Abstract
Divalent metal ions, such as Mg2+, are generally required for tertiary structure formation in RNA. Although the role of Mg2+ binding in RNA-folding equilibria has been studied extensively, little is known about the role of Mg2+ in RNA-folding kinetics. In this paper, we explore the effect of Mg2+ on the rate-limiting step in the kinetic folding pathway of the Tetrahymena ribozyme. Analysis of these data reveals the presence of a Mg2+-stabilized kinetic trap that slows folding at higher Mg2+ concentrations. Thus, the Tetrahymena ribozyme folds with an optimal rate at 2 mM Mg2+, just above the concentration required for stable structure formation. These results suggest that thermodynamic and kinetic folding of RNA are cooptimized at a Mg2+ concentration that is sufficient to stabilize the folded form but low enough to avoid kinetic traps and misfolding.
The folding of RNA molecules into compact tertiary structures requires the presence of positively charged ions to allow close packing of the negatively charged phosphate backbone. Positively charged ions can interact with RNA in two ways: either through nonspecific electrostatic interactions or through binding to specific sites that stabilize a particular folding motif (1). Although considerable progress has been made in understanding equilibrium Mg2+ binding to a variety of RNAs, less is known about the role of Mg2+ in folding kinetics. Studies on the Mg2+-induced kinetic folding of tRNA (2–4), a pseudoknot derived from the Escherichia coli α-mRNA (5), the Tetrahymena group I ribozyme (6–14), the group II ribozyme (15), and RNase P (16–18) have revealed that folding occurs through a variety of steps in which the rates are both Mg2+-dependent and Mg2+-independent, with multiple intermediates occurring on the folding pathways.
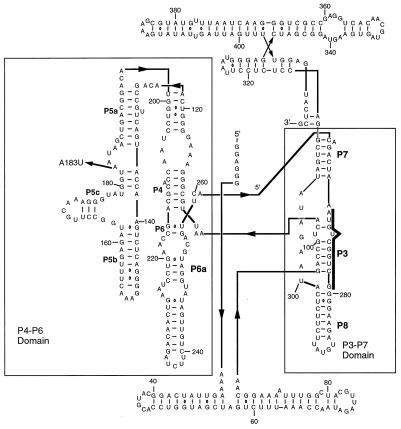
To elucidate principles of RNA-folding further, we study the Mg2+-induced kinetic folding pathway of the L-21 ScaI version of the Tetrahymena ribozyme. The ribozyme has a complex tertiary fold and is composed of two major domains. One domain is composed of helices P4, P5, and P6 (P refers to paired region), and the other domain is composed of helices P3, P7, and P8. The two domains are aligned by a triple helical scaffold (Fig. 1). These two domains make up the phylogenetically conserved catalytic core, which is stabilized by the nonconserved peripheral extensions P5abc, P2–2.1, and P9–P9.2. A model for the three-dimensional structure has been proposed (19, 20), and crystal structures have been solved for the P4–P6 domain (21) and for an RNA construct that contains the P4–P6 and P3–P9 domains and retains catalytic activity (22). Thermodynamic studies of ribozyme folding have led to a proposed equilibrium folding pathway in which folding proceeds hierarchically (23, 24). Secondary structure forms first and in the absence of divalent metal ions, and Mg2+ is required for formation of P5abc (0.6 mM), which is followed by the formation of the catalytic core at higher Mg2+ concentrations (0.9 mM). A kinetic folding pathway has also been proposed and is similar to the thermodynamic folding pathway (6, 9–14). The P5abc subdomain folds first (on the second time scale) perhaps collapsing around specifically bound Mg2+ ions (11, 25), followed by the P4–P6 domain, to form an intermediate (I2) in which P4–P6 is formed, but P3–P7 is not yet formed. A slow (on the minute time scale), Mg2+-independent unimolecular rearrangement follows that leads to the formation and/or stabilization of the P3–P7 domain. A subsequent rate-limiting step leads to the formation of the active ribozyme. Kinetic folding seems to be slowed by the presence of kinetic traps that are stabilized by native interactions and include nonnative secondary structure (7–9, 12). A series of mutations at positions that are important for stabilizing the P4–P6 domain accelerate the rate of P3–P7 folding (kP3–P7), and this acceleration was attributed to the destabilization of native structure in a kinetically trapped intermediate (12). Urea globally accelerates P3–P7 domain folding and/or overall folding in both mutant and wild-type ribozymes (7, 9), confirming the prevalence of kinetic traps.
Figure 1.
The L-21 Tetrahymena ribozyme. The major structural domains, P4–P6 and P3–P7, are delineated with rectangles. The P3 sequence targeted by complementary oligonucleotide probes in folding assays is highlighted with a bold line. The A183U mutation is indicated by an arrow on the left.
