Abstract
MLN64 is a protein that is highly expressed in certain breast carcinomas. The C terminus of MLN64 shares significant homology with the steroidogenic acute regulatory protein (StAR), which plays a key role in steroid hormone biosynthesis by enhancing the intramitochondrial translocation of cholesterol to the cholesterol side-chain cleavage enzyme. We tested the ability of MLN64 to stimulate steroidogenesis by using COS-1 cells cotransfected with plasmids expressing the human cholesterol side-chain cleavage enzyme system and wild-type and mutant MLN64 proteins. Wild-type MLN64 increased pregnenolone secretion in this system 2-fold. The steroidogenic activity of MLN64 was found to reside in the C terminus of the protein, because constructs from which the C-terminal StAR homology domain was deleted had no steroidogenic activity. In contrast, removal of N-terminal sequences increased MLN64’s steroidogenesis-enhancing activity. MLN64 mRNA was found in many human tissues, including the placenta and brain, which synthesize steroid hormones but do not express StAR. Western blot analysis revealed the presence of lower molecular weight immunoreactive MLN64 species that contain the C-terminal sequences in human tissues. Homologs of both MLN64 and StAR were identified in Caenorhabditis elegans, indicating that the two proteins are ancient. Mutations that inactivate StAR were correlated with amino acid residues that are identical or similar among StAR and MLN64, indicating that conserved motifs are important for steroidogenic activity. We conclude that MLN64 stimulates steroidogenesis by virtue of its homology to StAR.
The rate-limiting step in steroidogenesis is the conversion of cholesterol into pregnenolone, a side-chain cleavage reaction that takes place in the mitochondrial inner membrane catalyzed by cytochrome P450scc and its associated electron-transport chain (1). In the adrenal cortex and gonads, the translocation of cholesterol substrate to P450scc is under the control of corticotropin (ACTH) and luteinizing hormone (LH), respectively, through the intermediacy of the steroidogenic acute regulatory protein (StAR) (2). Individuals who are homozygous for mutations that inactivate StAR have a marked impairment in adrenal and gonadal steroidogenesis (3, 4). The C-terminal domains of StAR are essential for its steroidogenic activity, since deletion of C-terminal sequences, but not N-terminal sequences, ablates steroidogenesis-enhancing activity (5). Although StAR appears to be critical for steroidogenesis in the adrenals and gonads, tissues that do not express the StAR gene, including the placenta, synthesize large amounts of pregnenolone (6). Thus, there must be StAR-independent mechanisms for movement of cholesterol to the P450scc enzyme.
MLN64, a gene product of unknown function, was found to be highly expressed in certain breast cancers (7). Analysis of the deduced amino acid sequence of the mouse (GenBank accession no. X82457) and human (GenBank accession no. X80198) MLN64 proteins revealed that the N terminus of MLN64 contains four potential transmembrane domains and a C terminus with striking homologies to StAR (7). The latter finding raised the possibility that MLN64 has steroidogenic activity, an idea that is intriguing in view of recent evidence that certain breast cancers express P450scc and 3β-hydroxysteroid dehydrogenase, potentially rendering them competent to produce steroid hormones that could affect the growth of hormone-responsive tumors (8). The aims of the present study were to determine if MLN64 is capable of enhancing steroidogenesis and if the structural similarities between MLN64 and StAR delineate residues in StAR that are essential for its steroidogenic activity.
MATERIALS AND METHODS
cDNAs and Plasmids.
A full-length MLN64 cDNA was prepared by reverse transcriptase PCR from human placental RNA and cloned into pCMV5. cDNAs encoding the wild-type MLN64 and deletion mutants were cloned into the pAT4 expression vector containing the F domain of the human estrogen receptor to epitope tag the proteins (7). A plasmid expressing human P450scc and its electron transport chain as a fusion protein (9) was generously provided by Walter L. Miller (University of California, San Francisco). The human StAR cDNA in pCMV5 has been previously described (6). Mutations in the human StAR cDNA were produced by site-directed mutagenesis as previously described (5).
Assessment of Steroidogenic Activity.
