Abstract
Fractionation of the abundant small ribonucleoproteins (RNPs) of the trypanosomatid Leptomonas collosoma revealed the existence of a group of unidentified small RNPs that were shown to fractionate differently than the well-characterized trans-spliceosomal RNPs. One of these RNAs, an 80-nt RNA, did not possess a trimethylguanosine (TMG) cap structure but did possess a 5′ phosphate terminus and an invariant consensus U5 snRNA loop 1. The gene coding for the RNA was cloned, and the coding region showed 55% sequence identity to the recently described U5 homologue of Trypanosoma brucei [Dungan, J. D., Watkins, K. P. & Agabian, N. (1996) EMBO J. 15, 4016–4029]. The L. collosoma U5 homologue exists in multiple forms of RNP complexes, a 10S monoparticle, and two subgroups of 18S particles that either contain or lack the U4 and U6 small nuclear RNAs, suggesting the existence of a U4/U6⋅U5 tri-small nuclear RNP complex. In contrast to T. brucei U5 RNA (62 nt), the L. collosoma homologue is longer (80 nt) and possesses a second stem–loop. Like the trypanosome U3, U6, and 7SL RNA genes, a tRNA gene coding for tRNACys was found 98 nt upstream to the U5 gene. A potential for base pair interaction between U5 and SL RNA in the 5′ splice site region (positions −1 and +1) and downstream from it is proposed. The presence of a U5-like RNA in trypanosomes suggests that the most essential small nuclear RNPs are ubiquitous for both cis- and trans-splicing, yet even among the trypanosomatids the U5 RNA is highly divergent.
Keywords: trans-splicing/small nuclear RNA/spliced leader RNA
Trans-splicing was first discovered in trypanosomes as a novel RNA processing pathway linking a common 39-nt spliced leader (SL) to all mRNAs (1). Trans-splicing is a two-step transesterification reaction that involves two substrates, the SL RNA and a 3′ acceptor pre-mRNA. In the reaction, the two introns derived from the SL RNA and pre-mRNA form the Y-branched intermediate, analogous to the intron lariat intermediate of cis-splicing. The final product is an mRNA carrying the spliced leader at the 5′ end (1). This process has been shown to occur in a number of other organisms, such as nematodes, trematodes and Euglena, in conjunction with cis-splicing, whereas in kinetoplastidae it is the only RNA processing pathway (2).
In cis-splicing the small ribonucleoproteins (RNPs) play a dual role in juxtaposing the splice sites and in contributing part of the catalytic center (3). Like in cis-splicing, U2, U4, and U6 snRNAs have been characterized in trypanosomes and were shown to be essential for trans-splicing (4). However, despite their essential role in cis-splicing, U1 or U5 have not been identified (5). Without exception, all trypanosome small nuclear RNAs (snRNAs) have been shown to be shorter than their cis-splicing counterparts. The presence of a uniform 5′ splice site carried on the SL RNA may have changed the requirement for U1 RNA in trans-splicing. The ability of the SL RNA to substitute for the need of U1 RNA in cis-splicing supports the idea that SL RNA may bear some of the critical functions of U1 RNP (6).
Unique to trans-splicing is the need to bring in close proximity two independent RNA substrates. In the nematode system, it has been shown that the U6 RNA together with the U2 RNA provide the bridge between the SL RNA and the acceptor pre-mRNA (7). Crosslinking in vivo using the bifunctional reagent psoralen led to the discovery in Trypanosoma brucei of a novel small RNA that interacts with the SL RNA. The RNA was termed SLA (spliced leader-associated RNA) and was initially proposed to function as the T. brucei U5 homologue (8). Phylogenetic analysis of SLA disproved this hypothesis (9). The current role of SLA in trans-splicing is unknown but its ability to specifically associate with the SL exon may indicate an important function in trans-splicing.
With the growing evidence for the essential role of U5 RNA in cis-splicing in interacting with both 5′ and 3′ splice sites throughout the splicing process (10, 11), it became impossible to reconcile the absence of such a key snRNA in trypanosomes. The primary sequence of U5 RNA in different organisms is not conserved, apart from three types of sequence elements: a 9-nt element that is invariant in all U5 RNAs (5′-GCCUUUYAY-3′), the sequences that bind the U5-specific proteins and the RNA motif that binds the Sm antigens (12). Recently, the second abundant small RNA that is crosslinked in vivo to the T. brucei SL RNA was identified and termed SLA2. SLA2 possesses the conserved stem–loop 1 and the canonical U5 loop sequence and was also shown to crosslink with the free 5′ spliced leader exon (13). Independently, we have identified in the monogenetic trypanosomatid Leptomonas collosoma, a small RNA of 80 nt that, like the T. brucei SLA2, possesses the characteristic structural features of U5, such as stem–loop 1 (SL1) and the internal loop 1 (IL1) sequence elements. Interestingly, both the T. brucei and the L. collosoma loop 1 sequence deviates from the canonical U5 sequence in the second position. The L. collosoma U5 RNA homologue is longer and possesses a second stem–loop structure, indicating that this RNA is highly divergent even among the trypanosomatids. Base pair interaction between SL RNA and SLA2 located upstream and downstream to the 5′ splice site was proposed to explain the mechanism by which these RNAs associate during trans-splicing (13). Our data suggest that in L. collosoma the potential for base pairing between these molecules is more limited and lies in the vicinity of the 5′ splice site (positions −1 and +1) and downstream from it. The existence of a tRNA gene 98 nt upstream to the U5 that is encoded by the opposite strand may suggest that the U5 gene belongs to the same transcriptional regulation unit as the U3, U6, and 7SL RNA genes that have been shown to be transcribed by RNA polymerase III (14).