Here, we investigate the effect of Mg2+ on the overall folding rate (koverall) for the wild-type ribozyme and a mutant ribozyme (A183U) with an accelerated P3–P7 folding rate and demonstrate that koverall is decelerated by high Mg2+ concentrations (5–10 mM) in both ribozymes. Similar results were reported recently for the self-splicing version of the ribozyme, suggesting that this effect may be of general relevance (26). We propose that the deceleration is the result of a Mg2+-stabilized kinetic trap and show that an optimal [Mg2+] exists at which equilibrium folding is complete but Mg2+-stabilized kinetic traps are not populated. These results show that, although Mg2+ is required for formation of stable tertiary structure and can accelerate Mg2+-dependent folding steps, it can also stabilize kinetically trapped folding intermediates and slow folding. Therefore, the optimal [Mg2+] for fast, productive folding must balance these two effects.
Materials and Methods
Kinetic Oligonucleotide Hybridization Assay.
RNA transcripts were prepared, and folding experiments were performed as described (13). When folding was monitored at a temperature other than 37°C, the ribozyme was first equilibrated at 37°C for 3 min and then equilibrated at the folding temperature for an additional 3 min. When the measurement of 5-, 15-, and 30-s time points was necessary, each time point was measured in a separate reaction. Folding was initiated by adding 5 μl of annealed RNA to 5 μl of 2× folding buffer (1×: 50 mM Tris, pH 8.0/10 mM NaCl/1–10 mM MgCl2), and after the desired time, 5 μl was removed and added to 5 μl of 1× folding buffer containing oligonucleotide probe and RNase H. When folding was monitored at a Mg2+ concentration other than 10 mM, the Mg2+ concentration in the oligonucleotide/RNase H quench was adjusted such that the concentration in the quench was 10 mM.
Measurement of koverall.
koverall was measured by monitoring the fraction of substrate cleaved by the ribozyme after various folding times. The ribozyme was annealed (final concentration of 20 nM) in 15 μl of 10 mM Tris, pH 8.0/2 mM NaCl by heating to 90°C for 1 min and incubating at 37°C for 3 min. An equal volume of 2× folding buffer at the desired Mg2+ concentration was then added to initiate folding. Folding was allowed to proceed for various times before the endonuclease reaction was initiated by adding 15 μl of the folding reaction to 15 μl of substrate mix containing 5′ 32P-labeled substrate-CCCUCUAAAAA (final concentrations: 30 nM substrate, 0.5 mM GTP, 10 mM Mg2+, and 10 mM NaCl). When assays were performed at temperatures other than 37°C, the ribozyme was annealed by heating to 95°C for 1 min, incubated at 37°C for 3 min, and incubated at the folding temperature for 3 min. The endonuclease reactions were always performed for 1 min at 37°C.
Results
koverall for the Tetrahymena Ribozyme Is Decelerated by High Mg2+ Concentrations.
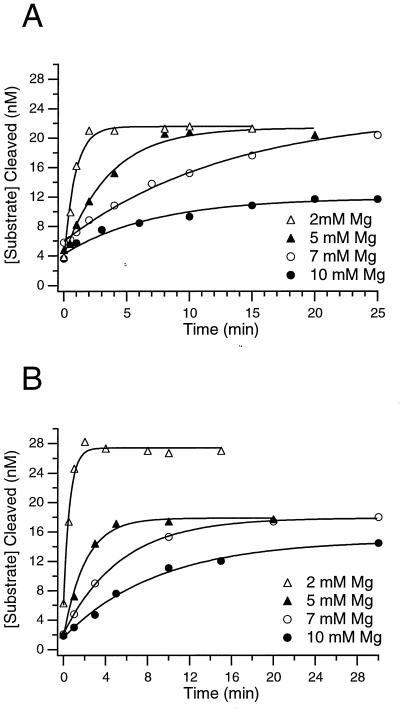
The rate of the slowest step on the Tetrahymena ribozyme folding pathway (koverall) can be measured by using a gain of activity assay (13). In this assay, the fraction of active ribozyme present at various times after addition of Mg2+ is determined by the magnitude of the kinetic burst that corresponds to the first turnover in a multiple-turnover cleavage reaction (Fig. 2). In previous experiments from our laboratory, the measured value for koverall was similar to the rate of P3–P7 folding (kP3–P7 = 0.6 min−1), leading to the proposal that P3–P7 folding is the slowest step on the folding pathway (13). However, recent experiments on a longer time scale provide a more accurate value for koverall that is slower (0.06 ± 0.01 min−1; ref. 9). Because koverall < kP3–P7, there must be an additional slow step on the folding pathway after P3–P7 formation.