COS-1 cells were transfected with the plasmid expressing the cholesterol side-chain cleavage enzyme and the plasmids expressing MLN64, MLN64 mutants, human wild-type StAR or StAR mutants, or the empty vectors by using Lipofectamine as previously described (3, 4, 6). Cultures were incubated in the absence or presence of (22R)-22-hydroxycholesterol (5 μg/ml) for 36 h. Pregnenolone production was determined by radioimmunoassay using an antibody kindly provided by Charles Strott (National Institutes of Health) (6).
Experiments were performed on at least three separate occasions with three dishes in each treatment group. Pregnenolone production in the absence of exogenous hydroxysterol was taken to reflect the steroidogenic activity of the expressed proteins. The normalization of values to pregnenolone production in the presence of hydroxysterol, which reflects the total cholesterol side-chain cleavage activity, controls for variations in expression of this enzyme (4).
Western Blotting.
To verify expression of MLN64, the mutant MLN64 proteins, StAR, and StAR mutants, whole-cell extracts of transfected COS-1 cells were subjected to Western blot analysis using standard methods (5). Extracts of human placenta, BeWo choriocarcinoma cells, and fetal adrenal cortex were also examined. A monoclonal antibody (2BE2F4) raised against a peptide corresponding to 16 amino acid residues in the C terminus of MLN64 (amino acid residues 369–384) was used to detect MLN64 (7). A polyclonal antibody was also generated in rabbits against the same peptide. Monoclonal antibody F3A6 was used to detect the epitope-tagged fusion proteins (7). A polyclonal antibody generated against human recombinant StAR was used to detect StAR in COS-1 cell extracts (10).
Northern Blotting.
Membranes containing poly(A)+ RNA isolated from human adult and fetal tissues were purchased from CLONTECH and probed with an MLN64 cDNA as previously described (6). Total RNA was isolated from human granulosa-lutein and thecal cells and subjected to Northern blotting. A housekeeping gene, RLP 32 cDNA, was used to probe blots to establish loading of the lanes.
Identification of MLN64 and StAR Homologs.
The align program (11) was used to quantify the similarity between protein sequences. For the analysis reported here, 10,000 random permutations were used for the statistical analysis and the Dayhoff matrix was used wth a bias of 6 and a gap penalty of 8.
The Feng–Doolittle algorithm (12) was used to construct the phylogenetic tree of MLN64, StAR, and their homologs. The method of Fitch and Margoliash (13) was then used to obtain the branching order for the sequences. Branch lengths are calculated by a linear regression analysis of the best fit of the pairwise distances and the branching order. The lengths of the branches are proportional to the relative distance between the sequences.
Statistical Analysis.
Values presented are means ± SE of the indicated number of separate experiments. The paired t test or Student–Newman–Keuls test was used to determine significant differences among treatment groups, using P < 0.05 as the level of significance.
RESULTS
MLN64 Stimulates Steroidogenesis.
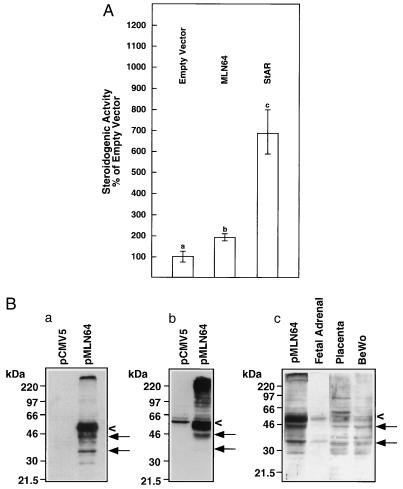
To test the steroidogenesis-enhancing activity of MLN64, we transfected monkey kidney COS-1 cells with the human cholesterol side-chain cleavage system and MLN64 and measured pregnenolone secretion. MLN64 caused an approximately 2-fold increase in pregnenolone secretion over COS-1 cells transfected with the cholesterol side-chain cleavage enzyme and empty plasmid vector (P < 0.01) (Fig. 1A). In contrast, human StAR increased pregnenolone production by nearly 7-fold. Thus, MLN64 demonstrated approximately 28% of the steroidogenic activity of StAR in this system.