MATERIALS AND METHODS
Growth and Extracts Preparation.
L. collosoma cells were grown as described (15). Extracts were prepared using nitrogen cavitation (15). Post-ribosomal supernatant (PRS) preparation and DEAE chromatography were as described (16). For sucrose fractionation, extracts were prepared as described above except that salt extraction and ultracentrifugation steps were omitted.
RNA Labeling, Sequencing, and Primer Extension.
RNA was labeled at the 5′ end with polynucleotide kinase after removing the phosphate termini with alkaline phosphatase (17). End-labeled RNA was partially digested with base-specific nucleases according to the supplier’s instructions (United States Biochemical). The alkaline partial hydrolysis ladder was produced by boiling the RNA in 50 mM NaOH and 1 mM EDTA for 2 min. Primer extension was as described (9, 18).
Probes.
The oligonucleotides used are as follows: (i) (5′-CTGCACTGCTTAGTAAAAGTCGCAAGCAATGATGC-3′) designed based on the RNA sequencing and complementary to nt 4–37; (ii) complementary to the 3′ end of U5 RNA (5′-CAAAAACCAAGGGTT-3′); (iii) complementary to U5 nt 46–67 (5′-TTCCCAAAATTTGACATGATAT-3′); (iv) complementary to 3′ end of U6 (5′-AGCTATATCTCTCGAA-3′); (v) complementary to U2 nt 84–103 (5′-GATCAAAGATTTCGTATTCC-3′); (vi) complementary to U4 nt 58–72 (5′-GTACCGGATATAGT-3′); and (vii) complementary to the 3′ end of the SL RNA (5′-GCCCGAAAGCTCGGTC-3′).
Northern Blot Analysis.
RNAs from column and sucrose gradient fractions were extracted and electrophoresed on 6% acrylamide 7 M urea gels, and the gels were electroblotted to Nytran paper (Schleicher & Schuell). Hybridization with oligonucleotides or RNA probes was as described (16).
Tagging the RNPs with Antisense Oligonucleotides.
Aliquots from sucrose gradient fractions were mixed with 50,000 cpm (0.5–1 pmols) of end-labeled oligonucleotide in binding buffer containing 20 mM Hepes⋅KOH (pH 7.9), 2 mM MgCl2, and 5 mM 2-mercaptoethanol and incubated for 1 h at 30°C. The samples were fractionated on 5% native gels [acrylamide/bisacrylamide (30:0.8)] buffered with 50 mM Tris-glycine (50 mM Trizama base/50 mM glycine, pH 8.8).
Cloning of the Gene Encoding U5 snRNA and Sequence Analysis.
To enrich the U5 RNA, flow-through column fractions were sedimentated on sucrose gradients and the RNA obtained from 10–20S particles was separated on a preparative denaturing gel. RNAs ranging in size between 67 and 80 nt were excised, eluted, and subjected to Northern blot analysis with oligonucleotide 1. The RNA samples that hybridized with the probe were 5′ end labeled and used to screen a phage genomic library (15). Three positive clones were obtained that hybridized with antisense oligonucleotide 1. A 1-kb Sau3A fragment was subcloned in to pBluescript SK plasmid. The gene was sequenced using the Sanger chain termination method using T3, T7, and oligonucleotide 2 (19).
RESULTS
Identification of a Novel 80-nt RNA.
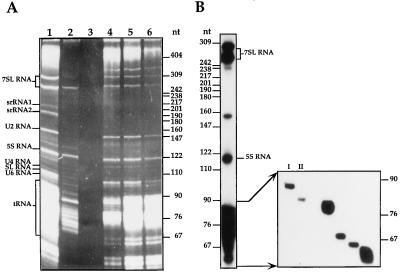
To characterize the L. collosoma trans-spliceosomal RNPs, we devised a purification scheme to enrich the RNPs. Whole-cell extracts were depleted of ribosomes by ultracentrifugation and the PRS was further fractionated on a DEAE-Sephacel column. The results presented in Fig. 1 indicate that whereas all the well-characterized U small nuclear RNPs (snRNPs) U2, U4, U6, and SL bound to the column and were eluted at 0.4 M KCl (Fig. 1A lanes 4–6) several small RNAs or RNPs were enriched in the flow-through fraction (Fig. 1A, lane 2). The latter preparations were analyzed further by fractionation on sucrose gradients, and the RNA was extracted from fractions carrying ≈10–20S RNPs and labeled at the 5′ end after removing the phosphate termini. The results (Fig. 1B) indicate that several RNAs were enriched in the sample, such as the two 7SL RNA molecules and different RNA molecules in the range of 60–90 nt. The individual end-labeled RNA molecules presented in Fig. 1B were subjected to enzymatic sequencing. The molecule designated I was identified as an 85-nt tRNA-like molecule that copurifies with the 7SL RNP (unpublished data). The enzymatic RNA sequencing of molecule II (80 nt), is presented in Fig. 2. Both the 80 and 85-nt RNAs could not be labeled at the 5′ end unless their phosphate termini were removed by alkaline phosphatase (results not shown), suggesting that these RNAs are not capped but possess a mono-, di-, or tri-phosphate at their termini. Inspection of the partial RNA sequence of molecule II revealed that this RNA can be folded into a stem–loop structure and that the loop sequence complies with the invariant U5 RNA loop sequence (5′-GCCUUUYAY-3′) except at the second position which is an A instead of C, as found in all U5 RNAs from yeast to humans (12). U5 particles, enriched in flow-through fractions, have sedimentation coefficients of ≈10–20S. These particles band in CsCl gradients in density of 1.3 g/ml, which is characteristic of spliceosomal RNPs (data not shown). Our results also indicate that the U5 homologue is highly susceptible to degradation, and it is degraded to one major RNA species of 70 nt (see below).