Figure 2.
The value for koverall decreases with increasing [Mg2+] in both the wild-type and A183U mutant ribozymes. All curves are fit to single exponentials. (A) Wild-type ribozyme folding at various [Mg2+]: ●, 10 mM, koverall = 0.06 ± 0.01 min−1; ○, 7 mM, koverall = 0.07 ± 0.01 min−1; ▴, 5 mM, koverall = 0.19 ± 0.01 min−1; ▵, 2 mM, koverall = 1.10 ± 0.08 min−1. (B) A183U ribozyme folding at various [Mg2+]: ●, 10 mM, koverall = 0.10 ± 0.02 min−1; ○, 7 mM, koverall = 0.18 ± 0.06 min−1; ▴, 5 mM, koverall = 0.53 ± 0.02 min−1; ▵, 2 mM, koverall = 1.50 ± 0.02 min−1.
Although Mg2+ is required for the formation of the catalytic core of the ribozyme, none of the observed folding steps have had rates that depend on the [Mg2+]. Folding of the P3–P7 domain is essentially [Mg2+]-independent, and folding of the P4–P6 domain is too rapid to measure accurately with our assay. Experiments monitoring P4–P6 folding with time-resolved hydroxyl radical footprinting have not explored the Mg2+ dependence of folding rates (11). To determine whether the rate of the additional slow step was [Mg2+]-dependent, koverall was measured at Mg2+ concentrations between 1 and 10 mM. Surprisingly, the value of koverall increased dramatically with decreasing concentrations of Mg2+ (Figs. 2 and 3). The sigmoidal dependence of koverall on the [Mg2+] (Fig. 3) suggested that an off-pathway intermediate is populated by the cooperative binding of multiple Mg2+ ions; as the [Mg2+] is increased and the off-pathway intermediate becomes populated, there is a corresponding reduction in the population of on-pathway species, which necessarily reduces koverall. In agreement with this model, the data are well described by the Hill binding equation (Fig. 3, see legend for thermodynamic constants).
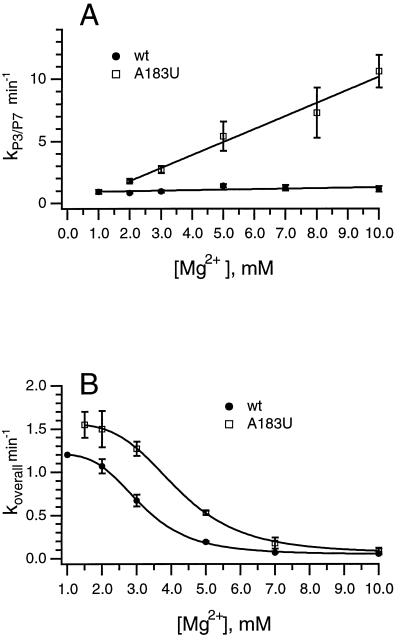
Figure 3.
[Mg2+] dependence of kP3–P7 for wild-type and A183U ribozymes. (A) [Mg2+] dependence of kP3–P7 at 37°C for the wild-type (wt, ●) and A183U (□) ribozymes as determined by kinetic oligonucleotide hybridization. (B) [Mg2+] dependence of koverall at 37°C for the wild-type (wt, ●) and A183U (□) ribozymes as determined by substrate cleavage. Curves were fit to the Hill binding equation. Thermodynamic constants: for wild type, [Mg2+]1/2 = 3.1 mM, and the Hill coefficient (n) = 4.3; for A183U, [Mg2+]1/2 = 4.2 mM, and n = 4.5.
In addition to affecting koverall for the wild-type ribozyme, the [Mg2+] affects the fraction of the ribozyme population that folds to the active structure. At [Mg2+] ≤ 7 mM, the entire ribozyme population folds to completion; however, at 10 mM Mg2+, only 80% of the ribozyme population is active after folding for 2 h (data not shown). The inactive population is either kinetically trapped or at equilibrium with the folded population, and it is unknown whether the inactive species is identical to the predominant Mg2+-stabilized intermediate described above. If these intermediates are in fact off-pathway, as Fig. 3 suggests, then their relevance to the productive folding pathway at low [Mg2+] is negligible; however, the data do not preclude possible on-pathway models (see Discussion).
Multiple [Mg2+]-Dependent Steps on the A183U Ribozyme Kinetic Folding Pathway.