Figure 1.
(A) Relative stimulation of pregnenolone production by MLN64 in COS-1 cells expressing the human cholesterol side-chain cleavage enzyme. COS-1 cells were transfected with a plasmid expressing the human cholesterol side-chain cleavage enzyme and the indicated plasmid. Relative steroidogenic activity (means ± SE) assayed as pregnenolone production normalized to the conversion of (22R)-22-hydroxycholesterol to pregnenolone is presented, taking the empty vector control as 100%. Values with a different letter are significantly different (P < 0.01 by the paired t test). (B) Expression of MLN64 in COS-1 cells and human tissues. (a) Western blot analysis was carried out on extracts of COS-1 cells transfected with empty vector or vector containing wild-type MLN64 cDNA. A monoclonal antibody recognizing an epitope in the MLN64 C terminus was used to probe the blot. A major band around 50 kDa is detected (<). Smaller immunoreactive proteins present in lesser quantities (←) may represent proteolytically processed MLN64. (b) Western blot analysis of transfected COS-1 cells probed with a polyclonal antibody raised against a C-terminal MLN64 peptide. (c) Western blot of extracts of transfected COS-1 cells, fetal adrenal cortex, placenta, and BeWo cells (50 μg of protein per lane) probed with a polyclonal antibody to the MLN64 C terminus.
MLN64 expressed in the COS-1 cells was detected as a predominant 50-kDa protein when a specific monoclonal antibody was used. Lower molecular mass proteins of about 42 and 33 kDa were also detected, probably representing proteolytic cleavage products (Fig. 1B a). MLN64 was not detected in the monkey kidney COS-1 cells transfected with empty vector, presumably because the monkey protein is not recognized by the monoclonal anti-MLN64 antibody. However, a polyclonal antibody detected MLN64 in the COS-1 cells transfected with empty vector, but at markedly lower levels than in cells transfected with the MLN64-expression vector (Fig. 1B b). To determine if the 50-kDa MLN64 and the smaller immunoreactive forms of the protein are present in tissues in vivo, we performed Western blot analysis on extracts of human placenta, BeWo choriocarcinoma cells, and fetal adrenal cortex. Fetal adrenal cortex contained 50-kDa and 33-kDa MLN64 species, whereas the placenta and BeWo extracts contained multiple lower molecular mass forms, including those of 42 and 33 kDa (Fig. 1B c). It should be noted that the anti-MLN64 antibodies do not crossreact with human StAR, nor do our anti-human StAR antibodies recognize MLN64 or the lower molecular mass MLN64 species.
The C Terminus of MLN64 Containing Sequences Homologous to StAR Is Required for Steroidogenic Activity.
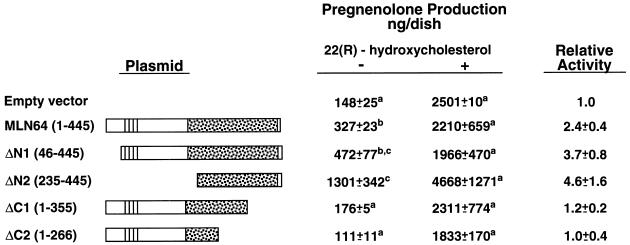
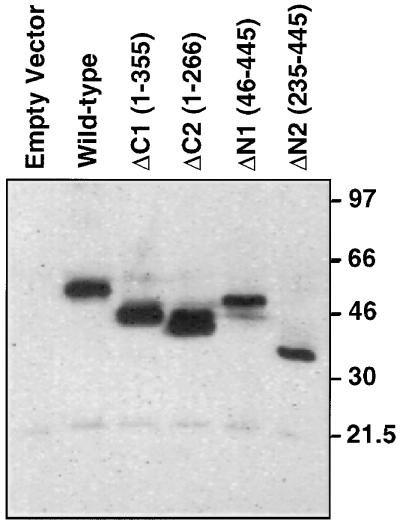
To identify the domains of MLN64 that are necessary for steroidogenic activity, we examined mutant proteins in which either N-terminal or C-terminal sequences were deleted (Fig. 2). Deletions of the C terminus containing sequences homologous to StAR resulted in the complete loss of steroidogenic activity. In contrast, removal of N-terminal sequences, which contain the putative N-terminal microsomal targeting sequence and the putative transmembrane domains, increased steroidogenesis-enhancing activity to levels that are about two-thirds of the level observed with wild-type human StAR (ΔN2 relative steroidogenic activity: 4.6; wild-type StAR relative steroidogenic activity: 6.7). Each of the mutant proteins was detectable in the transfected COS-1 cells at its expected molecular mass (Fig. 3).
