Abstract
Steroids, thyroid hormones, vitamin D3, and retinoids are lipophilic small molecules that regulate diverse biological effects such as cell differentiation, development, and homeostasis. The actions of these hormones are mediated by steroid/nuclear receptors which function as ligand-dependent transcriptional regulators. Transcriptional activation by ligand-bound receptors is a complex process requiring dissociation and recruitment of several additional cofactors. We report here the cloning and characterization of receptor-associated coactivator 3 (RAC3), a human transcriptional coactivator for steroid/nuclear receptors. RAC3 interacts with several liganded receptors through a mechanism which requires their respective ligand-dependent activation domains. RAC3 can activate transcription when tethered to a heterologous DNA-binding domain. Overexpression of RAC3 enhances the ligand-dependent transcriptional activation by the receptors in mammalian cells. Sequence analysis reveals that RAC3 is related to steroid receptor coactivator 1 (SRC-1) and transcriptional intermediate factor 2 (TIF2), two of the most potent coactivators for steroid/nuclear receptors. Thus, RAC3 is a member of a growing coactivator network that should be useful as a tool for understanding hormone action and as a target for developing new therapeutic agents that can block hormone-dependent neoplasia.
Steroids, thyroid hormones, vitamin D3, and retinoids are fat-soluble small molecules that control cell differentiation, embryonic development, and homeostasis, as well as adult organ physiology (1, 2). The diverse biological effects of these molecules are mediated by a large superfamily of steroid/nuclear hormone receptors which display substantial specificity in regulating gene expression (3–6). These receptors share a common domain structure, including an N-terminal DNA-binding domain (DBD), which binds to specific DNA sequences, and a C-terminal ligand-binding domain (LBD), which binds to the cognate hormones. Retinoic acid receptors (RARs), thyroid hormone receptors (TRs), vitamin D3 receptor (VDR), peroxisomal proliferator-activated receptors (PPARs), and several other orphan receptors form heterodimeric complexes with retinoid-X receptors (RXRs) (7, 8). Such receptor heterodimers bind to a broad range of response elements, which are composed of two related half-sites and activate target gene expression (9).
Transcriptional activation by steroid/nuclear receptors apparently involves at least two separate processes: derepression and activation (3). Repression is effected in part by association of unliganded receptors with nuclear receptor corepressor (N-CoR) and/or silencing mediator for RAR and TR (SMRT) (10, 11). Ligand binding triggers dissociation of these corepressors and recruitment of coactivators (12). To date, several putative receptor-associated coactivators (RACs) have been identified, including RIP-140 and RIP-160 (13, 14), ERAP-140 and ERAP-160 (15), TIF1 (16), SRC-1 (17, 18), TRIP1/SUG1 (19, 20), ARA70 (21), TIF2 (22), and CBP/p300 (17, 23, 24). Two of these potential coactivators, SRC-1 (steroid receptor coactivator 1) and TIF2 (transcriptional intermediate factor 2), are related proteins which can enhance transcriptional activation by several steroid/nuclear receptors (18, 22, 24, 25).
Hormone binding is thought to induce conformational changes in the receptor and in turn, activate the C-terminal ligand-dependent activation function (AF-2) of the receptor (2). At the extreme C terminus of the AF-2 domain, about 20 amino acids form an amphipathic helix (26). This helix is referred to as AF-2 activation domain (AF2-AD) (27), τC, or τ4 domain (28, 29). Deletion and several point mutations of this domain abolish the AF-2 function (30–32), and it can act alone as an activation domain when fused to a heterologous DBD (31). Comparison of LBD crystal structures of the unliganded RXRα (26) with liganded RARγ (27) and liganded TRα (33) reveals a striking difference in the relative position of the AF2-AD helix. It is proposed that, upon hormone binding, this helix rotates almost 180° and forms part of the hormone-binding surface. The hydrophobic residues of the helix face toward the ligand-binding cavity, while the charged residues extend into the solvent, possibly mediating protein–protein interactions with cofactors. AF2-AD also has been shown to be required for derepression by promoting dissociation of corepressors (11, 28). These findings suggest a possible mechanistic link among hormone binding, transcriptional repression, and activation by steroid/nuclear receptors through the AF2-AD.
