Abstract
The Oct-1 POU domain binds diverse DNA-sequence elements and forms a higher-order regulatory complex with the herpes simplex virus coregulator VP16. The POU domain contains two separate DNA-binding domains joined by a flexible linker. By protein–DNA photocrosslinking we show that the relative positioning of the two POU DNA-binding domains on DNA varies depending on the nature of the DNA target. On a single VP16-responsive element, the POU domain adopts multiple conformations. To determine the structure of the Oct-1 POU domain in a multiprotein complex with VP16, we allowed VP16 to interact with previously crosslinked POU-domain–DNA complexes and found that VP16 can associate with multiple POU-domain conformations. These results reveal the dynamic potential of a DNA-binding domain in directing transcriptional regulatory complex formation.
The human transcriptional activator protein Oct-1, which contains a DNA-binding POU domain, displays diversity in DNA-sequence recognition and coregulator association. The POU domain is a bipartite DNA-binding structure made up of two helix–turn–helix-containing DNA-binding domains: a POU-specific (POUS) domain joined by a flexible linker to a carboxy-terminal POU-homeo (POUH) domain (1–3). The Oct-1 POU domain displays exceptional sequence recognition flexibility and directs complex formation with viral and cellular coregulators, including the herpes simplex virus transactivator VP16 (Vmw65, αTIF) (reviewed in ref. 4).
DNA-binding studies of POU domains have revealed a high degree of structural flexibility in their interactions with DNA. For example, the x-ray crystal structures of the Oct-1 and Pit-1 POU domains bound to Oct-1- and Pit-1-responsive sites, respectively, differ: The Oct-1 POU domain binds to the octamer sequence ATGCAAAT as a monomer, with the POUS and POUH domains bound to opposite faces of the DNA (2), whereas the Pit-1 POU domain binds its site as a dimer with the POUS and POUH domains of each POU domain bound to the same face of the DNA and with the POUS domain in an inverted orientation to that found with Oct-1 (3). Furthermore, combined mutagenesis of the Oct-1 or related Brn-2 POU domains and their respective DNA targets has provided indirect evidence that even the same POU domain adopts different conformations on different cis-regulatory sites, providing a mechanism for divergent sequence recognition (5, 6).
To probe directly the conformational flexibility of the Oct-1 POU domain bound to DNA and in a higher-order complex, we adapted a site-specific protein–DNA photocrosslinking protocol (7) to map, individually, the locations of the POUS and POUH domains on DNA and in complex with VP16. Our results show that the Oct-1 POU domain adopts multiple conformations on a single binding site and in association with VP16.
MATERIALS AND METHODS
In Vitro Mutagenesis.
The site-specific protein–DNA photocrosslinking method we used (7) involves the attachment of the photoactivatable crosslinker 4-azidophenacyl bromide to an appropriately positioned unique cysteine in the DNA-binding protein. We removed the existing Oct-1 POU domain cysteines at positions 61 of the POUS domain and 50 of the POUH domain and introduced unique cysteines by oligonucleotide-mediated site-directed mutagenesis (6). To create unique cysteine (UC) derivatives, we made the following POUS mutations: L23C (POUS-UC23), G24C (POUS-UC24), F25C (POUS-UC25), G28C (POUS-UC28), D29C (POUS-UC29), and N54C (POUS-UC54); and the following POUH mutations: S7C (POUH-UC7), E9C (POUH-UC9), and T10C (POUH-UC10).
4-Azidophenacyl-Bromide Modification.
Oct-1 POU-domain variants were prepared in Escherichia coli as glutathione S-transferase (GST)-fusion proteins essentially as described (8). Purified POU domain was cleaved from the GST moiety (3.5 μM in 500 μl) with thrombin and treated for 6 hr at room temperature in the dark with 140 μM 4-azidophenacyl bromide (Sigma) and 1.7% dimethylformamide as described (7). Reaction mixtures were then dialyzed twice against 1 liter of 20 mM Tris⋅HCl, pH 8.0/200 mM KCl/5% glycerol/0.1 mM EDTA for a total of 14 hr.
POU Domain–DNA Photocrosslinking and Site-Specific Cleavage.
POU domain–DNA binding reactions (100 μl in flat-bottomed microtiter plate wells) contained 0.28 μM Oct-1 POU domain protein, 3 × 105 cpm of the appropriate singly end-labeled DNA probe generated by PCR amplification (see ref. 6) in 8 mM Na⋅Hepes, pH 7.9/60 mM NaCl/4 mM spermidine/2 mM EDTA/0.2 mM DTT/0.03% Nonidet P-40/0.1 mg/ml bovine serum albumin. Reaction mixtures were incubated at room temperature for 30 min in the dark before being irradiated for 1.5 min in a UV Stratalinker 2400 (Stratagene) with 312-nm UV bulbs. The irradiation time was determined empirically for optimal crosslinking. After UV irradiation, 5 μl of each binding reaction mix was removed, added to 5 μl of SDS loading buffer, heated to 90°C for 10 min, and subjected to 10% SDS/PAGE in the absence of a stacking gel. Levels of crosslinking (typically 3%) were determined by phosphorimager analysis (Fuji BAS1000). The remainder of the sample was used to map the sites of crosslinking as described (7). Purine cleavage patterns for each probe were generated as described (9). DNA cleavage products were analyzed on 20% polyacrylamide/7 M urea denaturing gels.