Figure 2.
Identification of the domain in MLN64 responsible for steroidogenesis-enhancing activity. COS-1 cells were transfected with a plasmid expressing the human cholesterol side-chain cleavage enzyme and the indicated plasmids. Cells were cultured in the absence (−) or presence (+) of (22R)-22-hydroxycholesterol (5 μg/ml). Pregnenolone production was determined by radioimmunoassay. Values presented are means ± SE from four separate experiments in which each treatment group contained triplicate cultures. Relative steroidogenic activities of the constructs are calculated normalizing pregnenolone production to total cholesterol side-chain cleavage activity as determined in the presence of (22R)-22-hydroxycholesterol with the normalized data expressed relative to the empty vector control. Values marked with a different superscript letter are significantly different from others in the column (P < 0.05) by the Student–Newman–Keuls test. Wild-type human StAR was transfected in some experiments. The mean relative steroidogenic activity of StAR was 6.7. The region of MLN64 homologous to StAR is stippled. The four putative transmembrane domains are indicated by solid lines.
Figure 3.
Expression of MLN64 deletion mutants in COS-1 cells. COS-1 cells were transfected with plasmids harboring the indicated MLN64 constructs in pAT4, each containing the F domain of the human estrogen receptor in the N terminus to tag the expressed protein so that it could be recognized by a monoclonal antibody to the F domain. The numbers in parentheses refer to the amino acid residues of MLN64 encoded by the construct.
MLN64 Is Widely Expressed in Human Tissues.
To determine the pattern of expression of MLN64, we analyzed Northern blots containing poly(A)+ RNA from various human tissues (Fig. 4). The major 2.1-kb and minor 3.4-kb MLN64 mRNAs were detected in all adult tissues. Of particular note was the expression of MLN64 mRNA in placenta and brain, tissues that produce steroid hormones. Human fetal brain, lung, liver, and kidney contained the 2.1- and 3.4-kb mRNAs and some larger transcripts. Northern analysis also revealed the presence of the 2.1-kb MLN64 message in human granulosa and thecal cells (data not shown).
Figure 4.
Expression of MLN64 mRNA in various human tissues. Northern blots containing poly(A)+ RNA (2 μg per lane) extracted from the indicated human adult (A) or human fetal (B) tissues were probed with a human MLN64 cDNA (Upper) or an RLP 32 cDNA (Lower).
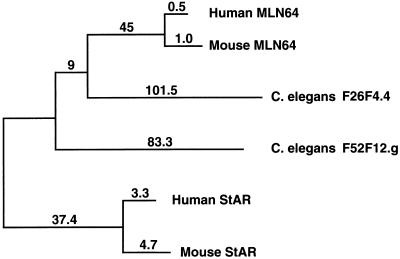
Homologs of MLN64 and StAR in Caenorhabditis elegans: Evolutionary Relationship of StAR and MLN64.