To understand the molecular mechanisms of transcriptional regulation by steroid/nuclear receptors, we screened a human brain cDNA library for RAR-interacting partners by using the yeast two-hybrid system (34). We describe here the cloning and characterization of a novel RAC. RAC3 displays properties of a transcriptional coactivator, including the capacity for ligand-dependent interactions with the receptors and direct transcriptional activation. Interestingly, RAC3 is highly related to SRC-1 and TIF2, suggesting the existence of a family of transcriptional coactivators for steroid/nuclear receptors. The identification of these three related coactivators has provided new insights into the fundamental mechanisms of gene activation by steroids/thyroid hormones, vitamin D3, and retinoids.
MATERIALS AND METHODS
Yeast Two-Hybrid Screen.
The plasmid vector pGBT-hRARα expressing Gal4 DBD fusion with full-length human RARα in yeast cells was constructed and used as bait for yeast two-hybrid screening. A human brain cDNA library in pGAD10 vector (CLONTECH) was screened for RAR-interacting proteins in the absence of ligand as previously described (34). After primary selection on synthetic dropout plates lacking tryptophan, leucine, and histidine, but supplemented with 50 mM 3-aminotriazole, we isolated 20 colonies, which were further tested for β-galactosidase expression by using a liquid assay as described (35). The library plasmids from positive clones that expressed both HIS3 and LacZ reporters were rescued and retransformed into yeast cells, together with the original bait and other constructs, for testing the specificity of protein–protein interaction. These analyses led to a single specific clone, RAC3.1, which was selected for further analysis in this study.
Transient Transfection Assay.
Monkey kidney-derived CV-1 cells and human lung carcinoma A549 cells were grown in phenol red-free Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% charcoal-stripped fetal bovine serum (GIBCO). One day before transfection, cells were seeded in 12-well plates at a density of 50,000 cells per well. A DNA mixture containing 0.5 μg of expression vector, 0.5 μg of internal control plasmid pCMX-βGal, 1 μg of luciferase reporter, and 1.5 μg of carrier DNA pGEM was prepared in a final volume of 30 μl. The DNA solution was mixed dropwise with 1 vol of 0.5 M CaCl2 and 2 vol of 2× BBS [50 mM N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid (Bes; Calbiochem)/280 mM NaCl/1.5 mM Na2HPO4, pH 6.95]. DNA precipitates were allowed to form at room temperature for 10 min and applied evenly to the culture cells. Transfection was allowed to continue for 12 hr, and the precipitates were removed by washing transfected cells twice with phosphate-buffered saline (PBS). Transfected cells were re-fed fresh medium containing either vehicle alone or vehicle plus ligands and were harvested 24–36 hr after treatment.
Luciferase and β-Galactosidase Assay.
Transfected cells in each well were lysed in 120 μl of cell lysis solution (35) and processed for luciferase and β-galactosidase assay. Fifty microliters of lysed cells was transferred into 96-well Microlite plates for luciferase assay and 96-well microtiter plates for β-galactosidase assay as described (35). The luciferase activities were determined with a MLX microtiter plate luminometer (Dynex) using 100 μl of assay solution (0.1 M potassium phosphate buffer at pH 7.8, 5 mM ATP, 10 mM MgCl2) and 100 μl of luciferin solution (0.01 M d-luciferin in 0.1 M potassium phosphate, pH 7.8). The luciferase activities were normalized to the β-galactosidase activity expressed from the cotransfected pCMX-βGal plasmid.
Isolation and Construction of Full-Length RAC3.
The cDNA insert of RAC3.1 clone was labeled with [32P]dATP by DECAprime II DNA labeling kit (Ambion), and used to screen a λgt11 HeLa cDNA library (CLONTECH). Three overlapping cDNA clones covering the full-length RAC3 coding region were identified and their sequences were determined by dideoxynucleotide sequencing using a T7 sequencing kit (Amersham). The sequence analysis and comparison were carried out with the GCG package. A full-length RAC3 expression vector (pCMX.F.RAC3) was constructed in the pCMX expression vector (9), containing a FLAG and a hemagglutinin epitope linked to the N terminus of RAC3.