Figure 1.
(A) RNA profile of PRS and DEAE Sephacel column fractions. PRS obtained from 5 × 1010 cells was fractionated on a DEAE-Sephacel column. The RNA was analyzed on a 6% denaturing gel and visualized by ethidium bromide staining. Lanes: 1, PRS; 2, DEAE flow-through fraction; 3, DEAE wash fraction; 4–6, DEAE elutions with 0.4 M KCl. Marker was pBR322 HpaII digest. (B) The 5′ end labeled RNA molecules enriched in a DEAE flow-through fraction. The flow-through fraction derived from 1010 cells was layered on a continuous 10–30% (wt/vol) sucrose gradient. Gradients were centrifuged at 4°C for 22 h at 35,000 rpm in Beckman SW 41 rotor. Fractions containing ≈20S RNPs were deproteinized. The RNAs were labeled at the 5′ end, and the profile of the RNAs is presented. End-labeled RNA molecules in the size range of 76–90 nt, derived from this sample were fractionated on a 10% denaturing gel. Molecules that were subjected to sequence analysis are designated. Marker was end-labeled pBR322 HpaII digest.
Figure 2.
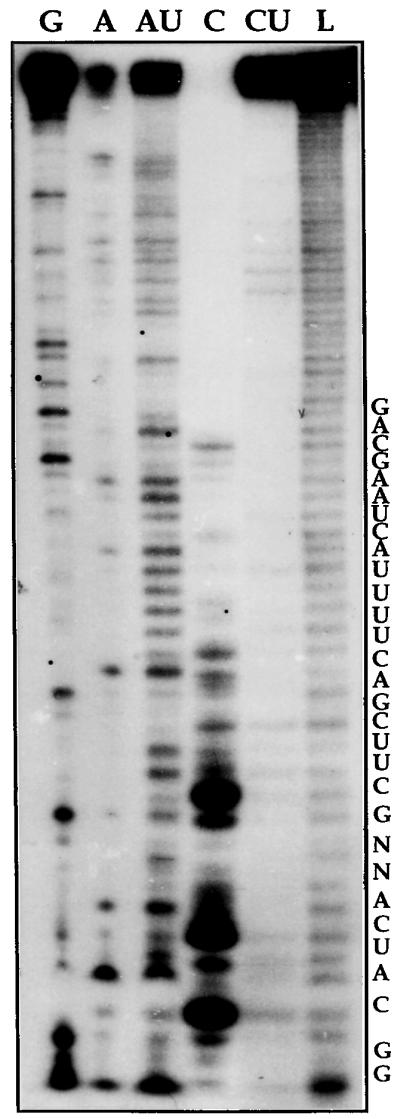
Partial enzymatic sequence determination of the U5 RNA. The RNA was prepared as described in Fig. 1B. Band II was subjected to partial digestion with base-specific RNases. Lanes: G, guanine-specific RNase T1 ladder; A, adenine-specific RNase U2 ladder; AU, adenine-uridine specific RNase phyM ladder; C, cytosine-specific RNase CL3 ladder; CU, cytosine-uridine specific RNase Bacillus cereus ladder; L, alkaline partial hydrolysis ladder. RNA sequence is indicated in the right.
The Gene Coding for the U5 RNA.
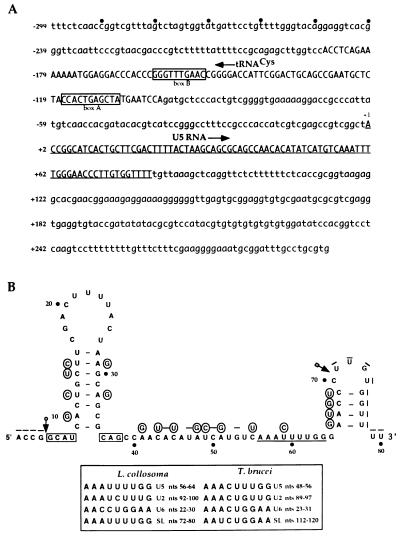
To obtain the complete sequence of the RNA, the gene coding for U5 was cloned from a L. collosoma genomic library. The primary sequence of the gene including 300-bp flanking sequences is presented in Fig. 3A. Searching GenBank with this sequence revealed the existence of sequence located 98 nt upstream to the U5 gene that shares 83% identity with tRNACys from Podocoryne carena (20) and 72% with mouse tRNACys (21). The exact position of the tRNACys was determined by folding the sequence to fit the secondary structure of the mouse tRNACys. The two boxes characteristic to tRNA are present in the sequence: box A (5′-TAGCTCAGTGG-3′) that fits the canonical box A, and box B (5′-GTTCAAACCC-3′) that agrees well with the consensus box B sequence. Except for the observed homology to tRNACys, no other relatedness was revealed between the L. collosoma U5 locus and data in GenBank.