Previous experiments measuring the temperature dependence of kP3–P7 in the mutant and wild-type ribozymes suggested that the nature of the rate-limiting step for P3–P7 formation was changed by the A183U mutation (9). Here, the effect of the mutation on the [Mg2+] dependence of kP3–P7 was examined. In the wild-type ribozyme, kP3–P7 is essentially [Mg2+]-independent (13), and escape from a kinetic trap is rate limiting (9, 12). Measurements of kP3–P7 showed a linear increase with [Mg2+] between 1 and 10 mM (Fig. 3A). The [Mg2+] dependence of kP3–P7 in the A183U mutant indicates that elimination of a kinetic trap can unmask additional elementary folding steps that are not observable in the wild type.
The A183U mutant was selected only for its ability to accelerate the rate of P3–P7 folding. Therefore, it is not surprising that similar values are observed for koverall of the wild-type and A183U ribozymes at 10 mM Mg2+ (9). To determine whether the similarity in rates is coincidental or whether the same step is being monitored, we measured the [Mg2+] dependence of koverall in both ribozymes, shown in Fig. 3B. In both ribozymes, koverall shows a sigmoidal dependence on [Mg2+], and the value for koverall decreases as [Mg2+] increases. The similarity of the two curves suggests that the same rate-limiting step is being monitored; however, koverall for the A183U mutant ribozyme is faster at all [Mg2+] tested. Additionally, the midpoint of the transition from fast to slow rates is shifted to a higher [Mg2+] in the A183U mutant (Fig. 3; for wild-type, [Mg2+]1/2 = 3.1 mM; for A183U, [Mg2+]1/2 = 4.2 mM). These results suggest the presence of a Mg2+-stabilized kinetic trap subsequent to P3–P7 formation on the folding pathway of both ribozymes. In addition, it seems that, in the A183U mutant, which has a destabilized P4–P6 domain, this trap is destabilized and requires a higher [Mg2+] than does the wild-type ribozyme to slow folding.
The Rate-Limiting Folding Step Is Determined by the Mg2+ Concentration.
In both the wild-type and the A183U mutant ribozymes at 10 mM Mg2+, there are two distinct slow steps on the folding pathway (kP3–P7 and koverall). However, when the Mg2+ concentration is lowered to 2 mM, koverall is increased and becomes similar to kP3–P7 in both ribozymes. To determine whether koverall and kP3–P7 are limited by the same folding step at 2 mM Mg2+, the temperature dependence of the folding rate constants was measured at 10 mM and 2 mM Mg2+, and the data were analyzed by using Eyring transition-state theory. Activation enthalpy (ΔH‡) and entropy (ΔS‡) values for folding (Fig. 4 and Table 1) were obtained by fitting plots of ln(kobs/T) vs. 1/T to Eq. 1, where T is the absolute temperature, kB is the Boltzmann constant, h is Planck’s constant, and R is the gas constant.
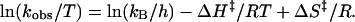 |
1 |
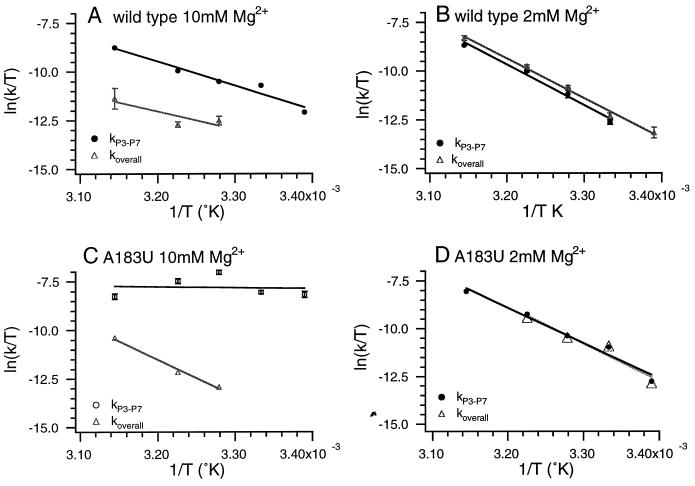
P3–P7 folding in the wild type at 10 mM Mg2+ is slow because of the presence of a kinetic trap with a large activation enthalpy (Table 1). The activation enthalpy for koverall is similar to that for kP3–P7, but the activation entropy is quite different (Table 1), providing additional evidence that different rate-limiting steps are involved. When the Mg2+ concentration is lowered to 2 mM, the temperature dependencies of koverall and kP3–P7 become nearly identical (Fig. 4B and Table 1), suggesting that, under these conditions, koverall and kP3–P7 are limited by the same folding step. Therefore, although at 10 mM Mg2+, koverall is much smaller than kP3–P7, at 2 mM Mg2+, koverall is increased and is limited by P3–P7 formation.
Figure 4.