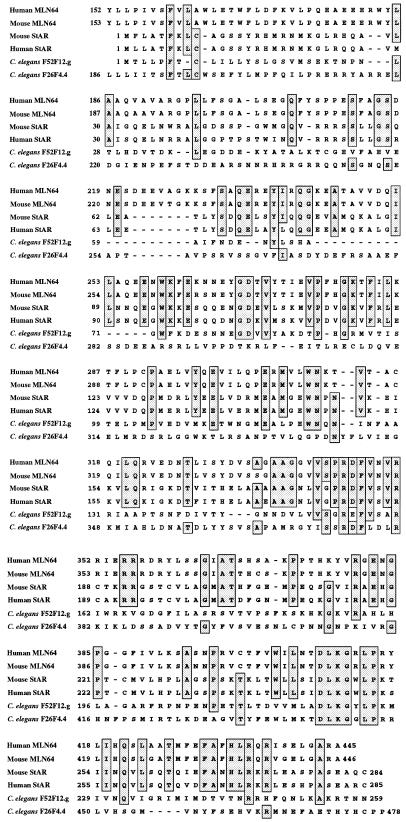
We previously reported that human MLN64 has a 478-residue putative homolog, F26F4.4, in C. elegans (7). To investigate further the origins of StAR, we did a series of blast searches of GenBank, which identified a 259-residue protein, F52F12.g (GenBank accession no. Z83228), that is homologous to human StAR based on an align analysis, which yielded a comparison score for the two proteins of 10.4 standard deviations higher than that for 10,000 comparisons of their randomized sequences. The probability of getting such a score by chance is 10−23, strongly supporting the common ancestry of C. elegans F52F12.g and StAR. Interestingly, the comparison of F52F12.g with the StAR domain on human MLN64 and C. elegans F26F4.4 yields scores of 6.3 and 5 standard deviations, respectively, indicating that F52F12.g is closer to StAR than to MLN64 and that the StAR domains on these two proteins have diverged substantially. Fig. 5 presents a multiple alignment of mouse and human MLN64, StAR, and the two C. elegans proteins.
Figure 5.
Multiple alignment of mouse and human MLN64 and StAR and their C. elegans homologs. Residues identical in at least four of the proteins are boxed.
We further investigated the evolutionary relationship between the mammalian StAR proteins and their C. elegans homologs by constructing a phylogenetic tree using the Feng–Doolittle algorithm. As seen in Fig. 6, the two mammalian StAR proteins cluster on one branch, while the two mammalian MLN64 proteins cluster on another branch. The branch containing C. elegans F52F12.g is closer to the StAR proteins than to the MLN64 proteins, in agreement with the align scores.
Figure 6.
Phylogenetic analysis of MLN64, StAR, and their C. elegans homologs. The numbers on the branches and lengths of the branches are proportional to the relative distance between sequences, which indicates the relationship of the sequences to each other.
The identification of the C. elegans 259-residue putative homolog of StAR and the 478-residue protein with a domain putatively homologous to StAR suggests that there was an ancient fusion of at least two genes to form MLN64. Considering that StAR and MLN64 arose over 600 million years ago, it is interesting that there are many positions with identical residues in either five or six StAR domains of the mammalian and invertebrate proteins (Fig. 5). As described below, the conserved motifs appear to be functionally important.
Mutations in Residues That Are Identical or Similar in StAR and MLN64 Result in the Loss of Steroidogenic Activity.
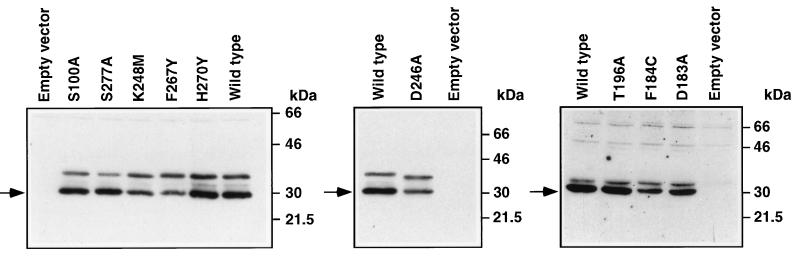
To determine if motifs conserved in StAR and MLN64 are important for functional activity, we categorized mutations that we previously described (3, 4, 5, 10) as well as nine newly created mutations according to whether they affected the steroidogenic activity of human StAR and if they were in residues that were identical, similar, or not conserved between StAR and MLN64. All but one of the mutations (H270Y) in human StAR residues that were identical or similar in MLN64 resulted in loss of steroidogenic activity (Table 1). The loss of activity ranged from complete to partial (100% to 19% inactivation). Although mutations in some residues that are conserved in StAR, MLN64, and the C. elegans proteins caused complete inactivation of StAR (e.g., R182L, ΔR272), mutations in other conserved residues (e.g., D246A and K248M) caused a lesser reduction in functional activity. In contrast, mutations of four different nonconserved residues had no significant impact on steroidogenesis-enhancing activity as did a four amino acid deletion of nonconserved residues in the C terminus. Western blot analyses of COS-1 cells transfected with constructs containing the newly created mutations that resulted in loss of steroidogenic activity demonstrated that the mutant proteins were expressed, with detection of the mature protein, indicating that the mutant StAR proteins can undergo mitochondrial import and processing (Fig. 7).