Figure 3.
DNA sequence analysis of the U5 gene locus. (A) The sequence of the 1-kb fragment is presented. The coding regions of the U5 and the tRNACys are indicated by upper-case lettering and the U5 RNA sequence is underlined. The +1 position of the U5 RNA gene is indicated and the direction of transcription is marked with an arrow. (B) The proposed secondary structure of the L. collosoma U5 RNA and the T. brucei U5 RNA structure (13). Circled are the T. brucei nucleotides that differ from the L. collosoma RNA and bars indicate gaps in the alignment. The arrows indicate the boundaries of deletions when compared with the T. brucei sequence. The putative core protein binding site is underlined and the IL1 elements that may serve as binding sites for U5 specific proteins are boxed. The putative core protein binding sites of T. brucei and L. collosoma snRNAs are listed.
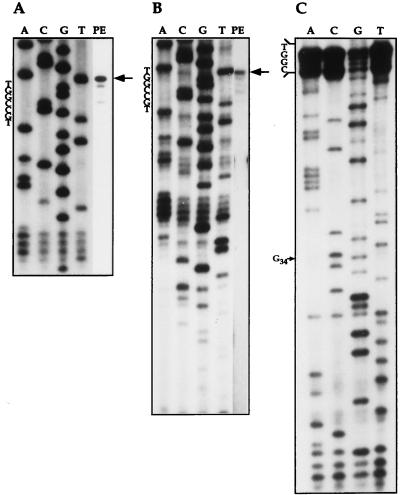
To determine the exact boundaries of the U5 RNA, primer extension was performed using an internal oligonucleotide (oligonucleotide 3) and an oligonucleotide complementary to the most 3′ end of the molecule (oligonucleotide 2). The results presented in Fig. 4 indicate that the terminal 5′ nucleotide of the U5 RNA observed in both experiments are identical (Fig. 4 A and B). Recently, the sequence of the T. brucei 62-nt U5 RNA was published (13). To verify that the L. collosoma RNA is longer than that of T. brucei by a 3′ end extension, a 3′ primer was designed to bind the RNA immediately downstream from the sequence that matches the most 3′ end of the T. brucei RNA (oligonucleotide 2). The ability of this oligonucleotide to extend the U5 RNA (Fig. 4-B and C), suggests that L. collosoma RNA is longer than the T. brucei RNA in 11 nt at the 3′ end of the molecule. Secondary structure modeling of the 3′ sequence suggests that this RNA can form a second stem–loop structure. The loop sequence of the L. collosoma RNA is 5′-CUUGU-3′ and related to the sequence of the second loop of the mammalian U5 RNA (5′-CUUG-3′). The staggered primer extension profile presented in Fig. 4 may suggest that the first 4 nt of the U5 RNA are modified. 2′-O-Methylation of the first 2 nt was detected in mammalian, Drosophila, plant, and fungi U5 RNA, and three modified nucleotides were found in dinoflagellates (22).
Figure 4.
Primer extension analysis of the L. collosoma U5 RNA. The primer extension reaction was separated next to sequencing reaction of the gene with the same primer. (A) Primer extension and sequencing using an internal oligonucleotide 3. (B) The same as in A but with a 3′ end oligonucleotide (oligonucleotide 2). (C) Primer extension sequencing using total L. collosoma RNA as template and oligonucleotide 2 as primer. The G in position 34 is indicated with an arrow. In all panels, the 5′ end of the U5 RNA is indicated with an arrow. The sequence of the cDNA in the vicinity to the 5′ end of the RNA is indicated.
The proposed secondary structure of the U5 RNA, based on the mfold program (23), is presented in Fig. 3B. The nucleotide differences between the T. brucei sequence and the L. collosoma RNA are circled. Only 55% overall sequence identity exists between these two RNAs. The only domains that are 100% conserved are loop 1 and the sequences flanking stem 1 (indicated with boxes) that were shown to serve as binding sites for U5-specific proteins (12). The C at position 34 creates a mismatch in the second nucleotide of stem 1. To verify that this mismatch exists in the U5 RNA, primer extension RNA sequencing was performed (Fig. 4C), which indicates that position 34 is indeed a C. Searching the U5 RNA sequence for a motif that may be involved in binding the trypanosome core proteins revealed a sequence in positions 56–64 (5′-AAAUUUUGG-3′) that is related to sequences found in other L. collosoma and T. brucei spliceosomal RNAs (listed in Fig. 3B). The only two spliceosomal RNAs that possess identical motifs are SL and U5 RNAs. This motif is always present in a single-stranded region that was previously implicated in binding the T. brucei snRNP core proteins (24).
U5 RNP Particles.