RESULTS
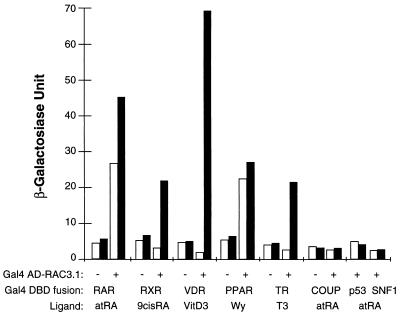
To understand the mechanisms of transcriptional regulation by steroid/nuclear hormone receptors, we screened a human brain cDNA library by the yeast two-hybrid system and identified one specific RAR-interacting protein designated RAC3. In addition to RAR, RAC3 also interacted with RXR, VDR, PPAR, and TR, but not with COUP-TFI or nonreceptor proteins such as p53 and SNF1 (Fig. 1). In the absence of ligand, RAC3 can interact with RAR and PPAR, and ligand treatment enhanced the RAR interaction by about 75% and PPAR interaction by about 10%. RAC3 also interacted with RXR, VDR, and TR, and these interactions were completely ligand dependent. These results suggest that RAC3 is a specific receptor-interacting protein which preferentially associates with ligand-bound nuclear receptors.
Figure 1.
RAC3 interacts with nuclear receptors. The protein–protein interactions between RAC3 and several steroid/nuclear receptors were analyzed by the yeast two-hybrid system. The original yeast two-hybrid clone, RAC3.1 in pGAD10 vector, was retransformed together with yeast expression vectors for Gal4-DBD fusion of selected hormone receptors into Y190 cells. The receptors used include RAR (full-length hRARα), RXR (full-length hRXRα), VDR (LBD of hVDR), PPAR (LBD of mPPARα), TR (LBD of hTRβ), and COUP (LBD of COUP-TFI) (h-, human; m-, murine). Three independent colonies from each transformation were selected and analyzed for expression of β-galactosidase activity by liquid o-nitrophenyl β-d-galactoside (ONPG) assay after treatment with 1 μM corresponding ligands (filled bars), or with an equal concentration of vehicle alone (open bars). Under the conditions used in these experiments, ligand treatment did not produce detectable β-galactosidase activity from the Gal4 DBD-receptor fusion alone. atRA, all-trans retinoic acid; VitD3, 1,25-dihydroxyvitamin D3; Wy, Wy 14,642; T3, 3,5,3′-triiodothyronine.
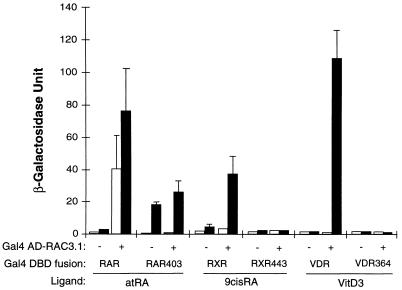
The ligand dependency of the interactions of RAC3 with the receptors suggests that a ligand-induced conformational change can be important for mediating protein–protein interaction with RAC3. Since the AF2-AD domain of the receptors is assumed to undergo a dramatic conformational change upon ligand binding (27), and since this domain is absolutely required for ligand-dependent transcriptional activation (32), we tested three AF2-AD deletion mutants for their abilities to interact with RAC3 in the presence or absence of hormones. Truncation of the C-terminal AF2-AD domain of RAR at residue 403 created the dominant-negative mutant RAR403, which acts as a constitutive repressor in mammalian cells (30, 36), but in yeast cells acted as a strong ligand-dependent transcriptional activator (Fig. 2). The mechanism underlying this difference between yeast and mammalian cells is unclear; however, a similar opposite activation effect by v-erbA in yeast cells was observed (37). Coexpression of RAR403 fusion with RAC3 did not further stimulate the reporter gene activity in either absence or presence of ligand, indicating that the interaction between intact RAR and RAC3 was abolished by deletion of AF2-AD. A similar conclusion was obtained by using mammalian culture cells (data not shown), in which a Gal4 DBD-RAR403 fusion did not activate transcription in response to ligand treatment (35). Truncation of the AF2-AD of RXR (RXR443) and VDR (VDR364), unlike that of RAR, did not create ligand-dependent transcriptional activators in yeast cells, while the abilities to interact with RAC3 were totally eliminated (Fig. 2). These results demonstrate that the AF2-ADs of RAR, RXR, and VDR are all absolutely required for interactions with RAC3; thus, RAC3 is an AF-2 dependent cofactor for nuclear receptors.
Figure 2.
AF2-AD is required for interaction with RAC3. The indicated Gal4-AD fusion (RAC3.1) and Gal4-DBD fusion were cotransformed into yeast Y190 cells, and the β-galactosidase activities from three independent colonies were determined. The yeast cells were treated with 1 μM indicated ligands (filled bars) or with an equal concentration of vehicle alone (open bars). RAR403: full-length hRARα truncated at amino acid 403; RXR443, full-length hRXRα truncated at amino acid 443; VDR364, LBD of hVDR truncated at amino acid 364.
