.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. pp. 352–355. [Google Scholar]
- 31.Lindner C, Stülke J, Hecker M. Microbiology. 1994;140:753–757. doi: 10.1099/00221287-140-4-753. [DOI] [PubMed] [Google Scholar]
- 32.Fabret C, Quentin Y, Chapal N, Guiseppi A, Haiech J, Denizot F. Microbiology. 1996;142:3089–3096. doi: 10.1099/13500872-142-11-3089. [DOI] [PubMed] [Google Scholar]
- 33.Duclos B, Marcandier S, Cozzone A J. Methods Enzymol. 1991;201:10–20. doi: 10.1016/0076-6879(91)01004-l. [DOI] [PubMed] [Google Scholar]
- 34.Martin-Verstraete I, Stülke J, Klier A, Rapoport G. J Bacteriol. 1995;177:6919–6927. doi: 10.1128/jb.177.23.6919-6927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saier M H, Jr, Chauvaux S, Deutscher J, Reizer J, Ye J J. Trends Biochem Sci. 1995;20:267–271. doi: 10.1016/s0968-0004(00)89041-6. [DOI] [PubMed] [Google Scholar]
- 36.Arnaud M, Débarbouillé M, Rapoport G, Saier M H, Jr, Reizer J. J Biol Chem. 1996;271:18966–18972. doi: 10.1074/jbc.271.31.18966. [DOI] [PubMed] [Google Scholar]
- 37.Stülke J, Martin-Verstraete I, Charrier V, Klier A, Deutscher J, Rapoport G. J Bacteriol. 1995;177:6928–6936. doi: 10.1128/jb.177.23.6928-6936.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miwa Y, Nagura K, Eguchi S, Fukuda H, Deutscher J, Fujita Y. Mol Microbiol. 1997;23:1203–1213. doi: 10.1046/j.1365-2958.1997.2921662.x. [DOI] [PubMed] [Google Scholar]
- 39.Gösseringer R, Küster E, Galinier A, Deutscher J, Hillen W. J Mol Biol. 1997;266:665–676. doi: 10.1006/jmbi.1996.0820. [DOI] [PubMed] [Google Scholar]
- 40.Reizer J, Sutrina S L, Wu L-F, Deutscher J, Reddy P, Saier M H., Jr J Biol Chem. 1992;267:9158–9169. [PubMed] [Google Scholar]