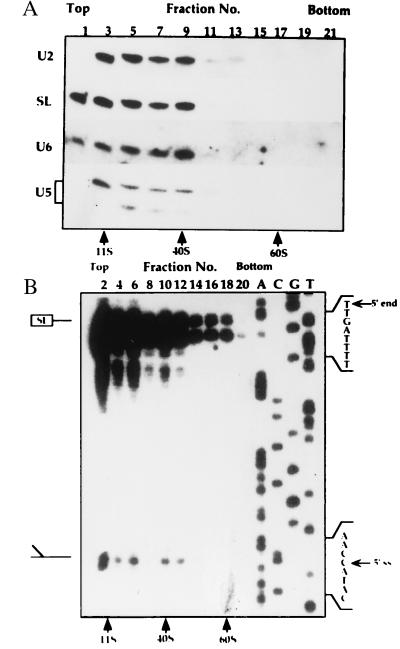
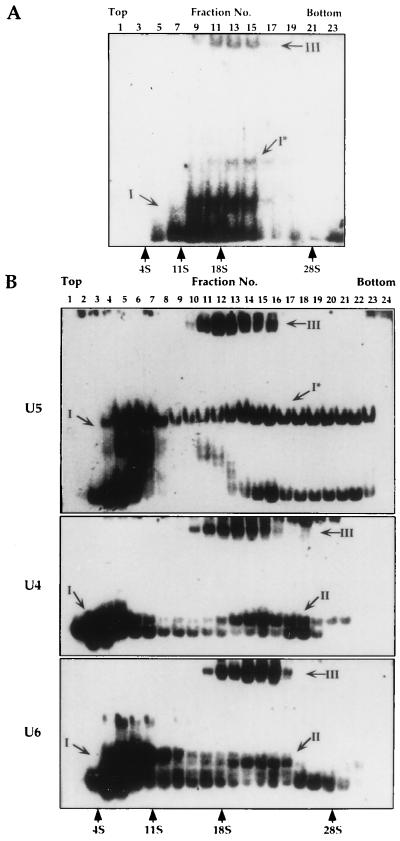
In mammalian cells the U5 RNA is present in at least three types of particles: (i) a 10S core particle, (ii) a 20S particle that contains the eight U5-specific proteins, and (iii) a 25S particle, a tri–snRNP complex that also contains the U4 and U6 RNPs (25). To examine whether homologous particles exist in L. collosoma, whole-cell extracts were fractionated on sucrose gradients, and the RNA was subjected to Northern blot analysis with probes specific to SL, U2, U6, and U5 RNAs. The results presented in Fig. 5A, indicate that two U5 transcripts exist in the cell, an 80-nt RNA and a 70-nt RNA that is the major degradation product of U5 RNA. Two peaks containing these RNPs can be seen, a ≈10S peak reflecting the monoparticles and a ≈40S particle that contains in addition all the other spliceosomal RNPs. To further characterize the 40S particle, the distribution of the Y structure intermediate was examined. The results presented in Fig. 5B indicate that the Y structure intermediate is enriched in a ≈40S particle, suggesting that this particle may represent the trans-spliceosome. To improve separation of the RNP complexes, especially the 10–30S particles, the extracts were fractionated for longer time, and the distribution of the U5 RNPs was examined by separating the RNPs from the sucrose gradient fractions on native gels. The RNPs were subjected to Northern blot analysis with antisense U5 RNA probe. The results presented in Fig. 6A indicate that the U5 RNA was found in at least three different RNPs: a ≈10S complex (complex I), and two ≈18S particles designated I* and III. To explore the composition of these different RNPs, the distribution of U5, U4, and U6 was examined by tagging the RNPs with antisense oligonucleotides. This method is very sensitive and allows the detection of particles that exist in minute amounts. The tagged RNPs were separated on native gels, and the results are presented in Fig. 6B. All the observed bands corresponding to tagged RNPs were specific, because they were competed by the unlabeled specific oligonucleotides but not with nonspecific oligonucleotides (results not shown). Two almost indistinguishable bands corresponding to the monomeric U4 and U6 ≈10S particles were observed, but two well-separated U5 bands were revealed for the U5 10S complex I. The two U5 bands correspond to the particles carrying either the 70- or 80-nt U5 RNAs, because when the RNPs were tagged with a 3′ end oligonucleotide (oligonucleotide 2), only the upper band, corresponding to RNPs carrying the 80-nt U5 RNA, was observed (results not shown). The results presented in Fig. 6B, like those presented in Fig. 6A, indicate that the U5 RNA exists in three subpopulations: (i) a 10S monoparticle, (ii) a ≈20S particle (marked I*), and (iii) an 18S particle that also contains the U4 and U6 RNPs. Although, three complexes were revealed in the experiments presented in Fig. 6, variation exists in the amount of these complexes, especially in the level of complex I*. The S value of complex I* is higher than that of the tri–snRNP complex, suggesting that it was not derived from it. Three types of U4 and U6 RNPs were observed: I, monoparticles; II, a dimeric particle containing the U4 and U6 RNAs; and III, the putative a tri–snRNP complex.
Figure 5.
Fractionation of spliceosomal RNPs on sucrose gradients. Low-salt whole-cell extracts were layered on a continuous 10–30% (wt/vol) sucrose gradient in the buffer as described (16), but containing 100 mM KCl. Gradients were centrifuged as in Fig. 1. but for 3 h. S values were determined using 40S and 60S ribosomes from HeLa cell extract and the enzyme catalase (11S). (A) RNA was extracted and separated on a 10% denaturing gel and was subjected to Northern blot analysis with oligonucleotides complementary to the SL, U2, U6, and U5 snRNAs. (B) Primer extension analysis was performed using an antisense SL RNA intron oligonucleotide (oligonucleotide 7), and the products were analyzed next to DNA sequence reactions of the L. collosoma SL RNA gene using the same primer. The positions of the full-length SL RNA and the Y structure intermediate are indicated. The fractions are numbered from top to bottom.