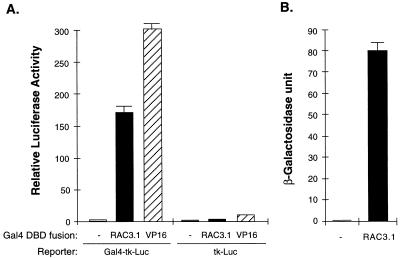
The above results suggest RAC3 as one of the components of a transcriptionally active complex of liganded receptors. Therefore, we further tested whether RAC3 can directly stimulate transcription when recruited to a specific promoter by linking with a heterologous DBD. Transient transfection of a Gal4 DBD-RAC3 fusion into CV-1 cells resulted in strong stimulation of gene expression from a luciferase reporter containing five copies of Gal4-binding sites, but not from a parental reporter without the Gal4-binding sites (Fig. 3A). The Gal4 DBD alone did not activate the reporter, while a Gal4 DBD fusion of VP16 activation domain strongly stimulated reporter gene expression. Similar experiments in yeast cells also demonstrated a RAC3-dependent transcription stimulation from a lacZ reporter in this organism (Fig. 3B). Thus, RAC3 itself contains a transcriptional activation domain functional in both mammalian and yeast cells.
Figure 3.
RAC3 contains a transcriptional activation domain. The ability of RAC3 to stimulate transcription was tested in both mammalian and yeast cells. (A) Mammalian cells. The Gal4 DBD alone (−) or its fusion with RAC3.1 or VP16 activation domain was expressed in CV-1 cells together with a luciferase reporter containing five copies of Gal4-binding sites (Gal4-tk-Luc) or the parental vector without Gal4-binding sites (tk-Luc). The relative luciferase activities are averages of three independent transfections normalized to β-galactosidase activity. (B) Yeast cells. Three independent colonies expressing Gal4 DBD fusion of RAC3.1 in Y190 cells were analyzed for their β-galactosidase activities. The average β-galactosidase units with standard deviation are shown. The control experiment (−) is β-galactosidase activity of Y190 cells transformed with Gal4 DBD alone.
Sequence analysis revealed that the yeast two-hybrid clone RAC3.1 encodes a polypeptide of 804 amino acids (Fig. 4A, in between arrows). Database search revealed a weak similarity of this polypeptide to the central domains of two recently identified steroid receptor coactivators, SRC-1 (17, 18, 38) and TIF2/GRIP1 (22, 39, 40). Full-length RAC3 cDNA was cloned and the nucleotide sequences were determined. The deduced amino acid sequences indicated that RAC3 is a 154.4-kDa protein containing 1417 residues (Fig. 4A). Sequence alignment of full-length RAC3, TIF2 (22), and human SRC-1 (38) revealed a striking homology at the N-terminal region of about 350 amino acids (Fig. 4B). This region contains a potential basic helix–loop–helix (bHLH) domain similar to many transcriptional regulators (41), and two Per-AhR-Sim (PAS) domains found in several nuclear proteins, including Period (Per), aryl hydrocarbon receptor (AhR) and its heterodimeric partner ARNT, the Single Minded protein (Sim), and the hypoxia-inducible factor HIF1α (42). Sequence comparison among these bHLH-PAS-containing proteins revealed that RAC3, TIF2, and SRC-1 share a higher degree of similarity than the other proteins (Fig. 4B), suggesting these three receptor-associated coactivators constitute a unique bHLH-PAS family.
Figure 4.
RAC3 is related to SRC-1 and TIF2. (A) Deduced amino acid sequences of full-length RAC3. The regions corresponding to the basic helix–loop–helix (bHLH) and Per-AhR-Sim (PAS “A” and “B”) domains are shown in boxes. The C-terminal glutamine-rich (Q-rich) domain is underlined. Six LXXLL or LLXXL motifs (i to vi) at the central region are shown in boxes. (L indicates hydrophobic residues including leucine, isoleucine, and valine). The starting and ending amino acids encoded by the two-hybrid clone RAC3.1 are shown within arrows. (B) Comparison of the bHLH and PAS domains of RAC3 with TIF2, SRC-1, and other bHLH-PAS domain proteins. The white letters are conserved residues determined when more than half of residues in all sequences are similar. (C) Sequence alignment of the LXXLL motifs. The starting and ending amino acids are shown at right in parentheses. The first six motifs are surrounded by highly charged residues, and motifs ii, iv, v, and vi were predicted to form α-helical structures. (D) Schematic diagram of the domain structures of full-length human RAC3, TIF2, and SRC-1. The starting and ending residues of indicated domains are shown. The domains encoded by RAC3.1, TIF2.1, GRIP1, and SRC-1(.8) clones are indicated (arrows). The RAR-binding and p300-binding domains defined in mSRC-1 are also indicated. The numbers at the right are the length of individual proteins and the percentage similarity. The pairwise similarities were calculated to be 65% between RAC3 and TIF2, 64% between TIF2 and SRC1, and 59% between RAC3 and SRC1.