Table 1.
Mutations in residues that are identical or similar in StAR and MLN64 cause loss of steroidogenic activity in human StAR
| Mutation | Residue type | Relative steroidogenic activity |
|---|---|---|
| S100A | N | 86 ± 5 |
| E169G | S | 13 ± 1* |
| E169K | S | 14 ± 2* |
| R182L | Id | 11 ± 1* |
| D183A | Id | 44 ± 5 |
| F184C | Id | 56 ± 6 |
| T196A | N | 119 ± 18 |
| A218V | S | 16 ± 4* |
| M225T | S | 49 ± 9‡ |
| D246A | Id | 63 ± 8 |
| K248M | Id | 51 ± 3 |
| F267Y | Id | 65 ± 1 |
| H270Y | Id | 81 ± 7 |
| ΔR272 | Id | 11 ± 1* |
| L275P | S | 24 ± 5* |
| S277A | N | 90 ± 16 |
| C285S | N | 105 ± 11† |
| ΔC285 | N | 110 ± 8† |
| ΔS282, ΔE283, ΔR284, ΔC285 | N | 102 ± 7† |
| Empty plasmid | 14 ± 2 |
COS-1 cells were transfected with the human cholesterol side-chain cleavage system, and expression plasmids for wild-type StAR, the indicated mutations, or empty plasmid. Steroidogenic activity was assessed by measuring pregnenolone production and is expressed relative to wild-type StAR as 100% normalized to (22R)-22-hydroxycholesterol metabolism as previously described (4). Id, identical; N, not conserved; S, similar. The footnote symbols indicate results taken from our previous publications: ∗, ref. 4; †, ref. 5; and ‡, ref. 10. All other values represent newly created mutations. Values are means ± SE from at least three separate experiments.
Figure 7.
Western blot analysis of StAR in COS-1 cells transfected with the indicated plasmids. Whole-cell extracts of COS-1 cells were analyzed using a polyclonal antibody raised against recombinant human StAR. Arrows indicate the mature form of StAR at ≈30 kDa. The higher molecular mass immunoreactive proteins represent StAR preproteins.
DISCUSSION
MLN64, a 50-kDa protein (7), has significant sequence similarity to StAR, and the present study demonstrates that MLN64 and StAR share steroidogenic activity. Moreover, removal of the C-terminal region of MLN64 that is homologous to StAR results in the complete loss of steroidogenic activity. Removal of the N-terminal domain of MLN64 (ΔN2 mutation) affects the distribution of the protein, changing the localization from endoplasmic reticulum-like membranous structures in the cytoplasm to a more uniform distribution throughout the cytoplasm (ref. 7 and unpublished observations). As with StAR, removal of the N-terminal sequences does not negatively affect steroidogenic activity. Indeed, steroidogenesis-enhancing activity was increased with the removal of the first 234 residues of N-terminal sequence. Our Western blot analysis of wild-type MLN64 expressed in COS-1 cells with a monoclonal antibody directed against a C-terminal epitope suggested the presence of lower molecular mass MLN64 species that may reflect the proteolytic processing of the protein, presumably the removal of N-terminal sequences. Lower molecular mass MLN64 proteins were also detected in human placenta, choriocarcinoma cells, and fetal adrenal cortex, suggesting that the lower molecular mass forms are not an artifact of the transfected COS-1 cell system. The ≈33-kDa MLN64 protein detected in Western blots of transfected COS-1 cells and human tissues could correspond to a MLN64 fragment similar to that encoded by the ΔN2 mutant which displayed the greatest steroidogenic activity. It remains to be determined if proteolytic processing is required for the production of a steroidogenically active MLN64 protein. Such a scenario would parallel the release of active transcription factors involved in regulating cholesterol and fatty acid homeostasis (SREBPs) through proteolytic cleavage of endoplasmic reticulum-associated precursor proteins (14).
