Abstract
The human androgen receptor (AR) is a ligand-activated transcription factor that regulates genes important for male sexual differentiation and development. To better understand the role of the receptor as a transcription factor we have studied the mechanism of action of the N-terminal transactivation function. In a protein–protein interaction assay the AR N terminus (amino acids 142–485) selectively bound to the basal transcription factors TFIIF and the TATA-box-binding protein (TBP). Reconstitution of the transactivation activity in vitro revealed that AR142–485 fused to the LexA protein DNA-binding domain was competent to activate a reporter gene in the presence of a competing DNA template lacking LexA binding sites. Furthermore, consistent with direct interaction with basal transcription factors, addition of recombinant TFIIF relieved squelching of basal transcription by AR142–485. Taken together these results suggest that one mechanism of transcriptional activation by the AR involves binding to TFIIF and recruitment of the transcriptional machinery.
The androgen receptor (AR) is a member of the steroid–thyroid hormone receptor superfamily and mediates the effects of the male sex hormones testosterone and dihydrotestosterone (for review see ref. 1). Mutations in the receptor proteins have been identified in disorders of male sexual differentiation (2, 3), X-chromosome-linked spinal bulbar muscular atrophy (4, 5), prostatic carcinoma (6, 7), and male breast cancer (8). Although there is good evidence that the AR binds to DNA response elements and activates gene expression, the underlying mechanisms are not well understood. The C-terminal steroid-binding domain and the central DNA-binding domain show significant homology between ARs of different species and also with other members of the nuclear receptor superfamily (ref. 1 and references therein). In contrast, the N terminus of the protein is more divergent and is characterized by homopolymer tracts of glutamine, glycine, and proline residues (ref. 9 and references therein). Regions within the N terminus of the human and rat receptors important for transactivation have been delineated by deletion analysis (10–13), the use of fusion proteins (14), and point mutations (15). These studies have highlighted the region between amino acids 142 and 370 (numbering for the human receptor), although sequences both N-terminal and C-terminal of this region appear to play an important role in the full activity of the wild-type AR and/or in promoter specific activity (see ref. 14).
Transcription of mRNA coding genes involves the concerted action of RNA polymerase II and a set of at least five general transcription factors (see refs. 16–19 for recent reviews). One mechanism by which gene regulatory proteins are thought to function is by recruiting one or more of the general transcription factors, and thus the polymerase, to the promoter (reviewed in refs. 17–19). This can be achieved by direct contact between the activator and the general transcription factors and/or interactions by means of coactivator proteins (refs. 17 and 19–21 and references therein).
In recent years a number of interactions have been described between members of the steroid–thyroid hormone receptor superfamily and basal transcription factors and co–activator proteins (see ref. 22 and references therein). However, very little is known concerning the identity of interacting proteins with the human AR. To better understand the mechanism of gene regulation by the human AR we have screened a panel of general transcription factors for binding to the receptor N-terminal transactivation domain and have reconstituted receptor-dependent activation under cell-free conditions. A region of the N terminus, containing the major transactivation activity, is capable of recruiting the general transcription machinery to a target promoter and shows selective binding to the general transcription factor TFIIF.
MATERIALS AND METHODS
AR Expression Constructs.
The DNA sequence coding for amino acids 142–485 of the human AR N terminus was amplified by using the Expand Long Template PCR system (Boehringer Mannheim) from plasmid pSVARo (a gift from A. O. Brinkmann, Erasmus University, Rotterdam, The Netherlands; see ref. 9). The primers used were ARN142, 5′-GCGCGCAGATCTCTGCCGCAGCAGCTGCCAGC-3′, and ARC485, 5′-GCGCGCGGATCCGCTTTCCTGGCCCGCCAGCCCC-3′. The PCR product was cleaved with BamHI and BglII and ligated into pET-19bm (23) previously digested with BamHI. The resulting plasmid, pET-AR4, expressed the AR142–485 domain fused to an N-terminal histidine tag. The insert was checked for orientation and sequenced (Dye terminator cycle sequencing, Perkin–Elmer). The expression plasmid pET-AR4-Lex was constructed by ligating a fragment encoding the LexA DNA-binding domain (LexADBD, amino acids 1–87) into the regenerated BamHI site, 3′ of the AR sequence in pET-AR4.