Within the RAC3.1 encoding region (amino acid residues 401-1024), we identified six specific motifs sharing a consensus sequence of LXXLL or LLXXL (Fig. 4A). These motifs and their neighboring residues are highly charged and well conserved among RAC3, TIF2, and SRC-1 (Fig. 4C, i to vi). Secondary structure prediction suggests that all these regions have the potential of forming helices, especially motifs iv, v, and vi. Because both transcriptional activation and receptor-interaction activities of RAC3 are located within the RAC3.1 fragment, it is possible that these conserved motifs may play a role in these functions. In addition, a similar motif was found at the C terminus of SRC-1, located within the originally identified receptor-interacting domain (Fig. 4C, vii) (18). At the C-terminal domain of RAC3, a glutamine-rich (Q-rich) domain was identified (Fig. 4A). This Q-rich domain is also conserved among RAC3, TIF2, and SRC-1 and, interestingly, a stretch of 26 consecutive glutamine residues was found only in RAC3. A schematic domain structure comparison among these three related proteins is shown in Fig. 4D.
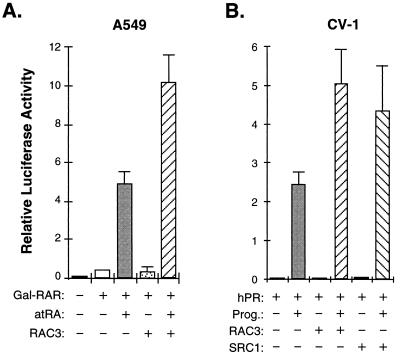
The ability of RAC3 to physically interact with liganded receptors in an AF-2-dependent manner and to directly stimulate transcription when recruited to a heterologous promoter, together with its sequence similarity to two of the most potent receptor coactivators, are properties which strongly suggest that RAC3 functions as a receptor-associated coactivator. To demonstrate the coactivator function of RAC3, we transiently transfected a mammalian expression vector for full-length RAC3 into human lung carcinoma A549 and/or monkey kidney CV-1 cells. Overexpression of RAC3 enhanced the ligand-dependent transcriptional activation by Gal4 DBD-RAR fusion on a Gal4-dependent luciferase reporter in A549 cells (Fig. 5A), as well as in CV-1 but not in HeLa cells, which express a high level of RAC3 (data not shown). We also tested the effect of overexpression of RAC3 on the transcriptional activation by the human progesterone receptor (hPRB) and found that RAC3 also enhanced transcription by wild-type hPRB from a natural mouse mammary tumor virus long terminal repeat (MMTV-LTR) promoter, close to the effect observed with full-length human SRC-1 under the test conditions (Fig. 5B). These results demonstrate that RAC3 is capable of enhancing transcriptional activation by liganded receptors in mammalian cells, suggesting that RAC3 is indeed a bona fide transcriptional coactivator for steroid/nuclear hormone receptors.
Figure 5.
RAC3 enhances transcriptional activation by steroid/nuclear receptors. (A) The pCMX-F.RAC3 construct expressing full-length RAC3 was transfected into A549 cells together with a construct expressing Gal4-DBD fusion of the LBD of hRARα (Gal-RAR) and a Gal4-tk-Luc reporter. The relative luciferase activities are averages from three independent experiments after normalization to β-galactosidase activity used as internal controls for transfection efficiency. Transfected cells were treated with vehicle alone (−) or with 100 nM of all-trans-retinoic acid for 24 hr after transfection. Overexpression of RAC3 does not have an effect on the luciferase reporter lacking Gal4-binding sites (not shown). (B) RAC3 enhances transcriptional activation by PR on MMTV-LTR promoter. Transfected cells were treated with or without 100 nM progesterone in the presence or absence of coexpressed RAC3 or SRC-1 in CV-1 cells.