Figure 6.
Fractionation of U4, U5 and U6 RNPs on sucrose gradients. (A) Extracts were prepared and separated on sucrose gradients as in Fig. 1. The sucrose gradient fractions were subjected to Northern blot analysis with antisense U5 RNA probe. (B) Tagging the U4, U5, and U6 snRNPs using antisense oligonucleotides. RNPs from the sucrose gradient fractions were incubated with end-labeled oligonucleotides to U4 (oligonucleotide 6), U5 (oligonucleotide 3), and U6 (oligonucleotide 4) and the RNPs were separated on native gels. The fractions are numbered from top to bottom. S values were determined relative to rRNA 28S, 18S, and 4S (GIBCO/BRL) and catalase (11S). The particles were designated as following: I, monomeric particles, except from the ≈18S U5 particle (I*); II, U4/U6 dimeric particle; and III, the tri–snRNP complex U4/U6⋅U5.
Possible Interaction of U5 with SL RNA.
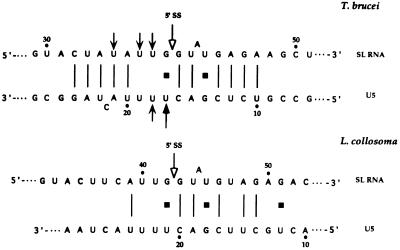
Evidence from cis-splicing systems of yeast (11) and mammals (26) suggests that base pairing may play a role in the interaction of the U5 and the 5′ splice site. Also in trans-splicing, psoralen crosslinking aligns the T. brucei U5 loop 1 sequence with the SL RNA such that the interaction domain is exactly analogous to the interaction of U5 with the 5′ splice site in cis-splicing (11). An extensive base pair interaction, located upstream and downstream to the 5′ splice site, was proposed as the mechanism that mediates the interaction between the T. brucei SL and U5 RNAs. Comparison between the potential for base pair interaction of U5 with SL RNA in T. brucei and L. collosoma suggests that the extensive base pair interaction proposed for T. brucei does not exist in L. collosoma (Fig. 7).
Figure 7.
Potential for possible base pairing interactions between SL and U5 RNAs of T. brucei and L. collosoma. Psoralen adducts mapped on the T. brucei SL and U5 RNAs are indicated with black arrows like in ref. 13, and the 5′ splice sites are indicated with open arrows.
DISCUSSION
In this paper we describe a small RNA of 80 nt that we suggest is the L. collosoma U5 homologue. This RNA was discovered based on the peculiar fractionation of its RNP, and independent of the T. brucei U5 RNA that was found due to its efficient crosslinking to the SL RNA (13). The recent finding of U5 RNA in trypanosomes solved a long debate in the last 10 years as to whether trans-splicing requires a U5 homologue (27). Realizing the crucial function of U5 in cis-splicing, it became impossible to envision how trans-splicing can operate without this essential snRNA. The data presented in this paper highlight the unique properties of the trypanosome U5 homologue. The trypanosome U5 RNA: (i) is the smallest U5 in nature and lacks stem Ic, IL2, and stem Ib (12), (ii) it lacks a TMG cap and has a 5′ end phosphate terminus, (iii) its invariant loop 1 sequence contains an A instead of a C in the second position, and (iv) the RNA lacks 2′-O-methyl modifications in the invariant loop, but the four most 5′ end nucleotides are modified. Two major differences exist between the L. collosoma and the T. brucei U5 RNAs are that the L. collosoma RNA is longer and possesses a second stem-loop structure, and its potential to interact with the SL RNA by base pairing is less extensive (Fig. 7).
The absence of a TMG cap is a unique property of U5 when compared with U5 RNAs from other organisms. However, this property is shared with at least another trans-spliceosomal RNA, the SLA (8). In other eukaryotes, a strong correlation exists between the TMG cap, binding of Sm proteins, and the polymerase that transcribes the RNA. In trypanosomes, such stringent rules do not exist as the core proteins are apparently common to all spliceosomal RNAs (28), even though these RNAs are transcribed by different polymerases—i.e., polymerase II for the SL RNA (29), and polymerase III for U2 and U6 (30). In addition, core proteins bind RNAs that harbor different cap structures: the U2 and U4 are TMG capped (5), the SL RNA possesses a 7 metG hypermodified “cap 4” (31), and the SLA (8) and the U5 RNA (ref. 13, and this study) have no caps. Despite the similarity between the proposed RNA binding sites among the U snRNAs and SL RNA (Fig. 3B), the only two sites that are identical and exactly like the canonical Sm binding sites are the sites of the L. collosoma SL and U5 RNAs. Common to these RNAs is the presence of 4 modified nt at the 5′ end. The absence of 2′-O-methyl modification from the trypanosome invariant U5 loop is surprising, because all U5 RNAs analyzed so far carry 2′-O-methyl modifications on the first G, the second U, and the last C (22), and these modifications as well as modifications in other snRNAs were shown to be clustered around functionally important regions of these RNAs (32).
tRNA genes that are divergently transcribed relative to the small RNA genes, were found upstream to the U3, U6, and 7SL RNAs (14, 30) and to U5 RNA (this study), suggesting that this transcriptional regulation is related neither to the cap structure of these RNAs nor to the identity of their binding proteins or even to their cellular compartmentalization. This genomic organization may therefore reflect a very archaic mode of transcriptional regulation with a potential to coordinate the synthesis of small RNAs that are essential for growth.