Expression and Purification of Recombinant Proteins.
AR142–485 with or without LexADBD was expressed in Escherichia coli strain BL21 plys by isopropyl β-d-thiogalactoside (IPTG; 1 mM) induction, and the recombinant proteins were purified from the soluble fraction by Ni2+–nitrilotriacetate (NTA) affinity chromatography. The bound protein was eluted with 200 mM imidazole and dialyzed against 25 mM Hepes, pH 7.6/100 mM sodium acetate/1 mM DTT/0.01% Nonidet P-40. Recombinant yeast TATA-box-binding protein (TBP) and human TFIIF (RAP30 and RAP74) were expressed in bacteria and partially purified as described previously (24). Protein concentrations were measured against BSA standards using the Bradford reagent (Bio-Rad).
Protein–Protein Interaction Assay.
The microtiter plate interaction assay was essentially as described previously (23, 24). Briefly, AR142–485 or BSA control in binding buffer [20 mM Hepes, pH 7.6/10% (vol/vol) glycerol/100 mM KCl/0.2 mM EDTA/5 mM MgCl2/5 mM 2-mercaptoethanol] were allowed to adsorb to the surface of a scintillation-microtiter plate (Wallac, Oy, Finland). Unoccupied surfaces were subsequently blocked with binding buffer containing 5 mg/ml BSA, and the wells were incubated with binding buffer containing 1 mg/ml BSA and radiolabeled human basal transcription factors, synthesized in a rabbit reticulocyte lysate system (Promega). After extensive washing with binding buffer + 1 mg/ml BSA, the bound radioactivity was measured directly in a micro β counter (Wallac, Oy, Finland), and bound proteins were recovered in SDS sample buffer.
In Vitro Transcription and Squelching Assays.
Preparation of yeast nuclear extracts for in vitro transcription, together with reporter genes and reaction conditions have all been described in detail previously (24–26).
RESULTS
The N-Terminal Region of the Human AR Contacts Basal Transcription Factors.
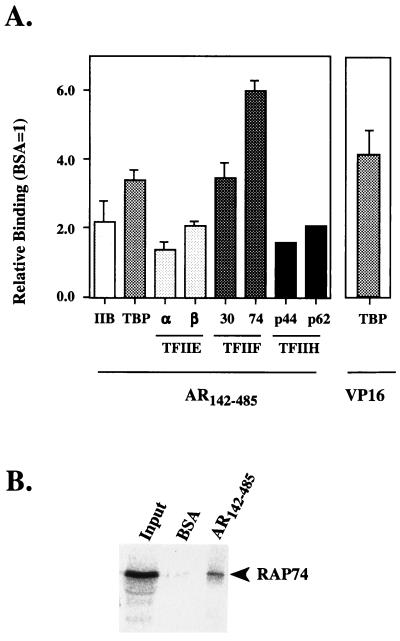
The N terminus of the AR has been shown to contain a complex transactivation function made up of multiple regions (see Introduction). In an attempt to understand the mechanism by which the AR activates transcription, a panel of basal transcription factors was screened for interactions with the receptor transactivation domain. A polypeptide containing amino acids 142–485 of the human receptor was expressed and purified by metal chelation chromatography (Fig. 1). The purified protein was allowed to adsorb onto the surface of a microtiter plate and incubated with 35S-labeled basal transcription factors TFIIB, TBP, TFIIEα and -β, TFIIF (RAP30 and RAP74), and two of the subunits of TFIIH (p44 and p62). AR142–485 interacted selectively with the RAP74 subunit of TFIIF and showed modest binding to the RAP30 subunit of TFIIF and to TBP (Fig. 2A). Little or no significant binding was observed with TFIIB, TFIIE, or the two subunits of TFIIH, as judged relative to the BSA control and binding to proteins lacking a transactivation function (Fig. 2A and data not shown; see also refs. 23 and 24). To evaluate the significance of these observations, we compared the binding of AR142–485 with previously reported interactions between the serum response factor (SRF) and RAP74 (27) and the potent viral activator VP16 and TBP (see ref. 16). The binding of AR142–485 to RAP74 was as strong, if not stronger, compared with SRF245–508 (see references 23 and 24; also data not shown). While the interaction with TBP was comparable to that seen with VP16 (Fig. 2A and refs. 23 and 24). Recovery of the bound material and analysis by SDS/PAGE confirmed that the measured radioactivity represented intact labeled RAP74 binding to AR142–485, while little or no labeled protein was recovered from the BSA-only control (Fig. 2B). Thus, a region of the AR N terminus containing the transactivation function bound selectively to the basal transcription factors TBP and TFIIF.