DISCUSSION
We identified RAC3 as a nuclear receptor-interacting protein on the basis of its ability to interact with RAR in a yeast two-hybrid screen. Several lines of evidence support the position that RAC3 is a transcriptional coactivator for steroid/nuclear hormone receptors. First, we showed that RAC3 interacts with ligand-activated receptors. Second, we demonstrated that the ligand-dependent interaction with RAC3 requires an intact AF2-AD on the receptor. Third, we found that RAC3 itself contains a transcriptional activation domain. Finally, we demonstrated that overexpression of RAC3 enhances ligand-stimulated transcriptional activation by the receptors. In summary, the biochemical properties of RAC3, together with its sequence similarity to SRC-1 and TIF2, identify this protein as a distinct third member of the RAC gene family.
At least two distinct activation functional domains (i.e., AF-1 and AF-2) have been identified in steroid/nuclear hormone receptors (43). The N-terminal AF-1 domain is constitutively active and its activity is not regulated by hormone. In contrast, the activity of the C-terminal AF-2 domain depends on ligand binding. The exact role of the C-terminal AF2-AD helix in recruitment of coactivators is unclear. It is known that the AF2-AD alone is not sufficient for interaction with RAC3 or other coactivators (unpublished data), indicating that other regions of the LBD are also required for recruitment of coactivators. Consistent with this hypothesis, several other activation motifs throughout the LBD of TR have been reported (28). Alternatively, the AF2-AD domain may simply stabilize ligand binding by forming part of the contacting surface for ligands (27, 33), which in turn induces additional conformational changes that allow association with coactivators.
RAC3 domains responsible for receptor interaction and transcriptional activation remain to be determined. Our data indicated that both domains are located within the central region of RAC3. The receptor-interacting domain of TIF2/GRIP1 has also been located to the central domains (22, 39), while a receptor-interacting domain of SRC-1 was originally found at the C terminus (18). Analysis of the murine SRC-1 has revealed an additional RAR-interacting domain located at the central region (44), where three of the six conserved LXXLL motifs were found. We have confirmed that the N-terminal 782 amino acids of human SRC-1, as well as the C-terminal SRC-1 fragment (amino acids 1245–1440), can interact with several steroid/nuclear hormone receptors in a ligand-dependent manner (unpublished results). However, the C-terminal domain of RAC3 (amino acids 1017–1417), which does not contain any LXXLL motif, cannot interact with liganded receptors (unpublished data). Similar motifs were also found in other potential nuclear receptor coactivators such as RIP-140 (13) and CREB-binding protein (CBP)/p300 (45, 46). These results are consistent with the model that these conserved regions are important for interaction with liganded receptors. Additional domain mapping and mutagenesis experiments should provide further insights into the mechanism of receptor–coactivator interaction. Whether or not some of these conserved motifs are involved in transcriptional activation is also unknown. It is noted that three of these LXXLL motifs are located within the p300-binding domain in mSRC-1 (44); thus, it is possible that these motifs may serve to recruit p300 and/or the basal transcriptional apparatus.
The high degree of sequence conservation within the bHLH-PAS region among RAC3, TIF2, and SRC-1, as compared with other bHLH-PAS proteins, indicates that these three coactivators belong to a distinct family. Intriguingly, the bHLH-PAS domain is dispensable for coactivator functions, including receptor interaction, transcriptional activation, and coactivation (18, 39, 40, 44). The bHLH domain of several transcriptional regulators has been shown to function as a distinct DNA-binding and dimerization motif (42, 47). Whether the bHLH domain in RAC3 and in TIF2 and SRC-1 can mediate protein–protein or protein–DNA interactions is currently unclear. The PAS domains have been shown to be involved in protein–protein interaction in ARNT and its heterodimeric partners (48); however, its role in receptor coactivators is unclear.