Temperature dependence of kP3–P7 and koverall at 10 mM and 2 mM Mg2+. (A) Wild-type ribozyme; 10 mM Mg2+; kP3–P7 (●), koverall (▴). (B) Wild-type ribozyme; 2 mM Mg2+; kP3–P7 (●), koverall (▴). (C) A183U; 10 mM Mg2+; kP3–P7 (●), koverall (▴). (D) A183U; 2 mM Mg2+; kP3–P7 (●), koverall (▴). All points represent the average of at least three experiments, and the error bars are less than or equal to the size of the markers unless indicated. Data were fit to the Eyring equation, and values for ΔH‡ and ΔS‡ are listed in Table 1.
Table 1.
Activation enthalpy and entropy values from temperature-dependence experiments
| Folding rate | [Mg2+], mM | Wild type
|
A183U
|
||
|---|---|---|---|---|---|
| ΔH‡, kcal/mol | ΔS‡, cal/mol·K | ΔH‡, kcal/mol | ΔS‡, cal/mol·K | ||
| kP3–P7 | 10 | 23 ± 0.6 | 8.0 ± 1.9 | 3.5 ± 1.1 | −73 ± 3.5 |
| koverall | 10 | 18 ± 6.0 | −13 ± 12 | 38 ± 2.2 | 51 ± 7.0 |
| kP3–P7 | 2 | 41 ± 2.0 | 66 ± 7.0 | 37 ± 2.6 | 53 ± 8.5 |
| koverall | 2 | 40 ± 1.3 | 63 ± 4.1 | 39 ± 5.0 | 60 ± 16 |
In the A183U mutant ribozyme at 10 mM Mg2+, kP3–P7 and koverall have a much different temperature dependence (Fig. 4C and Table 1). kP3–P7 is temperature-independent, whereas koverall has a fairly steep temperature dependence. In contrast, at 2 mM Mg2+, the temperature dependencies of kP3–P7 and koverall are nearly identical (Fig. 4D and Table 1). Therefore, as with the wild-type ribozyme at high Mg2+ concentrations, kP3–P7 > koverall; as the Mg2+ concentration is reduced, koverall is increased, and P3–P7 formation becomes the rate-limiting step. The main differences between the two ribozymes are that kP3–P7 for the A183U mutant is also [Mg2+]-dependent and that kP3–P7 increases, whereas koverall decreases, as the [Mg2+] is increased.
In both the wild-type and A183U ribozymes at 2 mM Mg2+, kP3–P7 and koverall seem to be monitoring the same step, which is consolidation of the P3–P7 domain. Interestingly, the ΔH‡ and ΔS‡ are strikingly similar for the wild-type and A183U ribozymes and different from the ΔH‡ and ΔS‡ measured for P3–P7 folding at 10 mM Mg2+ for either ribozyme. These results suggest that, at low [Mg2+], there is a change in the rate-limiting step for P3–P7 folding. It is possible that, at lower Mg2+ concentrations, a step that occurs before P3–P7 formation becomes rate limiting in both the wild-type and mutant ribozymes, but what such a transition might represent structurally is not clear at this time.
Discussion
Multiple Kinetic Traps on the Folding Pathway of the Tetrahymena Ribozyme.
Large complex RNAs, like the Tetrahymena ribozyme, tend to have complex kinetic folding pathways with multiple intermediates. Are these intermediates required for folding, or are they the result of kinetic traps? One way to discriminate between these possibilities is to vary folding conditions such as temperature or ion concentration or to make mutations that may destabilize the folding intermediates and to see how these changes affect folding rates. In the present study, the effects of [Mg2+] and temperature on the rates of P3–P7 formation (kP3–P7) and folding to the catalytically active structure (koverall) were compared for the wild-type Tetrahymena ribozyme and the A183U mutant ribozyme. Reducing the [Mg2+] leads to an increase in the value of koverall and reveals the presence of an additional kinetic trap on the folding pathway of both ribozymes. Interestingly, this trap is stabilized by high [Mg2+]. Recent studies with the self-splicing Tetrahymena group I intron pre-RNA, from which the L-21 ScaI ribozyme is derived, indicate that folding is also faster at low [Mg2+] (26). Thus, these effects are not specific to the L-21 ScaI ribozyme construct and may have in vivo relevance. Because the effects of high [Mg2+] on koverall can be suppressed by Na+ and NH4+ (ref. 26; D.K.T. and J.R.W, unpublished observations), the optimal [Mg2+] for in vivo folding will likely reflect the intracellular monovalent ion concentration.