The results presented in Fig. 5A indicate that the spliceosomal small RNPs including the U5 RNP exist in at least two forms, free particles (≈10S) and ≈40S particles that also contain the Y structure intermediate. Because the ≈40S particles contain both the spliceosomal RNPs and the splicing intermediate, it may represent the in vivo trans-spliceosome. Our results also indicate the existence of a tri–snRNP complex that contains the U4/U6⋅U5 RNPs. As expected, this complex migrates slower than the dimeric U4/U6 or the U5 complex on native gels, but its S value is ≈18S and not 25S as the mammalian tri–snRNP complex. This may suggest that proteins were dislodged from the tri–snRNP complex either in vivo or during extract preparation. The identity of complex I* is currently unknown; it may have dissociated from a higher S value complex and may contain spliceosomal RNAs other than the U4 and U6 snRNAs.
The trypanosome U5 is the shortest U5 RNA described so far. Although U5 RNAs from different organisms vary in length, the minimal U5 structure that is essential for its function, at least in yeast, consists of SL1 flanked by IL1 that is linked to the Sm protein binding site (12). The trypanosome U5 RNA obeys the minimal requirement for a U5 RNA. Despite the small size of the L. collosoma U5 it carries a second stem–loop structure downstream from the core protein binding site, but this domain is missing from the T. brucei RNA. Although it has recently been demonstrated that the second stem–loop structure is important for Sm binding and cap trimethylation in mammalian U5 RNA (33), this domain is missing from several U5 RNAs and is dispensable for growth in yeast (12).
As opposed to cis-splicing, where the 5′ and 3′ splice sites are imbedded in between sequences that vary in different genes, in trans-splicing the only 5′ splice site is within the SL RNA and is flanked by fixed sequences. Recognition of the 5′ splice site in trans-splicing may therefore differ from that of cis-splicing and may depend more on base pair interactions. The base pairing, suggested by the location of psoralen crosslinks of the T. brucei U5 RNA with the SL RNA (13) enables alignment of these sequences in a manner exactly like that proposed for cis-splicing (11). Thus, loop 1 residue 4 is placed opposite to the SL exon position −1 and the loop position 3 is placed opposite to intron position G1, but the potential can be extended (Fig. 7). However, in L. collosoma such an extended potential does not exist and only the interaction domain located downstream to the 5′ splice site is phylogentically conserved (Fig. 7). It has been recently shown that base-substitution in position +8 of the intron affects the utilization of the L. seymouri SL RNA in the reaction (34). According to the proposed SL–U5 interaction domain (Fig. 7), this phenotype may result from inefficient interaction of the SL RNA with U5 RNA, and, if so, a compensatory mutation introduced in the U5 RNA gene may change the phenotype of the SL RNA mutation. Using the system we have developed to study the structure and function of L. collosoma SL RNA (35), we should be able to genetically study the validity of the SL–U5 interaction proposed in Fig. 7.
An additional important base pair interaction of SL RNA is with SLA (8). Based on the interaction domain proposed for SL RNA and U5 (Fig. 7), it is possible to envision interaction among these three molecules that are not mutually exclusive. Although the U5 RNA directly interacts with the exon sequence (13), this interaction may be facilitated by base pairing with intronic sequences (Fig. 7), whereas the interaction with SLA is mostly with sequences located in the exon region (13). Part of the interaction domain of U5 with SL RNA overlaps with the intron sequence that were shown in cis-splicing to interact with the U1 RNA during the early stages of the cis-splicing reaction (3). This may suggest that the trypanosome U5 may carry some of the U1 RNA functions in cis-splicing. Only by following the sequential interaction of the SL RNA with the SLA, U6, and U5 will it be possible to dissect the complicated interactions that take place during the trans-splicing process. In the absence of an in vitro system, this question can be addressed by utilizing the permeable cell system that is amenable to transcribe and trans-splice newly synthesized SL RNA (4). The description of U5 RNA in trypanosomes suggests that, despite the unique features of trans-splicing, the core catalytic components are shared and conserved between cis- and trans-splicing. Regardless of its central role in splicing, the U5 RNA molecule is highly divergent even between two different trypanosomatid species.
Acknowledgments
We thank Igor Goncharov for his contribution to the initial stages of this work. This work was supported by the MINERVA Foundation (Munich), by the German–Israel Foundation and by the Leo and Julia Forchheimer Center for Molecular Genetics of the Weizmann Institute of Science.
ABBREVIATIONS
- SL RNA
spliced leader RNA
- RNP
ribonucleoprotein
- SLA
spliced leader associated
- TMG
trimethylguanosine
- PRS
post-ribosomal supernatant
- snRNA
small nuclear RNA
- snRNP
small nuclear RNP
- IL
internal loop
- SL1
stem loop 1
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF006632).