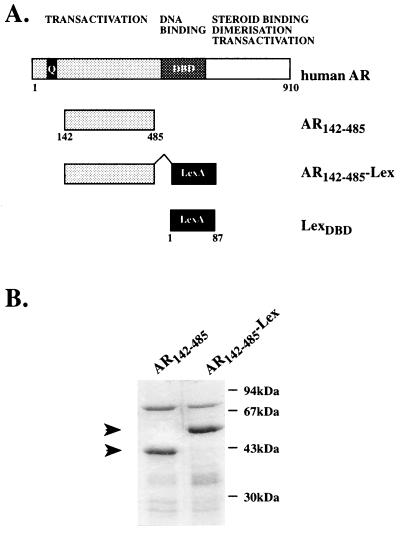
Figure 1.
(A) Schematic representation of the human AR showing the domain organization and the region of the N terminus used in the present study. (B) Purification of His-tagged AR142–485 and AR142–485-Lex proteins. Proteins were estimated to be at least 75% pure from the Coomassie blue-stained gel.
Figure 2.
(A) Screening of 35S-labeled human general transcription factors for binding to AR142–485. The measured radioactivity is plotted relative to wells containing BSA only, which was set at 1. The binding of TBP to the C-terminal transactivation domain of the herpes simplex viral activator protein VP16 is shown for comparison. The results represent the mean ± SD of at least four observations from two or more independent experiments, except TFIIH, subunits where the mean of two observations only is shown. (B) SDS/PAGE analysis of the bound RAP74 subunit of TFIIF to AR142–485. The input represents 5% of the material incubated per well.
Reconstitution of Receptor Transactivation Activity Under Cell-Free Conditions.
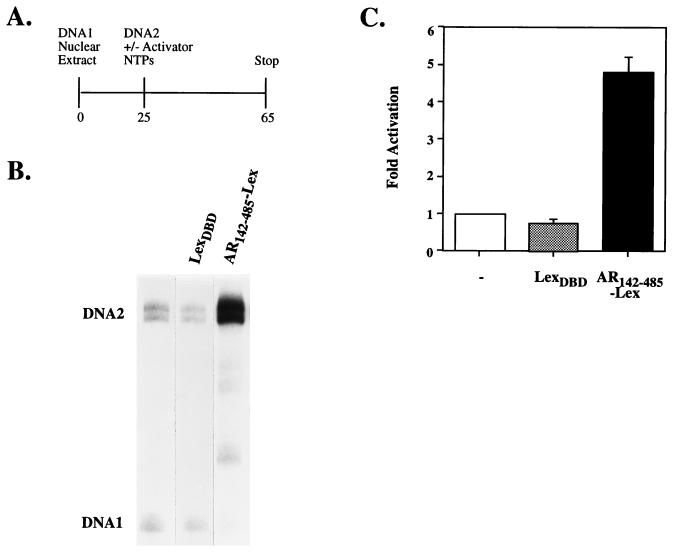
To study the mechanism of transcriptional activation and to evaluate the functional relevance of the observed interactions, the N-terminal transactivation activity was reconstituted under cell-free conditions. The AR142–485 region was fused to the heterologous DNA-binding domain of the bacterial LexA protein (LexADBD) and was expressed and purified (Fig. 1). Previously, a relatively crude preparation of full-length human AR, expressed using vaccinia virus, was shown to activate a reporter gene in HeLa nuclear extracts (28). However, the mechanism(s) of activation was not addressed in this study. To test whether AR142–485 could recruit factors to the promoter, the basal transcription machinery was allowed to assemble on a competing template, prior to addition of the reporter DNA and the receptor. In the absence of recruitment, transcription from the first template would be expected to occur preferentially. Therefore, nuclear extracts were preincubated with the AdMLΔ-50 basal promoter reporter gene (DNA 1), prior to the addition of the 1xLexCG reporter gene (DNA 2), NTPs, and activator (Fig. 3A). Under these conditions the levels of basal transcription from DNA 2 were 10–20% lower than observed in the absence of DNA 1 (Fig. 3B lane 1 and data not shown). Addition of AR142–485-LexADBD resulted in an approximately 5-fold activation of transcription relative to no added activator (Fig. 3 B and C; DNA 2). This activation was dependent on the presence of the AR142–485 polypeptide, as LexADBD alone showed no stimulation of transcription (Fig. 3 B and C; DNA 2). Furthermore, transcription from the AdMLΔ-50 template was essentially eliminated in the presence of AR142–485-LexADBD (Fig. 3B; DNA 1). Since the 1xLexCG reporter template (DNA 2) was added after preincubation of the competing template (DNA 1), these results suggest that the AR functions, at least in part, to recruit the general transcription machinery to a target promoter.