Crosslinking Interference Analysis.
POUS-UC54 or POUH-UC50 was allowed to bind to the (OCTA+)TAATGARAT probe and crosslinked to the DNA as described above except that, after UV-induced crosslinking, E. coli-expressed GST–VP16 and an in vitro-translated form of the VP16-induced complex accessory factor HCF (10) were added, and the reaction was incubated for 20 min at 37°C. The protein–DNA complexes were separated on a native 4% polyacrylamide/0.25× TBE gel, and bands corresponding to the VP16-induced and POU-domain complexes were cut out of the gel, soaked in 20 mM ammonium acetate, 0.1 mM EDTA, and 2% SDS at 4°C overnight and spun through Spin-X filters (Costar). The eluted protein/DNA was treated as described for cleavage at sites of crosslinking (7), and the cleavage products were mapped by denaturing PAGE.
RESULTS
Experimental Design.
Pendergrast et al. (7) have described a site-specific protein–DNA photocrosslinking method in which the photoactivatable crosslinking agent 4-azidophenacyl bromide is covalently attached to a unique cysteine side chain on the surface of a DNA-binding protein. The modified protein is allowed to bind to DNA, and the binding reaction is then subjected to UV irradiation, enabling the modified cysteine side chain to form a covalent bond with nearby DNA. Crosslinked protein–DNA complexes are then analyzed directly by SDS/PAGE or the sites of crosslinking mapped by site-specific crosslink-induced DNA cleavage (7). We created Oct-1 POU domains carrying either POUS- or POUH-domain-specific crosslinkers. These domain-specific crosslinkers allowed us to probe independently the positions of the two Oct-1 POU DNA-binding domains on cis-regulatory sites.
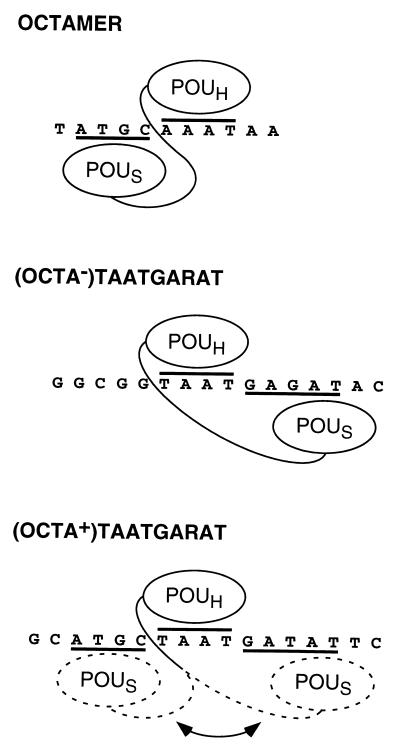
We examined Oct-1 POU-domain binding to three previously described sites (see ref. 6): the high-affinity octamer sequence ATGCAAAT from the human histone H2B promoter [for which the structure of the bound Oct-1 POU domain is known (2)] and two VP16-responsive TAATGARAT (R = purine) elements derived from the herpes simplex virus ICP0 [called (OCTA+)TAATGARAT] and ICP4 [called (OCTA−)TAATGARAT] immediate-early (IE) promoters. These two sites represent the two different kinds of TAATGARAT elements found in herpes simplex virus IE promoters (see ref. 11); only (OCTA+)TAATGARAT elements contain an overlapping octamer sequence (shown in bold: ATGCTAATGARAT).
Fig. 1A shows a representation of the crystal structure of the Oct-1 POU domain bound to the histone H2B octamer site (2), with the POUS domain considered 5′ of the POUH domain. The structure of the Oct-1 POU domain on a TAATGARAT site is not known. Mutational analysis has suggested that on an (OCTA−)TAATGARAT site, the POUS domain lies 3′ of the POUH domain (6), and high-resolution hydroxy-radical footprinting of an Oct-1 POU domain–(OCTA+)TAATGARAT complex revealed only protection of the TAAT sequence, suggesting an unusual POU-domain conformation on this site (12).