The amino acid residues that are identical or similar in the MLN64 and StAR C termini appear to be critical for steroidogenic activity, because mutation of these residues results in the loss of StAR’s steroidogenic activity. However, if the mutated residues are not conserved between these proteins, steroidogenic activity is retained. The findings on the new mutations in the present study are consonant with the recent genetic analysis of congenital lipoid adrenal hyperplasia (3, 4, 10, 15). Mutations that truncate the C terminus of StAR destroy the functional activity of StAR, and all known mutations that cause amino acid replacements that inactivate the protein are clustered in exons 5–7, which encode the StAR C terminus.
Some cells that do not express StAR, like the human placental syncytiotrophoblast and glia, are capable of producing pregnenolone (6). The basis for this StAR-independent steroid production is not understood. The finding that MLN64 is widely expressed in human tissues, including the placenta and brain, raises the possibility that StAR-independent steroidogenesis may be due, in part, to the presence of MLN64.
The abundant expression of MLN64 in certain breast cancers is of interest. Some breast cancer cells express cholesterol side-chain cleavage enzyme and 3β-hydroxysteroid dehydrogenase (8). The presence of MLN64 in these tumor cells could potentially enhance the formation of pregnenolone, which might be converted into progesterone and possibly other steroids. Although aromatase has been detected in breast cancers, P450c17 has not been found (8). Thus, it is unlikely that de novo synthesis of estrogens from cholesterol could take place in breast tumors. However, the local production of progestins through the action of the enzymes involved in the early steps of steroidogenesis could influence the behavior of tumor cells, as certain breast cancers express progesterone receptors. It is interesting to note that regions of chromosome 8 and 17 are frequently amplified in breast cancers. The 8 p11–p12 (16) and 17 q12–q21 (17) amplicons contain the StAR (6) and MLN64 genes (7), respectively. Whether this is a coincidence or a functionally significant association remains to be explored in studies correlating expression of MLN64 in breast cancers with the steroidogenic potential of the tumors.
The expression of MLN64 in tissues which do not produce steroid hormones is not unexpected, since mitochondria of many tissues possess other P450 enzymes involved in cholesterol oxidation, including P450c27 (18), which produces hydroxysterols that are precursors of bile acids as well as potent controllers of cholesterol synthesis and low density lipoprotein receptor expression (19).
The present findings do not shed light on the mechanisms by which proteins that contain StAR homology domains stimulate mitochondrial cholesterol metabolism. However, the discovery that the removal of N-terminal sequences that direct import of StAR into mitochondria does not impair steroidogenic activity strongly suggests that StAR acts either outside of mitochondria (i.e., in the cytoplasm) or on the mitochondrial surface membranes (5). The fact that MLN64 and the N-terminal truncated mutants of MLN64 that have increased steroidogenic activity contain no obvious mitochondrial import sequences is consistent with this idea.
In conclusion, the present experiments reveal a functional activity of a StAR homolog, MLN64. The ability of MLN64, and particularly its C terminus, to stimulate steroidogenesis may reflect only one of this protein’s roles; the others remain to be determined. We suggest that proteins containing StAR motifs may play key roles in controlling the intra-organelle movement of cholesterol.
Acknowledgments
We thank Dr. Walter L. Miller (University of California, San Francisco) for the gift of the expression plasmid for the human cholesterol side-chain cleavage system and Dr. Charles Strott (National Institutes of Health) for providing the anti-pregnenolone antiserum used in these studies. This work was supported by U.S. Public Health Service Grant HD-06274 (J.F.S.) and the Medical Scientist Training Program (C.B.K.), funds from the Institut National de la Santé et de la Recherche Médicale (M.C.R.), the Centre National de la Recherche Scientifique (M.C.R.), the Bristol–Myers Squibb Pharmaceutical Research Institute (M.C.R.), a grant from the Lalor Foundation (F.A.), and the Ligue National Contre le Cancer (C.M.L.).