Figure 3.
(A) Overview of the experimental protocol (time shown in minutes) followed to reconstitute the transactivation activity of the AR142–485 domain fused to the LexADBD in yeast nuclear extracts. DNA 1 represents the adenovirus major late basal promoter upstream of the 180-bp G-free cassette (AdMLΔ-50; ref. 29), and DNA 2 is the CYC1 basal promoter with one LexA binding site cloned upstream fused to the 380-bp G-free cassette (1xLexCG; ref. 23). (B) Representative experiment showing RNase T1-resistant transcripts derived from DNAs 1 and 2 in the absence or presence of 50 pmol of LexADBD or AR142–485-LexDBD. Note two specific transcripts are obtained from DNA 2 (see ref. 30). (C) Transcripts from DNA 2 were quantitated using a Bio-imaging analyzer BAS2000 (FujiFilm), and mean and SD (n = 3) are plotted relative to basal transcription (no added activator) set to 1.
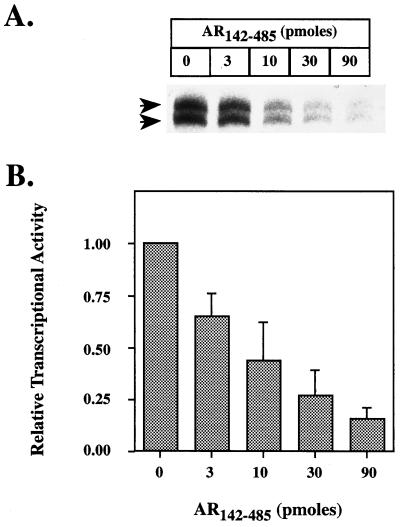
Previously, we have used a squelching assay to assess the role of protein–protein interactions in activator function (23–25), where the squelching is thought to result from the competition for a limiting pool of target factor(s) (31). Addition of AR142–485, in the absence of the DNA-binding domain, together with a basal reporter gene resulted in a concentration-dependent inhibition of transcription, consistent with the transactivation domain interacting with a component(s) required for basal transcription (Fig. 4). This is likely to be a specific effect, since the squelching occurred within the concentration range required for activation and was comparable to that seen with other activator proteins, VP16 (32, 33), glucocorticoid receptor (GR)-τ1 (25), c-myc (24), and the arylhydrocarbon receptor (AhR) and the AhR nuclear translocator protein (ARNT) (23) transactivation domains under similar conditions.
Figure 4.
(A) Representative experiment showing squelching of basal transcription from CYC1 basal promoter (pΔCG; ref. 30) by increasing amounts of AR142–485. (B) Quantitation of basal transcription in the presence of increasing amounts of AR142–485. The results were quantitated and plotted as described in legend to Fig. 3C for three independent experiments.