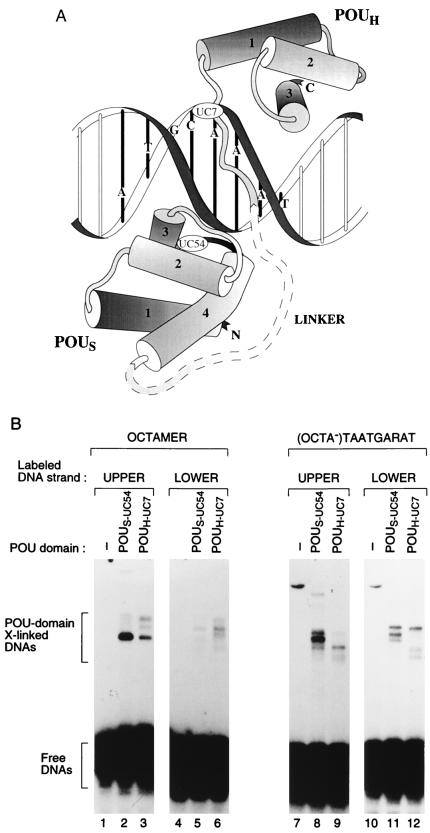
Figure 1.
Unique cysteines in the POUS and POUH domains effective for protein–DNA crosslinking. (A) Representation (adapted from refs. 2 and 4) of the Oct-1 POU domain–octamer site crystal structure with the positions of the unique cysteines used for crosslinking analysis indicated (UC54 in the POUS domain and UC7 in the POUH domain; the precise location of UC54 is hidden behind helix 2 on the turn between helices 3 and 4). (B) SDS/PAGE analysis of crosslinked protein–DNA complexes after UV irradiation of POU domain-binding reactions containing no protein (lanes 1, 4, 7, and 10), POUS-UC54 protein (lanes 2, 5, 8, and 11), or POUH-UC7 protein (lanes 3, 6, 9, and 12). Shown are crosslinked complexes formed on the upper and lower strands of the histone H2B octamer and the (OCTA−)TAATGARAT sites, as indicated.
To generate Oct-1 POU domains with unique cysteines suitable for protein–DNA crosslinking, we created the cysteine-free double mutant POUS-C61A/POUH-C50S by replacing the wild-type POUS-domain cysteine with an alanine and the wild-type POUH-domain cysteine with a serine. This cysteine-free Oct-1 POU domain binds like wild-type POU domain to the octamer, and (OCTA+) and (OCTA−) TAATGARAT sites (13). Directed by the structure of the Oct-1 POU domain–octamer site complex (2), we placed unique cysteines at a number of POUS- and POUH-domain positions that (i) were likely not to be important for DNA binding and (ii) whose α carbons were ≈9–13 Å from the DNA. The ability of these mutant proteins to crosslink to DNA was analyzed by SDS/PAGE. Of six POUS-domain derivatives tested (see Materials and Methods), only POUS-UC54 crosslinked to the octamer or (OCTA−)TAATGARAT sites effectively, and, of the four POUH-domain derivatives tested (see Materials and Methods), only POUH-UC7 and POUH-UC50 crosslinked effectively (data not shown).
Fig. 1B shows the results of crosslinking POUS-UC54 and POUH-UC7 to the octamer and the (OCTA−)TAATGARAT sites as analyzed by SDS/PAGE in the absence of a stacking gel. The slower migrating complexes probably represent POU-domain crosslinked DNAs because formation of these species depended on (i) added POU domain (Fig. 1B; a nonspecific slower migrating species in lanes 7 and 10 formed in the absence of POU domain), (ii) UV irradiation, (iii) modification of the protein with 4-azidophenacyl bromide, and (iv) an appropriately positioned cysteine (data not shown). Some crosslinked samples produced more than one band (lanes 3, 6, 8, 9, 11, and 12), which may reflect POU-domain crosslinking at different positions in the DNA.
The Oct-1 POU Domain Adopts Different Conformations on the Octamer and (OCTA−)TAATGARAT Sites.
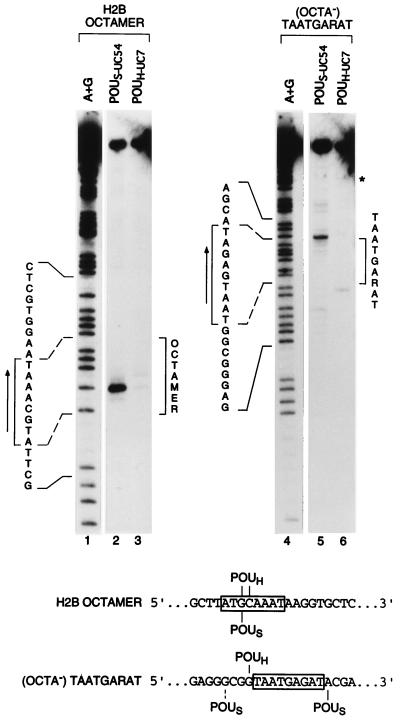
Although both strands of the octamer and (OCTA−)TAATGARAT sites could be crosslinked to the POU domain (Fig. 1B), only the upper strands of these probes were good substrates for site-specific DNA cleavage (data not shown). Fig. 2 shows the cleavage patterns that map sites of POUS- and POUH-domain crosslinking to the upper strands of these two sites. On the upper strand of the octamer probe, the primary site of crosslink-induced cleavage by the modified POUS-UC54 protein (lane 2) maps to the G at the third position of the octamer sequence (ATGCAAAT) and is consistent with the POU domain–octamer site crystal structure (2). The POUH-UC7 protein generated two cleavage products (Fig. 2, lane 3). These products map to the G and C at positions 3 and 4 (ATGCAAAT) and are also consistent with the POU domain–octamer site crystal structure (2).