SRC-1 has been shown to enhance transcriptional activation by several steroid/nuclear hormone receptors, including PR, growth hormone receptor (GR), estrogen receptor (ER), TR, PPAR, and RXR (18, 25, 49), and it can function synergistically with the common coactivator CBP/p300 (17, 24, 44). It has been proposed that SRC-1 can stabilize a functional interaction between AF-1 and AF-2 of the ER (25). TIF2/GRIP1 has also been shown to enhance transcriptional activities of several steroid/nuclear receptors (22, 40). Our results demonstrated that RAC3 can at least enhance transcriptional activation by RAR and PR. However, it is not clear why we observed only a 2-fold enhancement in both cases by RAC3, while a much higher level of enhancement was observed previously by SRC-1 (18). We reason that this might be due the fact that, in our experiments, an initial 175-fold induction was obtained in PR by hormone treatment in the absence of cotransfected RAC3 or SRC-1, which is in contrast to a 5-fold initial enhancement in an earlier report (18). Therefore, a saturation effect might have limited our ability to detect high level of enhancement. Furthermore, in these experiments we used an SRC-1 cDNA clone that contains the N-terminal bHLH-PAS domain, while in the earlier report a variant of SRC-1 lacking the bHLH-PAS domain was used. By direct comparison between RAC3 and SRC-1, it is clear that RAC3 has a similar degree of coactivation function like SRC-1 in CV-1 cells under our assay conditions. Further analysis of the effect of RAC3 on transcriptional activation by other steroid/nuclear receptors and a direct comparison with SRC-1 and TIF2 will be necessary to understand how these three related proteins work together to regulate gene activation by hormone receptors.
Acknowledgments
We thank Drs. E. Bresnick, N. Brown, K. Carlson, W. F. Greenlee, C. L. Peterson, and D. Waud, and other colleagues for stimulating discussion and critical reading of the manuscript. We are very grateful to Dr. K. Umesono for the MMTV-LTR luciferase reporter plasmid and Dr. R. M. Evans for plasmids originated from his laboratory. We also acknowledge Dr. B. O’Malley and M-J. Tsai for providing the hPRB and SRC-1 expression constructs. This work was supported by Institutional Grant IRG203 from the American Cancer Society and a Howard Hughes Pilot Grant for Genetic Research, both provided through the University of Massachusetts Medical School, and an American Society of Hematology Junior Faculty Scholar Award to J.D.C.
ABBREVIATIONS
- SRC-1
steroid receptor coactivator 1
- TIF2
transcriptional intermediate factor 2
- RAC
receptor-associated coactivators
- RAR
retinoic acid receptor
- RXR
retinoid-X receptor
- TR
thyroid hormone receptor
- VDR
vitamin D3 receptor
- PPAR
peroxisomal proliferator-activated receptor
- PR
progesterone receptor
- DBD
DNA-binding domain
- LBD
ligand-binding domain
- AF
activation function
- AF2-AD
AF-2 activation domain
- CBP
CREB-binding protein
- h
human
- m
murine
- bHLH
basic helix–loop–helix
- PAS
Per-AhR-Sim
Note Added in Proof
While this paper was in press, Torchia et al. (50) reported a CBP-interacting protein (p/CIP, GenBank accession no. AF000581) which is essential for transcriptional activation by steroid/nuclear receptors as well as by other CBP-dependent transcription factors. RAC3/pCIP shares about 78% identity, including a highly diverged C-terminal 103 amino acids, suggesting that p/CIP is an alternatively spliced form of the murine homologue of RAC3. In the same issue of Nature, Heery et al. (51) reported that the first three conserved LXXLL motifs (motifs i, ii, and iii) in SRC-1 are essential and sufficient to mediate protein–protein interactions with liganded receptors.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF010227).