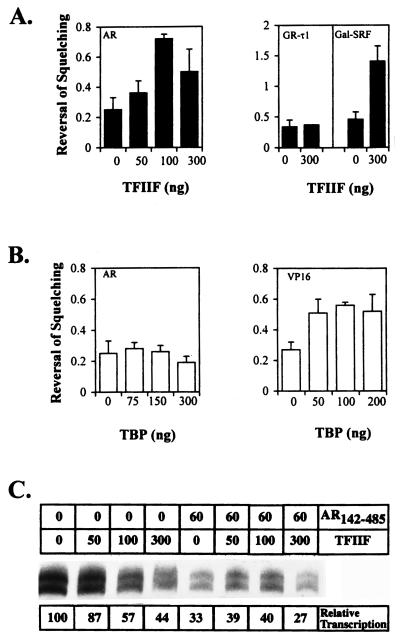
Recombinant TFIIF Reverses AR-Dependent Squelching.
The squelching assay provides a functional test for the significance of interactions identified in protein–protein binding studies, since the addition of putative target factors would be predicted to relieve the squelching. Fig. 5A and C shows that addition of partially purified human TFIIF resulted in an inhibition of basal transcription in the absence of AR142–485 but significantly relieved the squelching of transcription by AR142–485. Relief of squelching was measured as the increase in the ratio of relative transcription in the presence versus the absence of the activator, AR142–485, relative to the amount of TFIIF added (Fig. 5A Left). Recombinant TFIIF also relieved auto squelching by the SRF245–508 transactivation domain but not squelching by the GR-τ1 (Fig. 5A Right). Under similar conditions, recombinant TBP was unable to reverse AR142–485-dependent squelching, although there was an overall increase in the levels of transcription in the absence of AR142–485 (Fig. 5B and data not shown). In contrast, TBP successfully overcame squelching of basal transcription by VP16 (Fig. 5B Right) and the c-myc (24) transactivation domains. Taken together, these data strongly suggest that TFIIF is a target factor for the AR N-terminal transactivation function.
Figure 5.
(A) (Left) Relief of AR142–485-dependent squelching by the addition of recombinant TFIIF. Reversal of squelching was calculated as the ratio of the relative transcription in the presence (60 pmol) versus the absence (0 pmol) of AR142–485 and plotted against the amount of TFIIF added (ng of each subunit). Data for at least three experiments are shown. In the absence of TFIIF subunits, the mean of this ratio is 0.26 ± 0.08 (n = 6), and an increase in this ratio is indicative of reversal of squelching. (Right) Reversal of self-squelching by Gal-SRF245–508 (15 pmol) but not squelching of basal transcription by GR-τ1 (20 pmol) by 300 ng of TFIIF. (B) Addition of recombinant TBP fails to relieve AR142–485-dependent squelching (Left). Conditions as in A. In the absence of TBP the mean of the ratio is 0.25 ± 0.11 (n = 3). In contrast, TBP did reverse squelching of basal transcription by VP16 (2–4 pmol) (Right). Mean and SD for at least four experiments are shown. (C) Representative experiment showing the effect of recombinant TFIIF on transcription levels in the absence or presence of AR142–485. The amounts of AR142–485 (pmol) and TFIIF subunits (ng) added are shown above each lane.
DISCUSSION
Regulation of transcription by gene regulatory proteins involves both direct and indirect interactions with the general transcription machinery (see Introduction). In the present study we show that the N-terminal transactivation function of the human AR acts, at least in part, to recruit the transcriptional machinery to the promoter. Consistent with this, the AR selectively binds to the general transcription factor TFIIF, and this interaction is sufficient to relieve squelching of basal transcription under cell-free conditions. The inability of TBP to relieve squelching by AR142–485 suggests that, although this region of the receptor can bind to TBP in an interaction assay, in the context of a functional assay this interaction is secondary to the interaction with TFIIF. This lack of a functional correlation with TBP binding is not thought to be due to the use of recombinant yeast TBP in the functional assay, as yeast TBP was capable of reversing squelching by VP16 (Fig. 5B) and the c-myc (24) transactivation domains under similar conditions. Although no significant binding of the AR142–485 domain to other components of the basal transcription machinery was observed we cannot exclude the possibility that such interactions were masked or occur with a component not tested. Wilson and co-workers (1) have previously reported that the N terminus of the human AR (amino acids 1–503) squelched activation by the full-length receptor in transient transfection studies. However, the nature of the factor(s) involved remains to be determined in this system.