Figure 2.
The POUS domain is located on opposite sides of the POUH domain on octamer and (OCTA−)TAATGARAT sites. Shown are the cleavage products generated after crosslinking of POUS-UC54 and POUH-UC7 proteins to the upper strands of the octamer (lanes 2 and 3) and the (OCTA−)TAATGARAT (lanes 5 and 6) sites, as indicated. Lanes 1 and 4 (A+G), purine-cleavage pattern of the probe indicated. In lanes 2, 3, 5, and 6, uppermost bands represent uncleaved DNA. Brackets mark the limits of the two sequences on the autoradiographs, and arrows indicate the directionality of each consensus motif. The two binding sites are cloned into the same plasmid in opposite orientations. ∗, Cleavage product mapping to flanking polylinker sequence, probably resulting from non-octamer-site binding. (Lower) Summary of positions of crosslink-induced cleavage. Dashed line indicates lower-yield cleavage product.
On the (OCTA−)TAATGARAT site (Fig. 2, lanes 4–6), the primary POUH-UC7 protein crosslink-induced cleavage (lane 6) maps to the G just 5′ of the TAATGARAT sequence (GTAATGAGAT). The location of this crosslink suggests that the TAAT sequence serves as the POUH domain-binding site the same way the AAAT sequence does in the octamer sequence. In contrast, the major POUS-UC54 cleavage product maps to the opposite side of the TAAT POUH domain-binding site, at an A immediately 3′ of the GARAT sequence (TAATGAGATA). Together with the POUH-domain cleavage product, this predominant POUS-domain cleavage product provides physical evidence that the POU domain can adopt a conformation on an (OCTA−)TAATGARAT sequence that is very different from its conformation on an octamer sequence.
In addition to the major 3′ POUS-domain cleavage product, there are several minor cleavage products including one resulting from a crosslink to the G four positions 5′ of the TAAT sequence (GCGGTAATGAGAT). This crosslink does not map to the same 5′ position as on the octamer site (see Fig. 2 Lower) and may result from binding of the POUS domain in an inverted orientation as seen with Pit-1 (3) and suggested for Brn-2 (5). The relative levels of cleavage 5′ and 3′ of the TAATGARAT sequence are consistent with previous chemical modification interference analyses (6, 14, 15) and mutational analysis (6), which indicate that the POUS domain is principally positioned over the GARAT element when the Oct-1 POU domain binds to this (OCTA−)TAATGARAT sequence.
The Oct-1 POU Domain Adopts Multiple Conformations on an (OCTA+)TAATGARAT Site.
The (OCTA+)TAATGARAT site contains both octamer and (OCTA−)TAATGARAT sequences. Differences in Oct-1 POU domain footprinting patterns of (OCTA+)TAATGARAT sites with wild-type and mutant GARAT sequences have suggested that the Oct-1 POU domain adopts an unusual conformation on the wild-type site (12). We used domain-specific crosslinking to probe this unusual conformation.
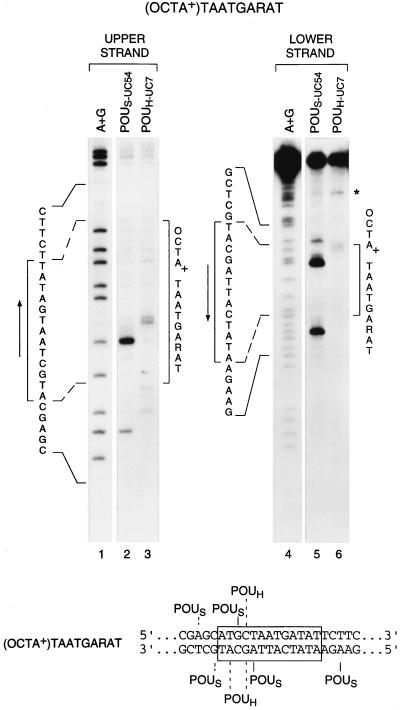
Fig. 3 shows the cleavage patterns resulting from crosslinking to both strands of the (OCTA+)TAATGARAT site. The POUH-UC7 protein crosslinked to both strands of this site just upstream of the TAAT sequence (lanes 3 and 6), suggesting that, on this site, the POUH domain binds uniquely to the TAAT sequence in the same orientation as it binds to the octamer and (OCTA−)TAATGARAT sites. In contrast, the POUS domain crosslinked to the (OCTA+)TAATGARAT site at several locations both 5′, as on an octamer site, and 3′, as on an (OCTA−)TAATGARAT site, of the TAAT sequence (Fig. 3, lanes 2 and 5, and Lower), suggesting that the Oct-1 POU domain adopts at least two very different conformations on this site. In the region 5′ of the TAAT POUH-domain-binding site, the POUS domain generates two crosslink-induced cleavages on each strand (Fig. 3, lanes 2 and 5, and Lower). The TAAT-proximal cleavages probably reflect Oct-1 binding in an octamer-site-like conformation; the other two cleavages may reflect an inverted POUS-domain orientation similar to that described for the Pit-1 POU domain (3).