References
- 1.Agabian N. Cell. 1990;61:1157–1160. doi: 10.1016/0092-8674(90)90674-4. [DOI] [PubMed] [Google Scholar]
- 2.Bonen L. FASEB J. 1993;7:40–46. doi: 10.1096/fasebj.7.1.8422973. [DOI] [PubMed] [Google Scholar]
- 3.Moore M J, Query C C, Sharp P A. In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 303–358. [Google Scholar]
- 4.Ullu E, Tschudi C. Cell. 1990;61:459–466. doi: 10.1016/0092-8674(90)90527-l. [DOI] [PubMed] [Google Scholar]
- 5.Mottram J, Perry K L, Lizardi P M, Lührmann R, Agabian N, Nelson R G. Mol Cell Biol. 1989;9:1212–1223. doi: 10.1128/mcb.9.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruzik J P, Steitz J A. Cell. 1990;62:889–899. doi: 10.1016/0092-8674(90)90264-f. [DOI] [PubMed] [Google Scholar]
- 7.Hannon G J, Maroney P A, Yu Y-T, Hannon G E, Nilsen T W. Science. 1992;258:1775–1780. doi: 10.1126/science.1465612. [DOI] [PubMed] [Google Scholar]
- 8.Watkins K P, Dungan J M, Agabian N. Cell. 1994;76:171–182. doi: 10.1016/0092-8674(94)90181-3. [DOI] [PubMed] [Google Scholar]
- 9.Palfi Z, Xu G L, Bindereif A. J Biol Chem. 1994;269:30620–30625. [PubMed] [Google Scholar]
- 10.Newman A J, Teigelkamp S, Beggs J D. RNA. 1995;1:968–980. [PMC free article] [PubMed] [Google Scholar]
- 11.Newman A J, Norman C. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 12.Frank D, Roiha H, Guthrie C. Mol Cell Biol. 1994;14:2180–2190. doi: 10.1128/mcb.14.3.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dungan J D, Watkins K P, Agabian N. EMBO J. 1996;15:4016–4029. [PMC free article] [PubMed] [Google Scholar]
- 14.Nakaar V, Dare A O, Hong D, Ullu E, Tschudi C. Mol Cell Biol. 1994;14:6736–6742. doi: 10.1128/mcb.14.10.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldring A, Karchi M, Michaeli S. Exp Parasitol. 1994;80:333–338. doi: 10.1006/expr.1995.1041. [DOI] [PubMed] [Google Scholar]
- 16.Michaeli S, Roberts T G, Watkins K P, Agabian N. J Biol Chem. 1990;265:10582–10588. [PubMed] [Google Scholar]
- 17.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 18.Patzelt E, Perry K L, Agabian N. Mol Cell Biol. 1989;9:4291–4297. doi: 10.1128/mcb.9.10.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Badder C D, Schuchert P. Nucleic Acids Res. 1992;20:5837. doi: 10.1093/nar/20.21.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood L S, Hatzenbuhler N, Peterson R, Vogeli G. Gene. 1991;98:249–252. doi: 10.1016/0378-1119(91)90181-a. [DOI] [PubMed] [Google Scholar]
- 22.Szkukalek A, Myslinski E, Mougin A, Lührmann R, Branlant C. Biochimie. 1995;77:16–21. doi: 10.1016/0300-9084(96)88099-0. [DOI] [PubMed] [Google Scholar]
- 23.Zuker M, Jaeger J A, Turner D H. Nucleic Acids Res. 1991;19:2707–2714. doi: 10.1093/nar/19.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cross M, Günzl A, Palfi Z, Bindereif A. Mol Cell Biol. 1991;11:5516–5526. doi: 10.1128/mcb.11.11.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bach M, Lührmann R. Biochim Biophys Acta. 1991;1088:139–143. doi: 10.1016/0167-4781(91)90164-h. [DOI] [PubMed] [Google Scholar]
- 26.Cortes J J, Sontheimer E J, Seiwert S D, Steitz J A. EMBO J. 1993;12:5181–5189. doi: 10.1002/j.1460-2075.1993.tb06213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steitz J A. Science. 1992;257:888–889. doi: 10.1126/science.1386941. [DOI] [PubMed] [Google Scholar]
- 28.Palfi Z, Günzl A, Cross M, Bindereif A. Proc Natl Acad Sci USA. 1991;88:9097–9101. doi: 10.1073/pnas.88.20.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito R M, Elgort M G, Campbell D A. EMBO J. 1994;13:5460–5469. doi: 10.1002/j.1460-2075.1994.tb06881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakaar V, Ullu E, Tschudi C. Parasitol Today. 1994;11:225–228. [Google Scholar]
- 31.Perry K L, Watkins K P, Agabian N. Proc Natl Acad Sci USA. 1987;84:8190–8194. doi: 10.1073/pnas.84.23.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu J, Patton J R, Shimba S, Reddy R. RNA. 1996;2:909–918. [PMC free article] [PubMed] [Google Scholar]
- 33.Hinz M, Moore M J, Bindereif A. J Biol Chem. 1996;271:19001–19007. doi: 10.1074/jbc.271.31.19001. [DOI] [PubMed] [Google Scholar]
- 34.Lücke S, Xu G-L, Palfi Z, Cross M, Bellofatto V, Bindereif A. EMBO J. 1996;15:4380–4391. [PMC free article] [PubMed] [Google Scholar]
- 35.Goldring A, Zimmer Y, Ben-Yehuda E, Goncharov I, Michaeli S. Exp Parasitol. 1996;84:28–41. doi: 10.1006/expr.1996.0087. [DOI] [PubMed] [Google Scholar]