TFIIF is composed of two subunits, RAP30 and RAP74, that were originally identified as RNA polymerase II-associated proteins (RAPs; reviewed in ref. 34). Furthermore, TFIIF has been identified as a component of the recently described holo-RNA polymerase/mediator complex found in both yeast (35, 36) and mammalian (37, 38) cells. TFIIF is unique in that it functions at two discrete steps in the transcription process: first, during transcription initiation, TFIIF is responsible for selective recruitment of the RNA polymerase II to promoter sequences (refs. 16 and 17 and references therein), and subsequently, TFIIF increases elongation efficiency by preventing pausing and so reducing arrest of the polymerase molecule (refs. 16, 17, and 39 and references therein). While the present results are compatible with a role for the AR in preinitiation complex assembly, it remains possible that the receptor also regulates gene expression at the level of elongation. Recently, a number of groups have demonstrated a role for activator proteins in elongation and correlated this to interactions with another of the basal transcription factors, TFIIH (40–43). Interestingly, preliminary results from kinetic studies of the AR-dependent squelching suggest that the receptor can act after preinitiation complex assembly (unpublished observations), and further investigation will be required to resolve these possible roles of the AR in transcription regulation.
Although many studies have focused on the general transcription factors TBP and TFIIB as possible targets for activator proteins, including nuclear receptors (refs. 16, 22, and 44 and references therein), a number of activators, including SRF (27), the protooncogenes c-myc (24), c-fos, and c-jun (45), and the arylhydrocarbon receptor and receptor transporter proteins (23), have been shown to interact with TFIIF subunits. While the functional significance of such interactions has not always been possible to demonstrate, the integrity of the RAP74 subunit was found to be critical for SRF-dependent transactivation of a reporter gene (27).
In recent years there have been a plethora of protein–protein interactions described involving members of the nuclear receptor superfamily and basal transcription factors, co-activator proteins, and proteins of unknown function (for review see ref. 22 and references therein). In the majority of cases these interactions have involved the generally weaker transactivation functions (AF-2, AF-τc) localized in the C-terminal ligand-binding domain (22). While many of the described interactions occur with several different receptors, Yeh et al. (46), using a two-hybrid screen, identified a protein (ARA70) that showed specificity for the androgen receptor steroid-binding domain. In the present study we demonstrate that a region of the N terminus, encompassing the AF-1 function of the human AR, acts to recruit the general transcription machinery to a target promoter. In addition, this region of the receptor selectively interacts with the subunits of the general transcription factor TFIIF. This report of an interaction involving the major transactivation function of the human AR indicates possible mechanisms whereby the DNA-bound receptor activates gene transcription.
Acknowledgments
We gratefully acknowledge the gift of plasmids from the following people: Drs. A. Berk (University of California at Los Angeles), A. O. Brinkmann (Erasmus University), Z. F. Burton (Michigan State University), J.-M. Egly (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Institut National de la Santé et de la Recherche Médicale), S. Hahn (Fred Hutchinson Cancer Research Center), M. Hampsey (Louisiana State University Medical Center), R. Kornberg (Stanford University), R. Prywes (Columbia University), D. Reinberg (University of Medicine and Dentistry of New Jersey–Robert Wood Johnson Medical School), and R. G. Roeder (Rockefeller University). We thank Drs. Anthony Wright and George Kuiper (Department of Biosciences, Karolinska Institute) for helpful discussions and critical reading of the manuscript, and we are grateful to Sara Windahl and Tova Almlöf (Department of Biosciences, Karolinska Institute) for reagents. This work was supported by a grant from the Swedish Medical Research Council (13X-2819).
ABBREVIATIONS
- AR
androgen receptor
- AR142–485
AR residues 142–485
- LexADBD
LexA DNA-binding domain (residues 1–87)
- TBP
TATA-box-binding protein
- SRF
serum response factor
Note Added in Proof
Recently, Snoek et al. (47) have used nuclear extracts derived from the prostate carcinoma cell line PC3, together with glutathione S-transferase (GST) fusion proteins to reconstitute rat AR function in vitro. In this system a recombinant protein encoding the receptor DNA-binding domain (DBD) and steroid-binding domain (SBD) activated a reporter gene more robustly than a fusion protein containing a region of the N terminus and DBD. Although this latter construct overlaps with the region used in the present study, a direct comparison is not possible, as we have used a heterologous DBD, to avoid the possible influence of DNA-binding efficiency, by the receptor DBD, on transactivation.