Figure 3.
The POU domain adopts multiple conformations on an (OCTA+)TAATGARAT site. Shown are the cleavage products generated from POU domain–DNA crosslinking to both strands of the (OCTA+)TAATGARAT site. Lanes 2 and 3, cleavage pattern generated on the upper strand. The POUH doublet may be due to an incomplete cleavage reaction. Lanes 5 and 6, cleavage pattern generated on the lower strand. Lanes 1 and 4 (A+G), purine-cleavage pattern of the strand indicated. In lanes 5 and 6, the uppermost bands represent uncleaved DNA. Brackets, arrows, and asterisk are as in Fig. 2. (Lower) Summary of positions of cleavage. Dashed lines indicate lower-yield cleavage products.
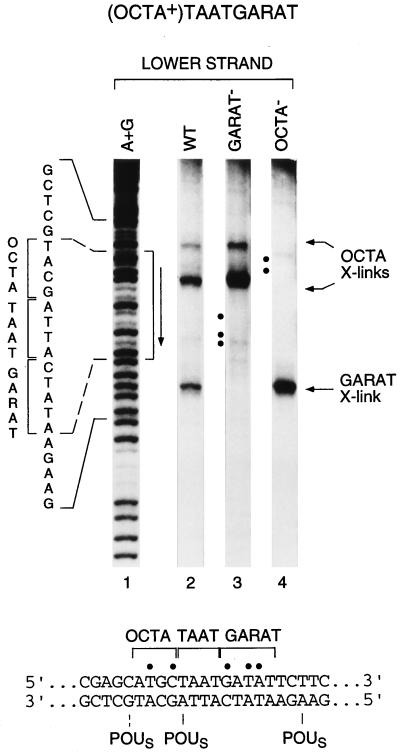
To examine whether the POUS domain can bind independently either 5′ or 3′ of the POUH domain-binding site of the (OCTA+)TAATGARAT sequence, we compared crosslinking of the POUS-UC54 protein bound to the wild-type (OCTA+)TAATGARAT sequence with crosslinking to two mutant forms of the site in which either the 5′ or 3′ POUS domain-binding site has been disrupted. As indicated in Fig. 4 by dots, we mutated residues in the POUS domain-binding sites but not the precise sites of crosslinking. Consistent with independent binding of the POUS domain to the 5′ ATGC element and the 3′ GARAT element, when the GARAT region of the (OCTA+)TAATGARAT sequence is mutated, we only detect cleavages in the upstream ATGC portion of the octamer site (compare lane 3 with lane 2). Likewise, when the upstream ATGC portion of the octamer site is mutated, we only detect cleavages near the GARAT sequence (compare lane 4 with lane 2). These results demonstrate that two functionally independent, but overlapping, POU domain-binding sites make up the (OCTA+)TAATGARAT element.
Figure 4.
The POUS domain binds to the wild-type half-site in mutant forms of the (OCTA+)TAATGARAT site. Shown are the cleavage products generated from crosslinking of the POUS domain to wild-type (lane 2) and mutant (lanes 3 and 4) (OCTA+)TAATGARAT sites. Lane 1 (A+G), purine-cleavage pattern of the wild-type probe; lane 3, (OCTA+)TAATGARAT site with the mutated 3′ GARAT sequence (underlined), … ATTAGCTTC… (mutations shown in bold); lane 4, (OCTA+)TAATGARAT site with the mutated 5′ OCTA sequence (underlined), … GCAAGGTA… (mutations shown in bold). Brackets and vertical arrows are as in Fig. 2. (Lower) Sequence with sites of mutations marked by dots and crosslinking sites indicated. Dashed line indicates lower-yield cleavage product.
These results may also explain why hydroxy-radical footprinting reveals a POUH-domain TAAT protection pattern but not a POUS-domain protection pattern when the POU domain is bound to an (OCTA+)TAATGARAT site (12). Whereas the POUH domain apparently binds stably to the TAAT sequence, the POUS domain can bind either upstream or downstream of the POUH domain, thus serving as an ineffective inhibitor of hydroxy-radical attack of the sequences flanking the POUH domain-binding site. Consistent with this hypothesis, when the GARAT sequence is mutated, the Oct-1 POU domain effectively protects the 5′ ATGC element (12). Together, the footprinting (12) and crosslinking (this study) results suggest that the Oct-1 POU domain structure is dynamic on an (OCTA+)TAATGARAT site.