References
- 1.Evans R M. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangelsdorf D J, Evans R M. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 4.Beato M, Herrlich P, Schültz G. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 5.Kastner P, Mark M, Chambon P. Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 6.Thummel C S. Cell. 1995;83:871–877. doi: 10.1016/0092-8674(95)90203-1. [DOI] [PubMed] [Google Scholar]
- 7.Yu V C, Delsert C, Anderson B, Holloway J M, Devary O V, Narr A M, Kim S Y, Boutin J M, Glass C K, Rosenfeld M G. Cell. 1991;67:1251–1266. doi: 10.1016/0092-8674(91)90301-e. [DOI] [PubMed] [Google Scholar]
- 8.Kliewer S A, Umesono K, Mangelsdorf D J, Evans R M. Nature (London) 1992;355:446–449. doi: 10.1038/355446a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Umesono K, Murakami K K, Thompson C C, Evans R M. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, Rosenfeld M G. Nature (London) 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 11.Chen J D, Evans R M. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 12.Fondell J D, Ge H, Roeder R G. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavaillès V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner P J, Parker M G. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavaillès V, Dauvois S, Danielian P S, Parker M G. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 16.Le Douarin B, Zechel C, Garnier J M, Lutz Y, Tora L, Pierrat B, Heery D, Gronemeyer H, Chambon P, Losson R. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 18.Oñate S A, Tsai S Y, Tsai M J, O’Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 19.Lee J W, Ryan F, Swaffield J C, Johnston S A, Moore D D. Nature (London) 1995;374:91–94. doi: 10.1038/374091a0. [DOI] [PubMed] [Google Scholar]
- 20.vom Baur E, Zechel C, Heery D, Heine M J, Garnier J M, Vivat V, Le Douarin B, Gronemeyer H, Chambon P, Losson R. EMBO J. 1996;15:110–124. [PMC free article] [PubMed] [Google Scholar]
- 21.Yeh S, Chang C. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voegel J J, Heine M J S, Zechel C, Chambon P, Gronemeyer H. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 23.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Nature (London) 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 24.Smith C L, Onate S A, Tsai M J, O’Malley B W. Proc Natl Acad Sci USA. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McInerney E M, Tsai M J, O’Malley B W, Katzenellenbogen B S. Proc Natl Acad Sci USA. 1996;93:10069–10073. doi: 10.1073/pnas.93.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Nature (London) 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 27.Renaud J P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Nature (London) 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 28.Baniahmad A, Leng X, Burris T P, Tsai S Y, Tsai M J, O’Malley B W. Mol Cell Biol. 1995;15:76–86. doi: 10.1128/mcb.15.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hollenberg S M, Evans R M. Cell. 1988;55:899–906. doi: 10.1016/0092-8674(88)90145-6. [DOI] [PubMed] [Google Scholar]
- 30.Damm K, Heyman R, Umesono K, Evans R M. Proc Natl Acad Sci USA. 1993;90:2989–2993. doi: 10.1073/pnas.90.7.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barettino D, Vivanco Ruiz M d M, Stunnenberg H G. EMBO J. 1994;13:3039–3049. doi: 10.1002/j.1460-2075.1994.tb06603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durand B, Saunders M, Gaudon C, Roy B, Losson R, Chambon P. EMBO J. 1994;13:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner R L, Apriletti J W, West B L, Baxter J D, Fletterick R J. Nature (London) 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 34.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 35.Chen J D, Umesono K, Evans R M. Proc Natl Acad Sci USA. 1996;93:7567–7571. doi: 10.1073/pnas.93.15.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai S, Collins S J. Proc Natl Acad Sci USA. 1993;90:7153–7157. doi: 10.1073/pnas.90.15.7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Privalsky M L, Sharif M, Yamamoto K R. Cell. 1990;63:1277–1286. doi: 10.1016/0092-8674(90)90423-c. [DOI] [PubMed] [Google Scholar]
- 38.Takeshita A, Yen P M, Misiti S, Cardona G R, Liu Y, Chin W W. Endocrinology. 1996;137:3594–3597. doi: 10.1210/endo.137.8.8754792. [DOI] [PubMed] [Google Scholar]
- 39.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong H, Kohli K, Garabedian M J, Stallcup M R. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murre C, McCaw P S, Baltimore D. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 42.Swanson H I, Bradfield C A. Pharmacogenetics. 1993;3:213–230. doi: 10.1097/00008571-199310000-00001. [DOI] [PubMed] [Google Scholar]
- 43.Tasset D, Tora L, Fromental C, Scheer E, Chambon P. Cell. 1990;62:1177–1187. doi: 10.1016/0092-8674(90)90394-t. [DOI] [PubMed] [Google Scholar]
- 44.Yao T P, Ku G, Zhou N, Scully R, Livingston D M. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arany Z, Seller W, Livingston D M, Eckner R. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 46.Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Nature (London) 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 47.Murre C, McCaw P S, Vaessin H, Caudy M, Jan L Y, Jan Y N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, Weintraub H, Baltimore D. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 48.Lindebro M C, Poellinger L, Whitelaw M L. EMBO J. 1995;14:3528–3539. doi: 10.1002/j.1460-2075.1995.tb07359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DiRenzo J, Söderström M, Kurokawa R, Ogliastro M-H, Ricote M, Ingrey S, Hörlein A, Rosenfeld M G, Glass C K. Mol Cell Biol. 1997;17:2166–2176. doi: 10.1128/mcb.17.4.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. Nature (London) 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 51.Heery D M, Kalkhoven E, Hoare S, Parker M G. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]