References
- 1.Zhou Z X, Wong C I, Sar M, Wilson E M. Recent Prog Horm Res. 1994;49:249–274. doi: 10.1016/b978-0-12-571149-4.50017-9. [DOI] [PubMed] [Google Scholar]
- 2.Quigley C A, De Bellis A, Marschke K B, el-Awady M K, Wilson E M, French F S. Endocr Rev. 1995;16:271–321. doi: 10.1210/edrv-16-3-271. [DOI] [PubMed] [Google Scholar]
- 3.Brinkmann A O, Jenster G, Ris-Stalpers C, van der Korput H, Bruggenwirth H, Boehmer A, Trapman J. Steroids. 1996;61:172–175. doi: 10.1016/0039-128x(96)00008-6. [DOI] [PubMed] [Google Scholar]
- 4.Trifiro M A, Kazemi-Esfarjani P, Pinsky L. Trends Endocrinol Metab. 1994;5:416–421. doi: 10.1016/1043-2760(95)92524-m. [DOI] [PubMed] [Google Scholar]
- 5.Brown T R. Prostate Suppl. 1996;6:9–12. [PubMed] [Google Scholar]
- 6.Klocker H, Culig Z, Hobisch A, Cato A C, Bartsch G. Prostate. 1994;25:266–273. doi: 10.1002/pros.2990250506. [DOI] [PubMed] [Google Scholar]
- 7.Bentel J M, Tilley W D. J Endocrinol. 1996;151:1–11. doi: 10.1677/joe.0.1510001. [DOI] [PubMed] [Google Scholar]
- 8.Lobaccaro J-M, Lumbroso S, Belon C, Galtier-Dereure F, Bringer J, Lesimple T, Heron J-F, Pujol H, Sultan C. Nat Genet. 1993;5:109–110. doi: 10.1038/ng1093-109. [DOI] [PubMed] [Google Scholar]
- 9.Brinkmann A O, Faber P W, van Rooij H C, Kuiper G G, Ris C, Klaassen P, van der Korput J A, Voorhorst M M, van Laar J H, Mulder E, Trapman J. J Steroid Biochem. 1989;34:307–310. doi: 10.1016/0022-4731(89)90098-8. [DOI] [PubMed] [Google Scholar]
- 10.Jenster G, van der Korput H A G M, van Vroonhoven C, van der Kwast T H, Trapman J, Brinkmann A O. Mol Endocrinol. 1991;5:1396–1404. doi: 10.1210/mend-5-10-1396. [DOI] [PubMed] [Google Scholar]
- 11.Simental J A, Sar M, Lane M V, French F S, Wilson E M. J Biol Chem. 1991;266:510–518. [PubMed] [Google Scholar]
- 12.Palvimo J J, Kallio P J, Ikonen T, Mehto M, Jänne O A. Mol Endocrinol. 1993;7:1399–407. doi: 10.1210/mend.7.11.8114755. [DOI] [PubMed] [Google Scholar]
- 13.Jenster G, de Ruiter P E, van der Korput H A, Kuiper G G, Trapman J, Brinkmann A O. Biochemistry. 1994;33:14064–14072. doi: 10.1021/bi00251a015. [DOI] [PubMed] [Google Scholar]
- 14.Jenster G, van der Korput H A, Trapman J, Brinkmann A O. J Biol Chem. 1995;270:7341–7346. doi: 10.1074/jbc.270.13.7341. [DOI] [PubMed] [Google Scholar]
- 15.Chamberlain N L, Whitacre D C, Miesfield R L. J Biol Chem. 1996;271:26772–26778. doi: 10.1074/jbc.271.43.26772. [DOI] [PubMed] [Google Scholar]
- 16.Conaway R C, Conaway J W. Annu Rev Biochem. 1993;62:161–190. doi: 10.1146/annurev.bi.62.070193.001113. [DOI] [PubMed] [Google Scholar]
- 17.Orphanides G, Lagrange T, Reinberg D. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 18.Roeder R G. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 19.Stargell L A, Struhl K. Trends Genet. 1996;12:311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- 20.Kaiser K, Meisterernst M. Trends Biochem Sci. 1996;21:342–345. [PubMed] [Google Scholar]
- 21.Verrijzer C P, Tjian R. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 22.McEwan I J, Wright A P H, Gustafsson J-Å. BioEssays. 1997;19:153–160. doi: 10.1002/bies.950190210. [DOI] [PubMed] [Google Scholar]