VP16 Associates with Multiple Oct-1 POU-Domain Conformations.
The multiple conformations of the Oct-1 POU domain on the (OCTA+)TAATGARAT site raise the issue of how the Oct-1 POU domain is structured in a VP16-induced complex. The structure of the VP16-induced complex is unclear, principally because VP16 binds DNA poorly in the absence of Oct-1 (16, 17). Thus, although the GARAT and neighboring 3′ sequences are specifically important for forming a VP16-induced complex (18–22), it is not known whether these sequences induce an Oct-1 POU-domain conformation compatible with recognition by VP16 (12) or create a VP16-binding site (see ref. 4).
To probe the conformation of the Oct-1 POU domain in a VP16-induced complex, we devised a crosslinking strategy analogous to chemical modification interference analysis. We first crosslinked either the POUS or POUH domain to the (OCTA+)TAATGARAT site and then selected those crosslinked species that could still form a complex with VP16 (see Materials and Methods). If a particular conformation of the POU domain is incompatible with VP16-induced complex formation, crosslinks resulting from that conformation should interfere with VP16-induced complex formation and be missing in the DNAs selected for VP16-induced complex formation. We refer to this strategy as “crosslinking interference analysis.”
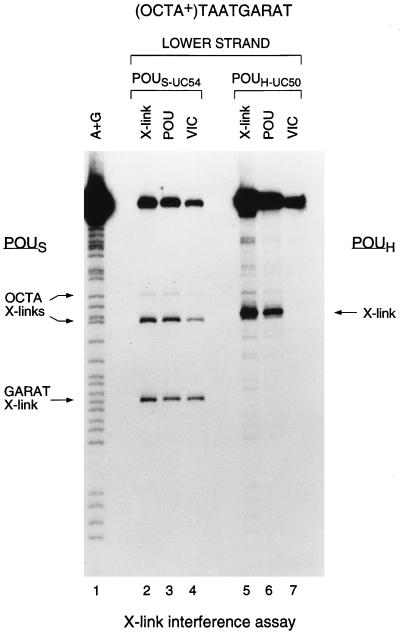
Fig. 5 shows the results of the crosslinking interference analysis with POUS- (POUS-UC54; lanes 2–4) and POUH- (POUH-UC50; lanes 5–7) specific crosslinkers. Because the level of POUH-UC7 cleavage on the (OCTA+)TAATGARAT site was not sufficient for the crosslinking interference analysis (see Fig. 3), we used the natural POUH-domain cysteine in POUH-UC50 for this analysis. Cysteine 50 lies in the DNA-recognition helix and contacts the DNA major groove in the octamer cocrystal structure (2); we did not use this crosslinker in the analyses described above to avoid crosslinker-induced changes in POUH-domain binding to the DNA.
Figure 5.
VP16 associates with multiple conformations of the Oct-1 POU domain. Shown are the results of a crosslinking interference analysis of two unique-cysteine derivatives of the POU domain, POUS-UC54 (lanes 2–4) and POUH-UC50 (lanes 5–7), crosslinked to the lower strand of the (OCTA+)TAATGARAT site. POUS crosslinks over the 5′ ATGC (OCTA X-links) or 3′ GARAT (GARAT X-link) sequences and POUH crosslinks (X-link) are indicated. In lanes 2–7, the uppermost bands represent uncleaved DNA. X-link (above lanes), standard crosslinking analysis of the POU domain alone to the site; POU, crosslinking pattern of the POU-domain complex after electrophoretic mobility retardation; VIC, crosslinking interference pattern of the VP16-induced complex after electrophoretic mobility retardation.
Because cysteine 50 is directed 3′ of the octamer sequence (2) in the Oct-1 POU-domain–octamer site crystal structure, we expected POUH-UC50 crosslinking to occur within the GARAT sequence. The observed crosslink-induced cleavage, however, maps 5′ of this region at the G residue in the C:G base pair at the fourth position within the octamer sequence (ATGCTAAT; Fig. 5, lane 5). This difference may reflect an alteration of the precise conformation of the POUH-UC50 domain on DNA induced by (i) the different binding sites [H2B octamer vs. (OCTA+)TAATGARAT] or (ii) the crosslinking moiety. Whichever the case, the POUH-UC50 crosslink interferes very strongly with VP16-induced complex formation (compare lanes 6 and 7). These results demonstrate that it is possible to interfere with VP16-induced complex formation by crosslinking and suggest that the conformation, precise position, or flexibility of the POUH domain on the (OCTA+)TAATGARAT site is critical for association of the Oct-1 POU domain with VP16.