ABBREVIATIONS
- cytochrome P450scc
cholesterol side-chain cleavage enzyme
- StAR
steroidogenic acute regulatory protein
References
- 1.Garren L D, Gill G N, Masui H, Walton G M. Recent Prog Horm Res. 1971;27:433–478. doi: 10.1016/b978-0-12-571127-2.50035-3. [DOI] [PubMed] [Google Scholar]
- 2.Stocco D M, Clark B J. Endocr Rev. 1996;17:221–224. doi: 10.1210/edrv-17-3-221. [DOI] [PubMed] [Google Scholar]
- 3.Lin D, Sugawara T, Strauss J F, III, Clark B J, Stocco D M, Saenger P, Rogol A S, Miller W L. Science. 1995;267:1828–1831. doi: 10.1126/science.7892608. [DOI] [PubMed] [Google Scholar]
- 4.Bose H, Sugawara T, Strauss J F, III, Miller W L. N Engl J Med. 1996;335:1870–1878. doi: 10.1056/NEJM199612193352503. [DOI] [PubMed] [Google Scholar]
- 5.Arakane F, Sugawara T, Nishino H, Liu Z, Holt J A, Pain D, Stocco D M, Miller W L, Strauss J F., III Proc Natl Acad Sci USA. 1996;93:13731–13736. doi: 10.1073/pnas.93.24.13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugawara T, Holt J A, Driscoll D, Strauss J F, III, Lin D, Miller W L, Patterson D, Clancy K P, Hart I M, Clark B J, Stocco D M. Proc Natl Acad Sci USA. 1995;92:4778–4782. doi: 10.1073/pnas.92.11.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moog-Lutz C, Tomasetto C, Regnier C H, Wendling C, Lutz Y, Muller D, Chenard M-P, Basset P, Rio M-C. Int J Cancer. 1997;71:183–191. doi: 10.1002/(sici)1097-0215(19970410)71:2<183::aid-ijc10>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 8.Sasano H, Nagura H, Harada N, Goukon Y, Kimura N. Hum Pathol. 1994;25:530–535. doi: 10.1016/0046-8177(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 9.Harikrishna J A, Black S M, Szkaz G D, Miller W L. DNA Cell Biol. 1993;12:371–337. doi: 10.1089/dna.1993.12.371. [DOI] [PubMed] [Google Scholar]
- 10.Nakae J, Tajima T, Arakane F, Sugawara T, Hanakai T, et al. Hum Mol Genet. 1997;6:571–576. doi: 10.1093/hmg/6.4.571. [DOI] [PubMed] [Google Scholar]
- 11.Dayhoff M O, Barker W C, Hunt L T. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- 12.Feng D, Doolittle R F. Methods Enzymol. 1990;183:375–387. doi: 10.1016/0076-6879(90)83025-5. [DOI] [PubMed] [Google Scholar]
- 13.Fitch W M, Margoliash E. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- 14.Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 15.Fujieda K, Tajima T, Nakae J, Sageshima S, Tachibana K, Suwa S, Sugawara T, Strauss J F., III J Clin Invest. 1997;99:1265–1271. doi: 10.1172/JCI119284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dib A, Adelaide J, Chaffanet M, Imbert A, Le Paslier D, Jacquemier J, Gaudray P, Theillet C, Birnbaum D, Pebusque M-J. Oncogene. 1995;10:995–1001. [PubMed] [Google Scholar]
- 17.Tomasetto C, Regnier C H, Moog-Lutz C, Mattei M G, Chenard M P, Liderau R, Basset P, Rio M C. Genomics. 1995;28:367–376. doi: 10.1006/geno.1995.1163. [DOI] [PubMed] [Google Scholar]
- 18.Anderson S, Davis D L, Dahlback H, Jornvall H, Russell D W. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 19.Sugawara T, Lin D, Holt J A, Martin K O, Javitt N B, Miller W L, Strauss J F., III Biochemistry. 1995;34:12506–12512. doi: 10.1021/bi00039a004. [DOI] [PubMed] [Google Scholar]