Generally, Mg2+ is thought to be important for rapid folding, because it can stabilize native folding motifs in RNA structure and allow packing of the negatively charged phosphate backbone; however, there are a number of ways in which Mg2+ could also slow folding. In principle, high Mg2+ concentrations can unfavorably reshape the folding pathway by changing the relative folding rates of individual structural motifs. Mg2+ can also slow folding by stabilizing kinetic traps. We propose that Mg2+ stabilizes an off-pathway intermediate that must unfold before completion of native structure formation. Our suggestion that the intermediate is off-pathway is based largely on the hyperbolic dependence of koverall on the [Mg2+], which is described by the Hill binding equation (Fig. 3). The folding pathway is complex and has different rate-limiting steps at high and low [Mg2+]; it is possible that the apparent hyperbolic relationship is fortuitous. As a result, our data do not preclude the possibility that the Mg2+-stabilized intermediate is on-pathway. In fact, recent studies on the self-splicing version of the ribozyme also showed an inverse dependence of koverall on the [Mg2+]. However, in these studies, the [Mg2+] dependence was exponential and not hyperbolic, leading to the proposal that Mg2+ slows folding by hyperstabilizing an on-pathway intermediate (26). The major difference between the two studies occurs at low [Mg2+], in which koverall is Mg2+-independent in our studies, but Pan et al. (26) observe further increases. These inconsistencies may be attributable to different RNA constructs or methodologies but most likely reflect the increased monovalent ion concentration used by Pan et al. (100 mM vs. 10 mM in our studies), which can have dramatic effects on RNA folding (ref. 26; D.K.T. and J.R.W, unpublished observations).
Why do kinetic traps predominate relatively late in the folding pathway? Kinetically trapped intermediates comprise both native and nonnative structures, and the native interactions can provide most of the stabilization energy (9, 12). Thus, as folding proceeds and native structure accumulates, it becomes increasingly likely that a nonnative structure will be stabilized. This sort of process is particularly probable in the ribozyme folding pathway in which the peripheral helices P2 and P2.1, which wrap around the catalytic core, and the P4–P6 domain fold early (11) and essentially form an RNA “cage” that surrounds the core. The stability of this cage likely restricts conformational searching in the interior and may explain the prevalence of kinetic traps.
Although the Mg2+-stabilized kinetic trap is populated in the A183U mutant, the koverall values are higher than those for wild type at all Mg2+ concentrations (Fig. 3). These increases can in part be explained by accompanying increases in kP3–P7 for A183U, which is partially or completely rate-limiting for koverall, particularly at low Mg2+ concentrations. In addition, the transition midpoint from high to low values of koverall is shifted to a higher [Mg2+] in A183U (Fig. 3), suggesting that the A183U mutation modestly destabilizes the kinetic trap. Thus, the A183U mutation seems to destabilize two kinetic traps on the folding pathway, suggesting that both traps are stabilized by native interactions that may limit conformational searching during formation of the interface between the P4–P6 and P3–P7 domains. A kinetic trap stabilized by domain–domain interactions has also been observed in the RNase P RNA, in which a misfolded structure results from presumably nonnative interactions at the domain interface (18). The role of native interactions in stabilizing this kinetic trap, however, is not clear.
The A183U mutation also changes the [Mg2+] dependence of kP3–P7. For the wild-type ribozyme, kP3–P7 is essentially [Mg2+]-independent. However, in the A183U mutant, the kP3–P7 increases linearly with increasing [Mg2+]. When the kinetic trap that slows P3–P7 folding in the wild-type ribozyme is removed, the effects of ion concentration on kP3–P7 are revealed.
An Optimal [Mg2+] for RNA Folding: Kinetic and Thermodynamic Folding Are Optimized at Similar Mg2+ Concentrations.
Mg2+ generally is required for the folding of complex RNA structures. The ability of high Mg2+ concentrations to compensate for mutations (24, 27) and for the absence of protein cofactors (28) shows the importance of Mg2+ for the stability of equilibrium structures, but how does Mg2+ affect the folding kinetics of large, complex RNA structures? Detailed studies on the Mg2+-induced kinetic folding of tRNA (2–4), a pseudoknot structure from the first ribosome initiation site in the E. coli α-mRNA (5), and the Tetrahymena ribozyme (6–14) give examples of three ways by which Mg2+ is known to affect RNA-folding kinetics: (i) Mg2+ binding can occur before a rate-limiting step, leading to a hyperbolic dependence on [Mg2+] (3); (ii) Mg2+ binding can occur after a rate-limiting step, leading to a Mg2+-independent rate (13); or (iii) Mg2+ binding can be coupled to folding, leading to the rate having a linear dependence on [Mg2+] (5). In tRNA, in the absence of Mg2+ and at low Na+ concentrations, an extended conformation with some nonnative base pairing interactions is adopted. When tertiary folding is initiated by the addition of Mg2+, folding occurs slowly, on the minute time scale, with two slow steps that involve rapid binding of Mg2+, followed by a slow conformational rearrangement. In this case, the folding rate increases hyperbolically with [Mg2+]. When folding is initiated from intermediate Na+ concentrations, folding is rapid (on the millisecond time scale), presumably because the correctly folded secondary structure is stabilized preferentially by the higher Na+ concentration. In a pseudoknot structure from the first ribosome initiation site in the E. coli α-mRNA, a Mg2+-dependent conformational change has been analyzed in which the rate-limiting step for the conformational change is coupled to Mg2+ binding and the rate of folding has a linear dependence on the [Mg2+]. Finally, in the folding of the Tetrahymena group I intron, although Mg2+ is required for the formation of the P3–P7 domain, the rate of P3–P7 folding is Mg2+-independent. Therefore, it has been proposed that P3–P7 forms via a conformational change, followed by rapid Mg2+ binding. In this study and that of Pan et al. (26), a fourth way in which Mg2+ can affect kinetic RNA folding is proposed: Mg2+ can slow folding by stabilizing folding intermediates.