- 23.Rowlands J C, McEwan I J, Gustafsson J-Å. Mol Pharmacol. 1996;50:538–548. [PubMed] [Google Scholar]
- 24.McEwan I J, Dahlman-Wright K, Ford J, Wright A P H. Biochemistry. 1996;35:9584–9593. doi: 10.1021/bi960793v. [DOI] [PubMed] [Google Scholar]
- 25.McEwan I J, Wright A P, Dahlman-Wright K, Carlstedt-Duke J, Gustafsson J-Å. Mol Cell Biol. 1993;13:399–407. doi: 10.1128/mcb.13.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McEwan I J, Almlöf T, Wikström A-C, Dahlman-Wright K, Wright A P H, Gustafsson J-Å. J Biol Chem. 1994;269:25629–25636. [PubMed] [Google Scholar]
- 27.Joliot V, Demma M, Prywes R. Nature (London) 1995;373:632–635. doi: 10.1038/373632a0. [DOI] [PubMed] [Google Scholar]
- 28.De Vos P, Schmitt J, Verhoeven G, Stunnenberg H G. Nucleic Acids Res. 1994;22:1161–1166. doi: 10.1093/nar/22.7.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawadogo M, Roeder R G. Cell. 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- 30.Lue N F, Flanagan P M, Sugimoto K, Kornberg R D. Science. 1989;246:661–664. doi: 10.1126/science.2510298. [DOI] [PubMed] [Google Scholar]
- 31.Ptashne M, Gann A A F. Nature (London) 1990;346:329–331. doi: 10.1038/346329a0. [DOI] [PubMed] [Google Scholar]
- 32.Berger S L, Cress W D, Cress A, Triezenberg S J, Guarente L. Cell. 1990;61:1199–1208. doi: 10.1016/0092-8674(90)90684-7. [DOI] [PubMed] [Google Scholar]
- 33.Kelleher R D, Flanagan P M, Kornberg R D. Cell. 1990;61:1209–1215. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- 34.Greenblatt J. Trends Biochem Sci. 1991;16:408–411. doi: 10.1016/0968-0004(91)90165-r. [DOI] [PubMed] [Google Scholar]
- 35.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 36.Koleske A J, Young R A. Nature (London) 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 37.Ossipow V, Tassan J P, Nigg E A, Schibler U. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 38.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. Nature (London) 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 39.Aso T, Conaway J W, Conaway R C. FASEB J. 1995;9:1419–1428. doi: 10.1096/fasebj.9.14.7589983. [DOI] [PubMed] [Google Scholar]
- 40.Yankulov K, Blau J, Purton T, Roberts S, Bentley D L. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 41.Blair W S, Fridell R A, Cullen B R. EMBO J. 1996;15:1658–1665. [PMC free article] [PubMed] [Google Scholar]
- 42.Blau J, Xiao H, McCracken S, O’Hare P, Greenblatt J, Bentley D. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yankulov K Y, Pandes M, McCracken S, Bouchard D, Bentley D L. Mol Cell Biol. 1996;16:3291–3299. doi: 10.1128/mcb.16.7.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zawel L, Reinberg D. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- 45.Martin M L, Lieberman P M, Curran T. Mol Cell Biol. 1996;16:2110–2118. doi: 10.1128/mcb.16.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeh S, Chang C. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snoek R, Rennie P S, Kaspar S, Matusik R J, Bruchovsky N. J Steroid Biochem Mol Biol. 1996;59:243–250. doi: 10.1016/s0960-0760(96)00116-1. [DOI] [PubMed] [Google Scholar]