In striking contrast to the strong interference caused by the POUH-UC50 crosslink, the POUS-UC54 crosslinks, either at or near the octamer ATGC sequence (OCTA X-links) or near the GARAT sequence (GARAT X-link), are compatible with VP16-induced complex formation (Fig. 5, compare lanes 3 and 4). An apparent slight decrease in crosslink-induced cleavages at the octamer ATGC sequence (compare lanes 3 and 4) may suggest a preference for POUS-domain binding to the GARAT sequence in a VP16-induced complex. Nevertheless, crosslinking of the POUS domain to the ATGC sequence is compatible with VP16-induced complex formation, demonstrating that VP16 can associate with multiple conformations of the Oct-1 POU domain.
DISCUSSION
We have probed the structure of the Oct-1 POU domain on different cis-regulatory sites and in a multiprotein complex. The structures of DNA-bound POU domains have been previously studied by x-ray crystallography (2, 3) and by mutagenesis (5, 6); crystallography, however, inherently provides only a static view of macromolecular interactions, and mutational analyses are indirect. Here, we have used protein–DNA photocrosslinking in a domain-specific manner to probe directly the conformation of the Oct-1 POU domain on DNA and in the VP16-induced complex. The photocrosslinking method we used (7) was particularly well suited for this analysis because the crosslinker is placed at a unique position on the protein and multiple sites of crosslinking to the DNA can be easily mapped with end-labeled probes. The results of our crosslinking studies of the two DNA-binding domains of the Oct-1 POU domain on different regulatory sites revealed the dynamic nature of Oct-1 binding to a single regulatory site—the (OCTA+)TAATGARAT site—and crosslinking interference revealed flexibility of the Oct-1 POU domain in VP16-induced complex formation.
We used three different Oct-1-binding sites in our studies—the high-affinity histone H2B octamer sequence and two VP16-responsive elements, an (OCTA−) and (OCTA+) TAATGARAT site—and each has revealed different arrangements of the Oct-1 POU domain as illustrated in Fig. 6. The pattern of crosslinking to the octamer site agrees with that predicted by the crystal structure (2), suggesting that, in solution, the POU-domain structure adopts the unique conformation elucidated crystallographically. Crosslinking to the (OCTA−)TAATGARAT site provides physical evidence for the unexpected structure of the Oct-1 POU domain revealed on this site by mutational analysis (6), with the POUS domain principally positioned over the GARAT element and thus to the opposite side of the POUH domain than when bound to the octamer sequence. The results of crosslinking to the (OCTA+)TAATGARAT site show that the Oct-1 POU domain can adopt either the octamer-like or (OCTA−)TAATGARAT-like conformation on the single binding site. These results lead to the view of a dynamic Oct-1 POU domain on an (OCTA+)TAATGARAT site: while the POUH domain remains fixed, the POUS domain swings from one POUS-domain-binding site to the next.
Figure 6.
Model for binding of the Oct-1 POU domain to three DNA targets, the octamer and the (OCTA−) and (OCTA+) TAATGARAT sites, highlighting conformational flexibility in Oct-1 POU DNA-binding domain arrangement.
The role of the Oct-1 POUS domain in VP16-induced complex formation—beyond its requirement for effective DNA binding by the Oct-1 POU domain (23)—is unclear, as is its position in the complex. It is known that the POUS domain is not essential for VP16-induced complex formation (16, 24); indeed, the POUS domain can be replaced by a completely different DNA-binding module, and the resulting chimeric “POU domain” can support efficient VP16-induced complex formation (25). In the native POU domain, however, the POUS domain may play a specific role, because mutations in the POUS domain can have a selective effect on VP16-induced complex formation on an (OCTA−)TAATGARAT site (6).
The crosslinking interference analysis described here with an (OCTA+)TAATGARAT site shows that, whether the POUS domain lies over the OCTA element 5′ of the POUH domain or the GARAT element 3′ of the POUH domain, the Oct-1 POU domain can associate with VP16. Consistent with these results even in a natural (OCTA+)TAATGARAT VP16-induced complex, the POUS domain can be crosslinked by UV irradiation to either the OCTA or GARAT sequences (unpublished results).
These results impact on our view of the VP16-induced complex. First, because the POUS domain need not be bound to the GARAT element to promote VP16-induced complex formation, the essential function of the GARAT element in VP16-induced complex formation (18–20) is probably not to provide a binding site for the Oct-1 POUS domain. Instead, it is more likely that the GARAT element is essential for VP16-induced complex formation because it provides an essential binding site for VP16. Second, the ability of the POUS domain to promote VP16-induced complex formation when crosslinked to the GARAT element suggests that VP16 and the POUS domain can co-occupy the GARAT element. We do not know how such co-occupancy may occur. The simple hypothesis that VP16 and the Oct-1 POUS domain bind to different faces of the DNA is probably not correct, because analysis of VP16-induced complex formation with inosine-substituted DNA probes suggests that VP16 and the Oct-1 POUS domain both recognize sequence determinants in the major groove of the DNA (13).
In conclusion, these studies reveal an extraordinary level of structural versatility and adaptability in how two macromolecules—a protein and DNA—interact with each other. As the DNA changes structure, as in the different sequences of the cis-regulatory sites, the protein—the Oct-1 POU domain—responds by also changing its structure.