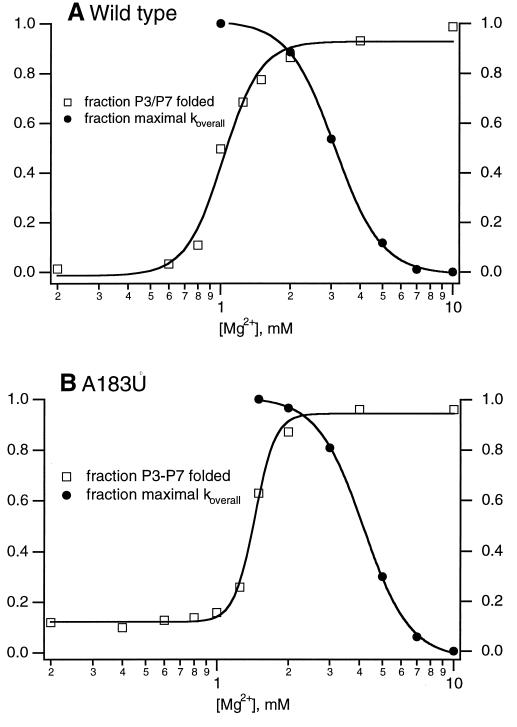
If Mg2+ is necessary for RNA folding but can also lead to slow folding, then there should be an optimal Mg2+ concentration at which native structure is stabilized but kinetic traps are largely avoided. When the temperature dependencies of kP3–P7 and koverall are measured at high and low [Mg2+], the effect of Mg2+ on the energy barriers of the two distinct folding steps can be measured. At 10 mM Mg2+, a concentration at which the value of koverall is small for both ribozymes, the activation enthalpy (ΔH‡) for koverall is different from the ΔH‡ for kP3–P7, and ΔH‡ for either step is different in the wild-type and the A183U ribozyme. These results show that kP3–P7 and koverall are distinct folding steps at high [Mg2+] and that the A183U mutation may alter multiple folding steps. However, when ΔH‡ is measured at 2 mM Mg2+, a concentration at which the value of koverall is similar to that of kP3–P7, the value of ΔH‡ is within error for both folding steps in both ribozymes. Therefore, at 2 mM Mg2+, kP3–P7 and koverall report on the same folding step in both the wild-type and A183U ribozymes, and 2 mM Mg2+ seems to be the optimal [Mg2+] for folding of the Tetrahymena ribozyme. When the fraction of P3–P7 folded at equilibrium and the value for koverall are plotted vs. [Mg2+], the optimal Mg2+ concentrations for both equilibrium and kinetic folding overlap (Fig. 5) for both the wild-type and A183U mutant ribozymes. These results show that there is a narrow window of [Mg2+] that allows stabilization of complex folds and avoids the population of kinetic traps.
Figure 5.
Cooptimal kinetic and thermodynamic folding occur over a narrow range of [Mg2+]. The fraction of P3–P7 folded at equilibrium and the fraction of the maximum koverall are coplotted as a function of [Mg2+]. In this range of [Mg2+], the fraction of P3–P7 folded accurately reflects the fraction of catalytically active ribozyme (D.K.T. and J.R.W., unpublished observation). (A) Wild-type ribozyme. (B) A183U mutant ribozyme. For thermodynamic constants for koverall vs. [Mg2+], see Fig. 3 legend. Thermodynamic constants for equilibrium P3–P7 folding: for wild type, [Mg2+]1/2 = 1.0 mM; for A183U, [Mg2+]1/2 = 1.4 mM.
Acknowledgments
We thank Paul Schimmel for critical comments on the manuscript. This work was supported by National Institutes of Health Grant GM53757.
Abbreviation
- Pn
paired region n
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Laing L, Gluick T, Draper D. J Mol Biol. 1994;237:577–587. doi: 10.1006/jmbi.1994.1256. [DOI] [PubMed] [Google Scholar]
- 2.Cole P E, Yang S K, Crothers D M. Biochemistry. 1972;11:4358–4368. doi: 10.1021/bi00773a024. [DOI] [PubMed] [Google Scholar]
- 3.Lynch D C, Schimmel P R. Biochemistry. 1974;13:1841–1852. doi: 10.1021/bi00706a012. [DOI] [PubMed] [Google Scholar]
- 4.Stein A, Crothers D M. Biochemistry. 1976;15:160–168. doi: 10.1021/bi00646a025. [DOI] [PubMed] [Google Scholar]
- 5.Gluick T C, Gerstner R B, Draper D E. J Mol Biol. 1997;270:451–463. doi: 10.1006/jmbi.1997.1119. [DOI] [PubMed] [Google Scholar]
- 6.Downs W D, Cech T R. RNA. 1996;2:718–732. [PMC free article] [PubMed] [Google Scholar]
- 7.Pan J, Thirumalai D, Woodson S A. J Mol Biol. 1997;293:7–13. doi: 10.1006/jmbi.1997.1311. [DOI] [PubMed] [Google Scholar]
- 8.Pan J, Woodson S. J Mol Biol. 1998;280:297–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- 9.Rook M S, Treiber D K, Williamson J R. J Mol Biol. 1998;281:609–620. doi: 10.1006/jmbi.1998.1960. [DOI] [PubMed] [Google Scholar]
- 10.Sclavi B, Woodson S, Sullivan M, Chance M R, Brenowitz M. J Mol Biol. 1997;266:144–159. doi: 10.1006/jmbi.1996.0775. [DOI] [PubMed] [Google Scholar]
- 11.Sclavi B, Brenowitz M, Woodson S. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 12.Treiber D K, Rook M S, Zarrinkar P, Williamson J R. Science. 1998;279:1943–1946. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- 13.Zarrinkar P P, Williamson J R. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- 14.Zarrinkar P P, Williamson J R. Nat Struct Biol. 1996;3:432–438. doi: 10.1038/nsb0596-432. [DOI] [PubMed] [Google Scholar]
- 15.Qin P Z, Pyle A M. Biochemistry. 1997;36:4718–4730. doi: 10.1021/bi962665c. [DOI] [PubMed] [Google Scholar]
- 16.Zarrinkar P P, Wang J, Williamson J R. RNA. 1996;2:564–573. [PMC free article] [PubMed] [Google Scholar]
- 17.Pan T, Sosnick T. Nat Struct Biol. 1997;14:931–938. doi: 10.1038/nsb1197-931. [DOI] [PubMed] [Google Scholar]
- 18.Pan T, Fang X, Sosnick T. J Mol Biol. 1999;286:721–731. doi: 10.1006/jmbi.1998.2516. [DOI] [PubMed] [Google Scholar]
- 19.Lehnert V, Jaeger L, Michel F, Westhof E. Chem Biol. 1996;3:993–1009. doi: 10.1016/s1074-5521(96)90166-0. [DOI] [PubMed] [Google Scholar]
- 20.Michel F, Westhof E. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 21.Cate J H, Gooding A R, Podell E, Zhou K, Golden B L, Kundrot C E, Cech T R, Doudna J A. Science. 1996;274:1678–1695. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 22.Golden B L, Gooding A R, Podell E R, Cech T R. Science. 1998;282:259–264. doi: 10.1126/science.282.5387.259. [DOI] [PubMed] [Google Scholar]
- 23.Celander D W, Cech T R. Science. 1991;251:401–407. doi: 10.1126/science.1989074. [DOI] [PubMed] [Google Scholar]
- 24.Laggerbauer B, Murphy F L, Cech T R. EMBO J. 1994;13:2669–2676. doi: 10.1002/j.1460-2075.1994.tb06557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cate J H, Hanna R L, Doudna J A. Nat Struct Biol. 1997;4:553–558. doi: 10.1038/nsb0797-553. [DOI] [PubMed] [Google Scholar]
- 26.Pan J, Thirumalai D, Woodson S A. Proc Natl Acad Sci USA. 1999;96:6149–6154. doi: 10.1073/pnas.96.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy F L, Cech T R. J Mol Biol. 1994;236:49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- 28.Weeks K M, Cech T R. Science. 1996;271:345–348. doi: 10.1126/science.271.5247.345. [DOI] [PubMed] [Google Scholar]