Acknowledgments
We thank N. Hernandez, L. Joshua-Tor, B. Stillman, and W. Tansey for critical readings of the manuscript, and J. Duffy and P. Renna for artwork. P.S.P. thanks N. Hernandez for support and encouragement. These studies were supported by Public Health Service Grant CA13106 from the National Cancer Institute.
ABBREVIATION
- UC
unique cysteine
References
- 1.Herr W, Sturm R A, Clerc R G, Corcoran L M, Baltimore D, Sharp P A, Ingraham H A, Rosenfeld M G, Finney M, Ruvkun G, Horvitz H R. Genes Dev. 1988;2:1513–1516. doi: 10.1101/gad.2.12a.1513. [DOI] [PubMed] [Google Scholar]
- 2.Klemm J D, Rould M A, Aurora R, Herr W, Pabo C O. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 3.Jacobson E M, Li P, Leon-del-Rio A, Rosenfeld M G, Aggarwal A K. Genes Dev. 1997;11:198–212. doi: 10.1101/gad.11.2.198. [DOI] [PubMed] [Google Scholar]
- 4.Herr W, Cleary M A. Genes Dev. 1995;9:1679–1693. doi: 10.1101/gad.9.14.1679. [DOI] [PubMed] [Google Scholar]
- 5.Li P, He X, Gerrero M R, Mok M, Aggarwal A, Rosenfeld M G. Genes Dev. 1993;7:2483–2496. doi: 10.1101/gad.7.12b.2483. [DOI] [PubMed] [Google Scholar]
- 6.Cleary M A, Herr W. Mol Cell Biol. 1995;15:2090–2100. doi: 10.1128/mcb.15.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pendergrast P S, Chan Y, Ebright Y W, Ebright R H. Proc Natl Acad Sci USA. 1992;89:10287–10291. doi: 10.1073/pnas.89.21.10287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai J-S, Cleary M A, Herr W. Genes Dev. 1992;6:2058–2065. doi: 10.1101/gad.6.11.2058. [DOI] [PubMed] [Google Scholar]
- 9.Maxam A M, Gilbert W. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 10.Wilson A C, LaMarco K, Peterson M G, Herr W. Cell. 1993;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- 11.apRhys C M J, Ciufo D M, O’Neill E A, Kelly T J, Hayward G S. J Virol. 1989;63:2798–2812. doi: 10.1128/jvi.63.6.2798-2812.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker S, Hayes S, O’Hare P. Cell. 1994;79:841–852. doi: 10.1016/0092-8674(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 13.Cleary M A. Ph.D. thesis. Stony Brook: Univ. of New York; 1995. [Google Scholar]
- 14.Baumruker T, Sturm R, Herr W. Genes Dev. 1988;2:1400–1413. doi: 10.1101/gad.2.11.1400. [DOI] [PubMed] [Google Scholar]
- 15.Preston C M, Frame M C, Campbell M E M. Cell. 1988;52:425–434. doi: 10.1016/s0092-8674(88)80035-7. [DOI] [PubMed] [Google Scholar]
- 16.Kristie T M, Sharp P A. Genes Dev. 1990;4:2383–2396. doi: 10.1101/gad.4.12b.2383. [DOI] [PubMed] [Google Scholar]
- 17.Stern S, Herr W. Genes Dev. 1991;5:2555–2566. doi: 10.1101/gad.5.12b.2555. [DOI] [PubMed] [Google Scholar]
- 18.Gerster T, Roeder R G. Proc Natl Acad Sci USA. 1988;85:6347–6351. doi: 10.1073/pnas.85.17.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Hare P O, Goding C R, Haigh A. EMBO J. 1988;7:4231–4238. doi: 10.1002/j.1460-2075.1988.tb03320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kristie T M, LeBowitz J H, Sharp P A. EMBO J. 1989;8:4229–4238. doi: 10.1002/j.1460-2075.1989.tb08608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang C C, Herr W. Mol Cell Biol. 1996;16:2967–2976. doi: 10.1128/mcb.16.6.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Misra V, Walter S, Yang P, Hayes S, O’Hare P. Mol Cell Biol. 1996;16:4404–4413. doi: 10.1128/mcb.16.8.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sturm R A, Herr W. Nature (London) 1988;336:601–604. doi: 10.1038/336601a0. [DOI] [PubMed] [Google Scholar]
- 24.Katan M, Haigh A, Verrijzer C P, van der Vliet P C, O’Hare P. Nucleic Acids Res. 1990;18:6871–6880. doi: 10.1093/nar/18.23.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pomerantz J L, Pabo C O, Sharp P A. Proc Natl Acad Sci USA. 1995;92:9752–9756. doi: 10.1073/pnas.92.21.9752. [DOI] [PMC free article] [PubMed] [Google Scholar]